Abstract
Improved understanding of the epidemiology of Streptococcus equi transmission requires sensitive and portable subtyping methods that can rationally discriminate between strains. S. equi is highly homogeneous and cannot be distinguished by multilocus enzyme electrophoretic or multilocus sequence-typing methods that utilize housekeeping genes. However, on sequence analysis of the N-terminal region of the SeM genes of 60 S. equi isolates from 27 strangles outbreaks, we identified 21 DNA codon changes. These resulted in the nonsynonymous substitution of 18 amino acids and allowed the assignment of S. equi strains to 15 distinct subtypes. Our data suggest the presence of multiple epitopes across this region that are subjected to selective immune pressure (nonsynonymous-synonymous substitution rate [dN/dS] ratio = 3.054), particularly during the establishment of long-term S. equi infection. We further report the application of SeM gene subtyping as a method to investigate potential cases of disease related to administration of a live attenuated S. equi vaccine. SeM gene subtyping successfully differentiated between the vaccine strain and field strains of S. equi responsible for concurrent disease. These results were confirmed by the development and application of a PCR diagnostic test, which identifies the aroA partial gene deletion present in the Equilis StrepE vaccine strain. Although the vaccine strain was found to be responsible for injection site lesions, all seven outbreaks of strangles investigated in recently vaccinated horses were found to be due to concurrent infection with wild-type S. equi and not due to reversion of the vaccine strain.
“Strangles,” caused by infection with the bacterium Streptococcus equi, remains one of the most commonly diagnosed and important infectious diseases of horses worldwide. The disease is characterized by pyrexia, followed by profuse nasal discharge and the formation of abscesses on the lymph nodes of the head and neck, which subsequently burst, discharging highly infectious pus. The swelling of the lymph nodes in the head and neck may, in severe cases, restrict the airway, and it is this clinical feature that gave the disease “strangles” its name (30). Approximately 10% of horses that recover from strangles become persistent carriers of S. equi, harboring the infectious agent in chondroids located in the guttural pouch. These carriers are capable of infecting other naïve horses and thereby continuing the spread of disease (3, 20, 21). A particular problem in the management of strangles outbreaks is the lack of a suitable assay to differentiate between S. equi strains and so to determine if an outbreak is likely to be due to the recurrence of an earlier infection or the introduction of a new S. equi strain. Furthermore, in the absence of methods for subtyping strains, it has not been possible to investigate whether S. equi strain variation may contribute to the different presentations of disease seen in different outbreaks.
The launch of Intervet's “Equilis StrepE” live attenuated strangles vaccine in 2004 has further highlighted the need to distinguish between different strains of S. equi (7, 8). The vaccine is recommended for use in horses at moderate to high risk of developing strangles, including horses located on premises with a known history of strangles. The symptomatic appearance of concurrent S. equi infection may by chance coincide with vaccination and may be misdiagnosed as an adverse reaction to the vaccine. Therefore, the ability to differentiate between the vaccine strain and wild-type S. equi would allow the differentiation of concurrent disease from vaccine-derived infection. Such information is valuable to the veterinary community, as it enables reliable assessment of vaccine safety in the field.
S. equi strains are known to be highly homogeneous (5, 9). To date, S. equi strains have been differentiated only by restriction fragment length polymorphism typing or repetitive PCR (1, 28). However, these techniques are poorly portable, and the variation that is indexed tends to change rapidly for unknown reasons. Multilocus enzyme electrophoresis (MLEE) analysis of 70 S. equi isolates placed 69 of them in a single MLEE type (9). Recently, the availability of the S. equi genome sequence enabled us to identify a number of housekeeping genes for the application of multilocus sequence typing (MLST) techniques. However, in agreement with the MLEE analysis, no variations in the sequences of the fba, gki, galU, xpt, and recA genes were identified across 70 clinical isolates of S. equi from disparate geographical sources (C. Robinson, unpublished results). The sequence typing of virulence genes or hypervariable genes has been utilized to enhance the discriminatory power of some MLST schemes (18, 25, 35). The short-term variability of virulence genes is particularly suited to studying local epidemiology (36) and allows the discrimination of different strains in the absence of variation in housekeeping genes (33).
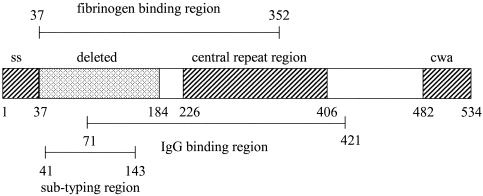
S. equi produces a novel M-like protein, SeM, which has been linked to its increased virulence over its evolutionary parent, Streptococcus zooepidemicus (4, 31). SeM actively binds fibrinogen and immunoglobulin G (IgG) and inhibits the deposition of C3b on the bacterial surface, resulting in an antiphagocytic action similar to that of the M proteins of group A streptococci (2, 16) (Fig. 1). Vaccination with recombinant SeM enhanced opsonization of S. equi in vitro (31) and protected mice against lethal challenge from S. equi (15). Despite these encouraging data, use of purified SeM was not found to confer significant protection on horses against subsequent challenge with S. equi (26). SeM was thought to be highly homogeneous because of the cross-reactivity of sera from a horse convalescent from strangles with a number of different S. equi isolates and the lack of variation in HindIII restriction patterns between different S. equi isolates on Southern blot analysis using a SeM gene probe (5). However, subsequent to early reports, differences from the published sequence of the SeM gene immediately after the N-terminal signal sequence have been observed by two independent researchers on sequencing a limited number of S. equi isolates (3, 15). Many of the changes identified altered the amino acid encoded, suggesting the presence of distinct SeM gene subtypes.
FIG. 1.
Schematic representation of the SeM protein of S. equi. Amino acids 37 to 352 are required for fibrinogen binding (16). Amino acids 1 to 37 contain the M-protein signal sequence (ss) (31). Amino acids 37 to 184 (deleted) were found to be absent from 24% of S. equi strains isolated from outwardly healthy horses (3). Amino acids 226 to 406 (central repeat region) contain the A and B repeat regions (31). Amino acids 482 to 534 contain the wall-spanning region and “LPSTG” cell wall anchor (cwa) (31). Amino acids 71 to 421 are required for IgG binding (17).
The extent of SeM gene variation was determined in order to evaluate the application of SeM gene sequencing to enhance the epidemiological analysis of disease transmission. These data were employed in the investigation of cases of strangles occurring shortly after vaccination with Equilis StrepE. The identity of the vaccine strain was confirmed with a diagnostic PCR test based on the knowledge that the Equilis StrepE strain is attenuated via a deletion in the aroA gene (http://www.biosafety.be/EMEA/Table_EquilisStrepT.htm).
(Part of this work was summarized as a letter to the Veterinary Record [22].)
MATERIALS AND METHODS
Bacterial strains.
Details of all of the isolates examined in this study are presented in Tables 1 and 2. Sixty S. equi isolates from 27 separate outbreaks were obtained from swabs of suspected cases of strangles sent to the Animal Health Trust's diagnostic laboratories from 1998 to 2005. A total of 15 of the isolates were from suspected adverse reactions caused by the Equilis StrepE vaccine. Of these, isolates 0223, R, and C were from lip injection site abscesses, while the other strains were from suspected cases of strangles following vaccination. The S. equi 4047 reference strain was isolated from a New Forest pony by the Animal Health Trust and is the focus of the S. equi genome-sequencing project at the Sanger Institute (http://www.sanger.ac.uk/Projects/S_equi/). The SeM gene sequences for strains TW, CF32, and A1 have been previously published and were included in our analysis (3, 15, 31). The TW928 strain was isolated following plating of reconstituted Equilis StrepE vaccine (Intervet) onto COBA Streptococcus selective agar (bioMérieux).
TABLE 1.
SeM gene sequence types of isolates from clinical cases of strangles and published literature
| SeM gene allele | Source | Outbreak no. | Isolate(s) | Date of isolationc |
|---|---|---|---|---|
| 1 | Netherlands | NAa | TW | Not known |
| 1 | Sussex | 1 | 6073 | 11/12/98 |
| 1 | Ireland | 2 | SA | 12/10/99 |
| 2 | New York | NA | CF32 | 1981 |
| 2 | Canadian | 3 | 303 | 8/11/99 |
| 3 | Hampshire | NA | 4047 | 1990 |
| 4 | Berwickshire | 4 | 0851, 0852 | 28/01/05 |
| 1458 | 22/02/05 | |||
| 2424 | 29/03/05 | |||
| 3731 | 11/05/05 | |||
| 5 | Leicestershire | 5 | 7325, 7326 | 8/12/03 |
| 6 | Suffolk | 6 | 7060, 7061, 7062, 7063, 7064, 7065, 7066 | 26/11/03 |
| 7094, 7098, 7099 | 27/11/03 | |||
| 7176 | 1/12/03 | |||
| 1610b | 25/02/04 | |||
| 6 | Gwent | 7 | 7329 | 7/12/03 |
| 6 | Sussex | 8 | 7331 | 8/12/03 |
| 6 | Wiltshire | 9 | 7350, 7352 | 9/12/03 |
| 6 | Aberdeenshire | 10 | 1321 | 16/02/04 |
| 6 | Kent | 11 | 1351 | 16/02/04 |
| 6 | Cheshire | 12 | 7132 | 26/11/03 |
| 6 | Hampshire | 13 | 1218 | 12/02/05 |
| 6 | Berwickshire | 4 | 1165 | 10/02/05 |
| 3446 | 29/04/05 | |||
| 4289 | 27/05/05 | |||
| 7 | Suffolk | 6 | 1931, 1932 | 05/03/04 |
| 2077 | 10/03/04 | |||
| 8 | Essex | 14 | 1350 | 17/12/03 |
| 8 | Buckinghamshire | NA | A1 | 1999 |
| 9 | Hampshire | 15 | 7344 | 8/12/03 |
| 10 | Hampshire | 16 | 7140 | 28/11/03 |
| 10 | Shetland | 17 | 7171 | 28/11/03 |
| 11 | Suffolk | 18 | 7364 | 10/12/03 |
| 12 | Suffolk | 19 | 3154 | 15/04/04 |
| 13 | Suffolk | 19 | 3155 | 15/04/04 |
| 14 | Suffolk | 19 | 3156 | 15/04/04 |
| 15 | Australia | 20 | 181063 | 27/09/99 |
NA, not applicable.
Isolate 1610 had gained a 6-amino-acid duplication, SEIAII, at codon 56.
Day/month/year.
TABLE 2.
SeM gene types of isolates from vaccinated horses
| SeM gene allele | Source | Outbreak no. | Isolate(s) | Date of isolationb | aroA gene PCR (bp) |
|---|---|---|---|---|---|
| 1 | Equilis StrepE vaccine | NAa | TW928 | 20/01/05 | 432 |
| 1 | Cambridgeshire | 21 | 8689 | 23/12/04 | 432 |
| 1 | Berwickshire | 4 | 0223 | 11/01/05 | 432 |
| 1 | Berkshire | 22 | R, C | 24/05/05 | 432 |
| 3 | Berwickshire | 4 | 199 | 8/01/05 | 1364 |
| 4 | Berwickshire | 4 | 0347 | 13/01/05 | 1364 |
| 7 | Sussex | 23 | 1974 | 11/03/05 | 1364 |
| 9 | Cambridgeshire | 21 | 0353, 0354 | 13/01/05 | 1364 |
| 9 | Greater Manchester | 24 | 1293 | 16/02/05 | 1364 |
| 9 | Dorset | 25 | 3081 | 19/04/05 | 1364 |
| 9 | Surrey | 26 | 3526 | 05/05/05 | 1364 |
| 10 | Surrey | 27 | 3679, 3680, 3682 | 10/05/05 | 1364 |
NA, not applicable.
Day/month/year.
Identification of S. equi and DNA isolation.
S. equi strains were isolated from clinical swabs as beta-hemolytic colonies on COBA strep select plates (bioMérieux). Their identities were confirmed by a lack of fermentation of trehalose, ribose, and sorbitol in Purple broth (Becton Dickinson). A single colony of S. equi was resuspended in 200 μl gram-positive lysis solution (GenElute kit; Sigma) containing 250 units/ml mutanolysin and 2 × 106 units/ml lysozyme and incubated for 1 h at 37°C to allow efficient cell lysis. The DNA was then purified using GenElute spin columns according to the manufacturer's instructions (all Sigma).
PCR and sequencing of the SeM gene.
The forward primer ASW73 (5′-CAGAAAACTAAGTGCCGGTG) and the reverse primer ASW74 (5′-ATTCGGTAAGAGCTTGACGC) were used to PCR amplify 541 bp of the N-terminal region of the SeM gene unique to S. equi (Fig. 1) using Vent DNA polymerase (New England BioLabs) with 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 1 min. The PCR products were purified on QIAquick spin columns (QIAGEN), and the sequences of both strands of the PCR fragments were determined using an ABI3100 DNA sequencer with BigDye fluorescent terminators and the primers used in the initial PCR amplification.
PCR and sequencing of the aroA gene.
Amplification across the aroA gene deletion present in the Equilis StrepE vaccine strain was performed using the forward primer aroa1 (5′-TTGCTGAGCTAATGCTGGTG) and the reverse primer aroa2 (5′-AACTGTCGTCTTGCCAACTC). These primers generated a 1,364-bp fragment from wild-type S. equi using Vent DNA polymerase (New England BioLabs) with 30 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 3 min. To determine the nature of the aroA gene deletion in the Equilis StrepE vaccine, the sequences of truncated PCR products generated from cultures of the vaccine strain were determined as described above, using the aroa1 and aroa2 primers. To determine the 5′ sequence of the full-length aroA gene, the primer aroa5 (5′-AACTCCTGACAGCCCTTTAC) was used instead of the aroa2 primer to generate a product of 503 bp, which included the first 464 bp of the aroA gene, and it was sequenced as described above, using the aroa1 and aroa5 primers.
Analysis of sequence data.
Sequence data were assembled using SeqMan 5.03 (DNAstar Inc.), and high-quality double-stranded-sequence data were used for further analysis. Different SeM gene alleles were assigned to SeM gene sequences that differed from each other by one or more nucleotide differences. To determine the level of selective pressure on the SeM gene, the nonsynonymous-synonymous substitution rate (dN/dS) ratio was calculated by the Nei-Gojobori method with Jukes-Cantor correction (19) using MEGA (11). A maximum likelihood tree was generated by PAUP* version 4.0 (beta 10) (27) using likelihood settings from the best-fit model (HKY+G) selected by Modeltest 3.6 (24), and visualized using TreeView (23).
RESULTS
PCR analysis of the SeM gene.
PCR of the SeM genes of 60 clinical isolates of S. equi and the reference strains TW928 and 4047 generated products of 541 bp in all but one of the strains. Strain 1610 was isolated 3 months after a strangles outbreak from a pony with prolonged S. equi infection and generated a PCR product of 559 bp.
Sequence analysis of the SeM gene.
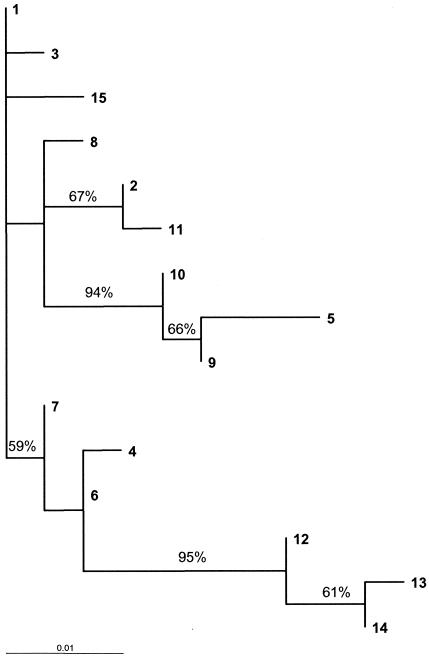
Analysis of the SeM gene sequences from 60 clinical isolates of S. equi and comparison with published SeM gene sequences identified 15 alleles (Table 3). Of the 21 base changes identified across the 15 alleles, 18 resulted in an altered amino acid sequence. The dN/dS ratio is used as a measure of selective pressure at the protein level, with a ratio of >1 indicating positive selection. For this region of the SeM gene, the dN/dS ratio was 3.054. A maximum likelihood tree of the SeM gene alleles is shown in Fig. 2.
TABLE 3.
SeM gene sequence types of S. equi
| Allele | No. of isolates (no. of strangles outbreaks)a | Amino acid at codon no.b:
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 41 | 54 | 58 | 62 | 63 | 65 | 78 | 90 | 92 | 104 | 107 | 108 | 110 | 122 | 125 | 127 | 143 | ||
| 1 | 2 (2)c | Ser | Asn | Glu | Ser | Arg | Ala | Ala | Leu | Ser | Asn | Met | His | Ser | Arg | Ser | Ala | Ser |
| 2 | 1 (1) | * | * | Asp | * | Gly | * | * | * | * | * | * | * | * | * | * | * | Arg |
| 3 | 1 (1) | * | * | * | * | * | * | * | * | * | * | Val | * | * | * | * | * | * |
| 4 | 6 (1) | * | * | * | * | * | * | * | Phe | * | * | * | Arg | * | Gly | * | * | * |
| 5 | 1 (1) | Arg | * | Asp | Asp | * | * | Alaf | * | * | Asng | * | Arg | * | * | * | Thr | * |
| 6 | 22 (9)d | * | * | * | * | * | * | * | Phe | * | * | * | Arg | * | * | * | * | * |
| 7 | 4 (2) | * | * | * | * | * | * | * | Phe | * | * | * | * | * | * | * | * | * |
| 8 | 1 (1) | * | * | Asp | * | * | * | * | * | Pro | * | * | * | * | * | * | * | * |
| 9 | 6 (5) | * | * | Asp | Asp | * | * | * | * | * | Asng | * | * | * | * | * | Thr | * |
| 10 | 5 (3) | * | * | Asp | Asp | * | * | * | * | * | * | * | * | * | * | * | Thr | * |
| 11 | 1 (1) | * | * | Glue | * | Gly | * | * | * | * | * | * | * | * | * | * | * | Arg |
| 12 | 1 (1) | * | Gly | * | * | * | Val | * | Phe | * | * | * | Arg | * | * | Asn | Ser | * |
| 13 | 1 (1) | * | Ser | * | * | * | Val | * | Phe | * | * | * | * | Pro | * | Asn | Ser | * |
| 14 | 1 (1) | * | Gly | * | * | * | Val | * | Phe | * | * | * | * | Pro | * | Asn | Ser | * |
| 15 | 1 (1) | * | * | Glue | * | * | * | * | * | * | * | * | Gln | * | * | * | * | * |
Some outbreaks contained multiple S. equi subtypes.
Amino acids differing from the majority are shown in boldface type. The asterisks indicate that the same codon is used as in allele 1.
Does not include isolates of the Equilis StrepE vaccine.
Does not include isolate 1610, which had a 6-amino-acid insertion into its MT6 sequence.
Glu is encoded by GAG in MT11 and MT15 and by GAA in other MTs with a Glu codon.
Ala is encoded by GCT in MT5 and by GCC in other MTs with an Ala codon.
Asn is encoded by AAC in MT5 and MT9 and by AAT in other MTs with an Asn codon.
FIG. 2.
Maximum likelihood tree showing the relationships of the 15 different SeM gene sequence types of S. equi generated using PAUP* version 4.0 (27). Bootstrap values are shown for bipartitions supported by >50% of replicate trees (1,000 replicates were performed).
The most frequently identified SeM gene alleles were 6 and 9, which were present in nine and five strangles outbreaks, respectively. Initial isolates from individual outbreaks were generally found to have the same SeM gene allele. However, an outbreak of strangles in three horses in Suffolk during 2004 generated three similar alleles: 12, 13, and 14 (Table 1). These three alleles group together as a distinct branch of the phylogenetic tree (Fig. 2).
The SeM gene sequence of the CF32 S. equi strain (31) shared the allele 2 sequence with Canadian field isolate 303 (Table 1). The A1 S. equi strain SeM gene (3) was found to be allele 8, a sequence shared with field isolate 1350. The TW strain (15) was found to have a SeM gene allele 1 also found in isolates 6073 and SA (Table 1).
Analysis of isolates taken either early or late during an outbreak of strangles in Suffolk demonstrated alteration of the SeM gene sequence for the later isolates. Thus, all strains initially isolated from 11 different horses suffering an outbreak of strangles in Suffolk shared allele 6. However, the strains 1610, 1931, 1932, and 2077 isolated 3 months later from three of these horses in the absence of clinical disease had different SeM gene sequences (Table 1). Although the SeM gene allele of 1610 remained similar to the initial isolates from this outbreak, the strain contained a duplication of 18 bp, resulting in the insertion of a tandem repeat of the amino acids SEIAII at amino acid position 56. Strains 1931, 1932 (both from the same horse), and 2077 all contained identical G-to-A substitutions at position 323, resulting in an arginine-to-histidine amino acid change at codon 108 and leading to an allele switch from 6 to 7.
Characterization of postvaccination isolates.
The Equilis StrepE vaccine and strains 0223, R, and C isolated from lip injection site reactions approximately 2 weeks postvaccination with Equilis StrepE were found to share SeM gene allele 1 (Table 2). None of the isolates of S. equi from recently vaccinated or in-contact horses suffering from strangles were found to have SeM gene allele 1 on sequencing, with the exception of isolate 8689. However, isolate 8689 was obtained from a nasal swab, and subsequent swabs from the discharging lymph nodes of this horse (0353 and 0354) indicated the presence of a different S. equi strain that had SeM gene allele 9 (Table 2), which differs at four nucleotide positions (Table 3).
Strain 0347 isolated from a case of strangles in a horse stabled next to a vaccinated horse was found to have SeM gene allele 4 (Table 2), matching strains 0851 and 0852 isolated from a suspected S. equi carrier located on the same premises (Table 1). The carrier animal was sampled regularly by nasal swab over the following 3 months, and the strains isolated (1458, 2424, 1165, 3446, 3731, and 4289) were found to have either allele 4 or 6, suggesting a mixture of infecting strains. These two alleles differ by a G-to-A substitution at position 364, which results in a glycine-to-arginine amino acid change at codon 122. Unfortunately, at the time of analysis, only a pure culture from a single colony of each sample was available, and so we were unable to determine the relative levels of these two strains at any one time in this animal. S. equi strain 199, isolated from a vaccinated pony also suffering from strangles on these premises, had a SeM gene allele 3 originating from an as-yet-unidentified source.
PCR of the aroA gene of the 4047 S. equi genome-sequencing strain generated a product of 1,364 bp. In contrast, aroA gene PCR of DNA generated from the Equilis StrepE vaccine yielded a 432-bp product. On sequencing the 432-bp product, it was apparent that the Equilis StrepE strain lacks a region of the aroA gene from positions 46 to 978.
PCR analysis of the aroA gene of clinical isolates taken from recently vaccinated or in-contact horses that had subsequently developed clinical signs of strangles was in agreement with the SeM gene subtyping results. Thus, all of the isolates, with the exception of 8689, yielded wild-type products of 1,364 bp, whereas 8689 yielded the 432-bp deleted aroA gene product, consistent with this strain being derived from the vaccine strain (Table 2). All isolates of S. equi from horses suffering from lip injection site reactions following recent vaccination (0223, R, and C) generated an aroA PCR product of 432 bp (Table 2). Sequencing of aroA PCR products showed that they exactly matched either the full-length aroA gene of the 4047 genome-sequencing strain or the Equilis StrepE aroA deletion, in full agreement with the presence of a wild-type S. equi or the vaccine strain. No sequence variation in the aroA gene was observed.
DISCUSSION
SeM is a potent immunogenic and opsonogenic determinant in S. equi (2, 29). A large proportion of antibody responses generated in convalescent horses are directed toward the N terminus (32). This region of SeM is responsible for fibrinogen and IgG binding, both important antiphagocytic activities (16, 17). S. equi is believed to have evolved from a subtype of S. zooepidemicus, and the acquisition of the SeM gene is believed to have been an important step in this process (31). Searches of recent S. zooepidemicus genome sequence data have identified the presence of a SeM homologue distinct from those previously identified in S. zooepidemicus (http://www.sanger.ac.uk/Projects/S_zooepidemicus). The H70 S. zooepidemicus SzM has the least sequence homology between amino acids 36 and 186 of the SeM protein (Fig. 3). Therefore, rather than the acquisition of the entire SeM gene, it may be the replacement or modification of this N-terminal domain that is the evolutionary step. It is interesting that deletion of regions of this domain up to amino acids 37 to 183 was observed in S. equi variants obtained from outwardly healthy horses (3). Sequencing of disparate strains of S. zooepidemicus is now required to determine the extent of SzM gene variation and to ascertain if this region is subjected to selective pressure from the immune system during prolonged S. zooepidemicus colonization.
FIG. 3.
Alignment of S. equi 4047 SeM and S. zooepidemicus H70 SzM amino acid sequences. The dashes indicate a gap inserted to optimize sequence alignment. The asterisks indicate identical amino acid residues. The colons indicate closely related nonidentical amino acid residues, and the periods indicate similar amino acid residues. The signal sequence of S. equi is shown in boldface, the repeat domain is in italics, and the LPSTG cell wall anchor motif is underlined.
We have identified virulent S. equi strains that contain 15 different combinations of SeM gene sequence variations. Of the 21 base changes identified in the SeM genes across 58 S. equi isolates, 18 resulted in amino acid substitutions. This generated a dN/dS ratio of 3.054, indicating that the SeM gene is under diversifying selection. An earlier report suggested that there was no variation in the immunoreactivity of SeM extracts from 21 different S. equi isolates (5). It will be interesting to ascertain if the strains in that study have different SeM gene alleles.
It is possible that the amino acid variations identified may affect SeM function. Although determination of this was beyond the scope of this paper, SeM gene alleles 1, 2, and 8 have all been previously documented to bind fibrinogen (3, 15, 31). It should also be noted that strains of each allele subtype were responsible for producing disease in naturally infected horses, although no direct comparison of the relative virulence of each strain subtype was possible.
The SeM amino acid sequence was seen to change over time in sequential isolates taken from one outbreak in Suffolk, in which three horses suffered prolonged S. equi infection. These results agreed with earlier observations that deletion of sections of the SeM gene encoding the fibrinogen binding domain occur in 24% (4/17) of S. equi isolates from persistently infected horses compared with only 0.6% (1/167) of S. equi isolates from strangles cases (3). Such deletions were found to result in decreased resistance to phagocytosis in vitro and were postulated to lead to attenuation (3). However, a strain containing a truncated SeM gene remained virulent in Welsh mountain ponies (N. Chanter, unpublished results). S. equi strains that alter their SeM gene sequences may achieve a selective advantage in the host environment. Persistent S. equi infections are believed to continually stimulate host tissues (20, 21, 34) and are likely to be subjected to continued selective pressure. None of the S. equi isolates from four persistently infected horses identified in this study contained SeM gene deletions (isolate 1610 had actually gained a 6-amino-acid duplication), and all probably retained their full virulence. Indeed, one carrier in Berwickshire was identified as a likely source of allele 4 disease in a recently vaccinated horse. The carrier was found to have a persistent sinus infection from which we isolated S. equi strains with SeM gene alleles 4 and 6 that probably existed as a mixed infection.
One of the outbreaks sampled in this study (3154, 3155, and 3156) generated three similar SeM gene allele subtypes, 12, 13, and 14, in the initial stages of disease from three different horses. It is possible that this outbreak may have started through contact of these horses with a long-term strangles infection involving at least three SeM gene allele subtypes probably originating from a single S. equi strain.
One model for the sequence variability observed for SeM is that the alteration of one or more SeM epitopes in carrier animals may be an important step in the establishment of a persistent S. equi infection. Further studies that determine sequential SeM gene sequence changes in a larger number of horses during the development of long-term infection will yield important information regarding the establishment of the carrier state and the targets of host selection. It is likely that other S. equi surface proteins (6, 10, 12, 13, 14) will be subjected to similar immune pressure during the establishment of the carrier state. These may also have varied amino acid sequences that could be used as part of a multivirulence locus sequence-typing system to further discriminate between S. equi isolates.
The Equilis StrepE vaccine strain (TW928) was found to have SeM gene allele 1, shared by strain TW isolated from The Netherlands (8), which was used to generate the vaccine strain. Subsequent application of our subtyping methods enabled the characterization of potential adverse reactions following vaccination with Equilis StrepE. The TW928 strain can clearly persist in the lip injection site (0223, R, and C) and nasopharynx (8689) shortly after vaccination. At the injection site, this can lead to inflammation and the formation of abscesses, as demonstrated by the three cases examined here. However, none of the seven strangles outbreaks investigated to date in vaccinated or in-contact horses appear to have been caused by S. equi strains with the same SeM gene allele as the vaccine strain, and all cases had a full-length aroA gene. To determine if the deleted aroA gene could have been repaired by recombination with S. zooepidemicus, we sequenced the first 464 bp of the full-length aroA gene in these virulent strains. S. equi is a clonal population, as determined by MLEE (9) and MLST analyses of housekeeping genes (Robinson, unpublished), while the aroA gene sequence of S. zooepidemicus strain H70 differs from that of S. equi 4047 at eight codons across this 464-bp coding section. Therefore, these or similar changes in the aroA gene sequence should be present if the Equilis StrepE strain had repaired its aroA gene by recombination with S. zooepidemicus. All of these isolates contained a full-length aroA gene, the sequence of the first 464 bp of which exactly matched that of S. equi 4047, indicating that repair of the aroA gene deletion by recombination with the common equine commensal S. zooepidemicus was unlikely to have occurred. We conclude that the strangles in these vaccinated or in-contact horses was most likely the result of concurrent disease, which had not been prevented or caused by vaccination. Therefore, there is currently no evidence that the Equilis StrepE vaccine strain readily reverts to virulence in the field. These data suggest that the deletion of part of the aroA gene in the Equilis StrepE vaccine strain is both a stable and an effective method for attenuation of S. equi.
Variations in the amino acid sequences of surface proteins may influence their effectiveness as strangles vaccines (33). An advantage of live attenuated vaccines, such as Equilis StrepE, is that potentially protective immune responses to a number of other cell surface proteins will be generated. However, it will be interesting to determine if horses vaccinated with SeM gene allele 1 Equilis StrepE vaccine remain protected from challenge with more disparate S. equi strains. The Arnica S. equi strain used as a potential heterologous challenge in earlier trials of the vaccine (8) has not yet been assigned a SeM gene allele.
The assignment of an S. equi strain type based on the sequence of its SeM gene may assist veterinarians to rationally differentiate between S. equi isolates and identify the source and transmission of a particular outbreak. Submission of sequence data to generate a SeM gene allele profile and all of the data presented in this study can be accessed at http://pubmlst.org/szooepidemicus/seM/. It is hoped that such information will improve the implementation of appropriate disease control and treatment strategies to reduce the risk of subsequent strangles outbreaks and to learn more about the epidemiology of disease transmission.
REFERENCES
- 1.Al-Ghamdi, G. M., V. Kapur, T. R. Ames, J. F. Timoney, D. N. Love, and M. A. Mellencamp. 2000. Use of repetitive sequence-based polymerase chain reaction for molecular epidemiologic analysis of Streptococcus equi subspecies equi. Am. J. Vet. Res. 61:699-705. [DOI] [PubMed] [Google Scholar]
- 2.Boschwitz, J. S., and J. F. Timoney. 1994. Inhibition of C3 deposition on Streptococcus equi subsp. equi by M protein: a mechanism for survival in equine blood. Infect. Immun. 62:3515-3520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chanter, N., N. C. Talbot, J. R. Newton, D. Hewson, and K. Verheyen. 2000. Streptococcus equi with truncated M-proteins isolated from outwardly healthy horses. Microbiology 146:1361-1369. [DOI] [PubMed] [Google Scholar]
- 4.Galan, J. E., and J. F. Timoney. 1987. Molecular analysis of the M protein of Streptococcus equi and cloning and expression of the M protein gene in Escherichia coli. Infect. Immun. 55:3181-3187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Galan, J. E., and J. F. Timoney. 1988. Immunologic and genetic comparison of Streptococcus equi isolates from the United States and Europe. J. Clin. Microbiol. 26:1142-1146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Harrington, D. J., I. C. Sutcliffe, and N. Chanter. 2002. The molecular basis of Streptococcus equi infection and disease. Microbes Infect. 4:501-510. [DOI] [PubMed] [Google Scholar]
- 7.Hartford, O. M., T. J. Foster, and A. A. C. Jacobs. April 1999. United States Patent 5,895,654.
- 8.Jacobs, A. A., D. Goovaerts, P. J. Nuijten, R. P. Theelen, O. M. Hartford, and T. J. Foster. 2000. Investigations towards an efficacious and safe strangles vaccine: submucosal vaccination with a live attenuated Streptococcus equi. Vet. Rec. 147:563-567. [DOI] [PubMed] [Google Scholar]
- 9.Jorm, L. R., D. N. Love, G. D. Bailey, G. M. McKay, and D. A. Briscoe. 1994. Genetic structure of populations of beta-haemolytic Lancefield group C streptococci from horses and their association with disease. Res. Vet. Sci. 57:292-299. [DOI] [PubMed] [Google Scholar]
- 10.Karlstrom, A., K. Jacobsson, M. Flock, J. I. Flock, and B. Guss. 2004. Identification of a novel collagen-like protein, SclC, in Streptococcus equi using signal sequence phage display. Vet. Microbiol. 104:179-188. [DOI] [PubMed] [Google Scholar]
- 11.Kumar, S., K. Tamura, and M. Nei. 1994. MEGA: Molecular Evolutionary Genetics Analysis software for microcomputers. Comput. Appl. Biosci. 10:189-191. [DOI] [PubMed] [Google Scholar]
- 12.Lannergard, J., L. Frykberg, and B. Guss. 2003. CNE, a collagen-binding protein of Streptococcus equi. FEMS Microbiol. Lett. 222:69-74. [DOI] [PubMed] [Google Scholar]
- 13.Lindmark, H., and B. Guss. 1999. SFS, a novel fibronectin-binding protein from Streptococcus equi, inhibits the binding between fibronectin and collagen. Infect. Immun. 67:2383-2388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lindmark, H., P. Jonsson, E. Engvall, and B. Guss. 1999. Pulsed-field gel electrophoresis and distribution of the genes zag and fnz in isolates of Streptococcus equi. Res. Vet. Sci. 66:93-99. [DOI] [PubMed] [Google Scholar]
- 15.Meehan, M., P. Nowlan, and P. Owen. 1998. Affinity purification and characterization of a fibrinogen-binding protein complex which protects mice against lethal challenge with Streptococcus equi subsp. equi. Microbiology 144:993-1003. [DOI] [PubMed] [Google Scholar]
- 16.Meehan, M., D. A. Muldowney, N. J. Watkins, and P. Owen. 2000. Localization and characterization of the ligand-binding domain of the fibrinogen-binding protein (FgBP) of Streptococcus equi subsp. equi. Microbiology 146:1187-1194. [DOI] [PubMed] [Google Scholar]
- 17.Meehan, M., Y. Lynagh, C. Woods, and P. Owen. 2001. The fibrinogen-binding protein (FgBP) of Streptococcus equi subsp. equi additionally binds IgG and contributes to virulence in a mouse model. Microbiology 147:3311-3322. [DOI] [PubMed] [Google Scholar]
- 18.Meinersmann, R. J., R. W. Phillips, M. Wiedmann, and M. E. Berrang. 2004. Multilocus sequence typing of Listeria monocytogenes by use of hypervariable genes reveals clonal and recombination histories of three lineages. Appl. Environ. Microbiol. 70:2193-2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nei, M., and T. Gojobori. 1986. Simple methods for estimating the numbers of synonymous and nonsynonymous nucleotide substitutions. Mol. Biol. Evol. 3:418-426. [DOI] [PubMed] [Google Scholar]
- 20.Newton, J. R., J. L. Wood, K. A. Dunn, M. N. DeBrauwere, and N. Chanter. 1997. Naturally occurring persistent and asymptomatic infection of the guttural pouches of horses with Streptococcus equi. Vet. Rec. 140:84-90. [DOI] [PubMed] [Google Scholar]
- 21.Newton, J. R., K. Verheyen, N. C. Talbot, J. F. Timoney, J. L. Wood, K. H. Lakhani, and N. Chanter. 2000. Control of strangles outbreaks by isolation of guttural pouch carriers identified using PCR and culture of Streptococcus equi. Equine Vet. J. 32:515-526. [DOI] [PubMed] [Google Scholar]
- 22.Newton, R., A. Waller, and A. King. 2005. Investigation of suspected adverse reactions following strangles vaccination in horses. Vet. Rec. 156:291-292. [DOI] [PubMed] [Google Scholar]
- 23.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 24.Posada, D., and K. A. Crandall. 1998. MODELTEST: testing the model of DNA substitution. Bioinformatics 14:817-818. [DOI] [PubMed] [Google Scholar]
- 25.Reid, S. D., C. J. Herbelin, A. C. Bumbaugh, R. K. Selander, and T. S. Whittam. 2000. Parallel evolution of virulence in pathogenic Escherichia coli. Nature 406:64-67. [DOI] [PubMed] [Google Scholar]
- 26.Sheoran, A. S., S. Artiushin, and J. F. Timoney. 2002. Nasal mucosal immunogenicity for the horse of a SeM peptide of Streptococcus equi genetically coupled to cholera toxin. Vaccine 20:1653-1659. [DOI] [PubMed] [Google Scholar]
- 27.Swofford, D. L. 2003. PAUP*. Phylogenetic analysis using parsimony (*and 15 other methods), version 4. Sinauer Associates, Sunderland, Mass.
- 28.Takai, S., T. Anzai, H. Yashiro, C. Ishii, S. Tsubaki, R. Wada, and J. F. Timoney. 2000. Detection of DNA restriction fragment polymorphisms in Streptococcus equi. Vet. Rec. 146:159-161. [DOI] [PubMed] [Google Scholar]
- 29.Timoney, J. F., and D. Eggers. 1985. Serum bactericidal responses to Streptococcus equi of horses following infection or vaccination. Equine Vet. J. 17:306-310. [DOI] [PubMed] [Google Scholar]
- 30.Timoney, J. F. 1993. Strangles. Vet. Clin. N. Am. Equine Pract. 9:365-374. [DOI] [PubMed] [Google Scholar]
- 31.Timoney, J. F., S. C. Artiushin, and J. S. Boschwitz. 1997. Comparison of the sequences and functions of Streptococcus equi M-like proteins SeM and SzPSe. Infect. Immun. 65:3600-3605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Timoney, J. F., S. C. Artiushin, and J. Wang. 1999. Regions of the protective M-like protein of Streptococcus equi recognized by serum and mucosal antibodies of convalescent horses, p. 88-94. In U. Wernery, J. F. Wade, J. A. Mumford, and O. R. Kaaden (ed.), Equine infectious diseases VIII. Proceedings of the Eighth International Conference, Dubai, United Arab Emirates, 23 to 26 March 1998. R&W Publications, Newmarket, United Kingdom.
- 33.van Loo, I. H., K. J. Heuvelman, A. J. King, and F. R. Mooi. 2002. Multilocus sequence typing of Bordetella pertussis based on surface protein genes. J. Clin. Microbiol. 40:1994-2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Verheyen, K., J. R. Newton, N. C. Talbot, M. N. de Brauwere, and N. Chanter. 2000. Elimination of guttural pouch infection and inflammation in asymptomatic carriers of Streptococcus equi. Equine Vet. J. 32:527-532. [DOI] [PubMed] [Google Scholar]
- 35.Zadoks, R. N., Y. H. Schukken, and M. Wiedmann. 2005. Multilocus sequence typing of Streptococcus uberis provides sensitive and epidemiologically relevant subtype information and reveals positive selection in the virulence gene pauA. J. Clin. Microbiol. 43:2407-2417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang, W., B. M. Jayarao, and S. J. Knabel. 2004. Multi-virulence-locus sequence typing of Listeria monocytogenes. Appl. Environ. Microbiol. 70:913-920. [DOI] [PMC free article] [PubMed] [Google Scholar]