Abstract
Typing of human papillomaviruses (HPV) by DNA hybridization procedures, such as reverse line blot (RLB) assay, is sensitive and well validated. However, the application of these assays to high-throughput analyses is limited. Here, we describe the development of multiplex human papillomavirus genotyping (MPG), a quantitative and sensitive high-throughput procedure for the identification of multiple high- and low-risk genital HPV genotypes in a single reaction. MPG is based on the amplification of HPV DNA by a general primer PCR (GP5+/6+) and the subsequent detection of the products with type-specific oligonucleotide probes coupled to fluorescence-labeled polystyrene beads (Luminex suspension array technology). Up to 100 different HPV types can be detected simultaneously with MPG, and the method is fast and labor saving. We detected all 22 HPV types examined with high specificity and reproducibility (the median interplate coefficient of variation was below 10%). Detection limits for the different HPV types varied between 100 and 800 pg of PCR products. We compared the performance of MPG to an established RLB assay on GP5+/6+-PCR products derived from 94 clinical samples. The evaluation showed an excellent agreement (kappa = 0.922) but also indicated a higher sensitivity of MPG. In conclusion, MPG appears to be highly suitable for large-scale epidemiological studies and vaccination trials as well as for routine diagnostic purposes.
Cancer of the uterine cervix (cervical cancer) is the second most common cancer among women world-wide, with about 470,000 newly diagnosed cases and almost 250,000 deaths every year (19). Human papillomaviruses (HPV) are a necessary prerequisite for cervical cancer development (3, 15, 21, 25). More than 100 HPV types are known. HPV of the Alphapapillomavirus genus preferentially infect the oral or anogenital mucosa, and 29 types within this genus have been divided into three groups based on their association to cancer: 15 high-risk types (16, 18, 31, 33, 35, 39, 45, 51, 52, 56, 58, 59, 68, 73, and 82), 3 putative high-risk types (26, 53, and 66), and 11 low-risk types primarily found in genital warts and low-grade cervical lesions (6, 11, 40, 42, 43, 44, 54, 61, 70, 72, and 81) (15).
The current cornerstone for detection of cervical cancer precursor lesions is the Papanicolaou-stained (Pap) cytological smear. However, the identification of high-risk HPV types prompted the development of new methods for early cancer screening. Given its high sensitivity, testing for DNA of high-risk HPV types has been investigated in primary screening in combination with the Pap test but also as a stand-alone test (6, 13, 17). These studies have demonstrated that adjunct HPV DNA testing can identify women at increased risk for cervical cancer, in particular if persistent infection by individual high-risk HPV types is diagnosed (2, 3, 15, 16). Furthermore, HPV genotyping is also valuable for investigation of the clinical behavior and epidemiology of particular types, for the characterization of study populations in HPV vaccination trials, and for monitoring the efficacy of HPV vaccines.
Various techniques are in use for HPV DNA detection: (i) direct probe methods, such as Southern blotting and in situ hybridization, (ii) signal amplification methods, such as the hybrid capture 2 (HC2) assay, and (iii) target amplification performed by a variety of PCR-based techniques. For genotyping, PCRs are being followed by signal read-out methods, such as sequence analysis, restriction fragment length polymorphism analyses, or hybridization with type-specific probes by different formats, such as membrane-based reverse line blot (RLB) assay. Recently, several RLB assays based on different PCR protocols (MY09/11 [11], SPF [14], or GP5+/6+ [20]) have been developed and validated.
GP5+/6+-RLB employs the general primer GP5+/bio6+ primer set (the backward GP6+ primer is biotinylated) to amplify a large variety of mucosotropic HPV types. After PCR amplification, HPV sequences are detected by enzyme immunoassay (EIA), and subsequent typing is performed by hybridization of the biotinylated PCR products to type-specific oligonucleotides immobilized on membranes followed by their detection using an enhanced chemiluminescence reaction. Due to the format of the line blot strips, current assays are restricted to a maximum of about 40 oligonucleotide probes per hybridization reaction and depend upon visual read-out of the signal.
Luminex (xMAP) suspension array technology is based on polystyrene beads with a diameter of 5.6 μm that are internally dyed with various ratios of two spectrally distinct fluorophores. Thus, an array of 100 different bead sets with specific absorption spectra is created. Different molecules, such as individual oligonucleotide probes, can be coupled to different bead sets. These sets are combined to a suspension array and, due to their unique absorption spectra, allow up to 100 different probes to be measured simultaneously in a single reaction (multiplexing). The technology is capable of performing both protein- and nucleic acid-based analyses. Regarding protein analyses, the development of multiplex HPV serology has been recently described by our laboratory (23). Detection of nucleic acids using Luminex technology has been described previously (e.g., rRNA gene of bacterial pathogens [9] and genotyping of single-nucleotide polymorphisms [1, 8, 24]). Recently, the technology has been employed for the genotyping of 45 HPV types (22) using PCR products generated with PGMY09/11-PCR (10). This assay, however, showed reduced sensitivity for identifying high-risk HPV types in cervical lesions compared to the HC2 assay (60.9% versus 81.8%).
We have developed multiplex HPV genotyping (MPG), a simple bead-based high-throughput hybridization method that allows the simultaneous detection and genotyping of up to 100 HPV types. We describe here its application to 15 high-risk HPV types, 1 putative high-risk HPV type, and the 6 most prevalent low-risk HPV types. In comparison to RLB, MPG appears to be more sensitive for the detection of HPV in GP5+/6+-PCR products from clinical samples, and it is suitable for epidemiologic and also diagnostic applications.
(Parts of this work have been presented as a poster at the 22nd International Papillomavirus Conference, Vancouver, Canada, May 2005.)
MATERIALS AND METHODS
Online software.
Alignments of DNA sequences were performed with T-COFFEE, which combines information for both local and global homologies (18).
Plasmid clones.
For evaluation of sensitivity, reproducibility, and specificity of the MPG method, plasmid clones of the following HPV types were used: 6, 11, 16, 18, 31, 33, 35, 39, 42, 43, 44, 45, 51, 52, 56, 58, 59, 66, 68, 70, 73, and 82. For references see Table 1.
TABLE 1.
Oligonucleotide probes used for multiplex HPV genotyping of GP5+/6+-PCR products
| HPV type or probe namea | Sourceb | Accession no.c | Probe sequence (5′-3′) | Length |
|---|---|---|---|---|
| 6 | dV | X00203 | TCC GTA ACT ACA TCT TCC A | 19-mer |
| 11 | dV | M14119 | TCT GTG TCT AAA TCT GCT AC | 20-mer |
| 16d | dV | K02718 | TAC CTA CGA CAT GGG GAG | 18-mer |
| 18 | dV | X05015 | TGC TTC TAC ACA GTC TCC T | 19-mer |
| 31 | AL | J04353 | GCA ATT GCA AAC AGT GAT AC | 20-mer |
| 33 | GO | M12732 | TGC ACA CAA GTA ACT AGT GA | 20-mer |
| 35 | AL | M74117 | CTG CTG TGT CTT CTA GTG A | 19-mer |
| 39d | GO | M62849 | TAC ATT ATC TAC CTC TAT AGA | 21-mer |
| 42d | GO | M73236 | GCC ACT GCA ACA TCT GGT G | 19-mer |
| 43 | AL | AJ620205 | TCT ACT GAC CCT ACT GTG | 18-mer |
| 44 | AL | U31788 | TAC TAG TGA ACA ATA TAA GCA | 21-mer |
| 45 | GO | X74479 | TAA TTT AAC ATT ATG TGC CTC | 18-mer |
| 51 | dV | M62877 | TGC TGC GGT TTC CCC AA | 17-mer |
| 52 | WL | X74481 | GAA TAC CTT CGT CAT GGC | 18-mer |
| 56d | AL | X74483 | GAT GCA CGA AAA ATT AAT CAG | 21-mer |
| 58 | TM | D90400 | TAT GCA CTG AAG TAA CTA AG | 20-mer |
| 59d | TM | X77858 | AGA ATA TGC CAG ACA TGT G | 19-mer |
| 66d | GO | U31794 | CGT GAA ATC AAT CAA TAC CTT C | 22-mer |
| 68 | GO | AJ831568 | CTG AAT CAG CTG TAC CAA A | 19-mer |
| 70d | GO | U22461 | TTT ACA TTG TCT GCC TGC A | 19-mer |
| 73d | dV | NC_006165 | GTA TGC CAA CTC WAA TTT TAAe | 21-mer |
| 82d | TM | AB027021 | ACT CCA RCA AAC TTT AAG CAG Tf | 22-mer |
| Universald | GiC ATG iiG ARG AAT ATG Ag | 19-mer |
As published previously (20).
HPV plasmid DNA was kindly provided by dV (E.-M. de Villiers; DKFZ, Heidelberg, Germany), GO (G. Orth; Institut Pasteur, Paris, France), AL (A. Lörincz; Digene Corp., Gaithersburg, MD), TM (T. Matsukura; National Institute of Infectious Diseases, Tokyo, Japan), and WL (W. Lancaster; Wayne State University, Detroit, MI).
Sequence used as a reference for the corresponding HPV genotype.
Redesigned probes.
W, A/T.
R, A/G.
i, Inosin.
Clinical specimens.
During the course of an ongoing population-based cervical screening trial, cervical scrapings were subjected to GP5+/bio6+-PCR followed by EIA and RLB genotyping (4, 5). A representative set of GP5+/bio6+-PCR products of 88 EIA-positive samples containing a broad range of HPV types, as determined by RLB, and 6 HPV-negative samples were selected and blindly reanalyzed by MPG in this study.
HPV sequences from GenBank.
HPV sequences of different isolates and complete genomes with the following accession numbers were obtained from GenBank and used for probe redesign (complete genome sequences marked with an asterisk are the reference for the corresponding HPV prototype): HPV-39, M62849*, AF548857, AF548856, M38185, U45899, U45900, U45901, U45902, U45903, U45904, and U45905; HPV-42, M73236* and A28090; HPV-56, X74483*, S40273, DQ007188, DQ007182, DQ007183, DQ007184, DQ007185, DQ007186, and DQ007187; HPV-59, X77858*, AF374230, U45930, U45931, U45932, U45933, and U12496; HPV-66, U31794*, NC_001695, U12498, and AY147908; HPV-70, U22461*, U21941, and NC_001711; HPV-73, NC_006165*, X94165, and Y12222; and HPV-82, AB027021*, U45939, U12484, U12482, U12488, U12483, AJ831565, U01532, U12481, and AF293961. The novel HPV-16 probe completely matched to 72 of 72 HPV-16 GenBank sequences containing the GP5+/6+ L1 fragment in an uninterrupted reading frame.
GP5+/6+-PCR.
Dilutions of HPV plasmid DNA in a background of 100 ng human placental DNA were subjected to GP5+/6+-PCR as previously described (7, 12, 20). Briefly, amplification of the ∼150-bp-long fragment of the viral L1 open reading frame was performed in 50 μl PCR mixture containing 50 mM KCl, 0.8 g/liter Nonidet P40, 10 mM Tris HCl, pH 8.8 (MBI Fermentas GmbH, St. Leon Roth, Germany), 200 μM of each deoxynucleoside triphosphate, 3.5 mM MgCl2, 1 U of DNA AmpliTaq polymerase (Roche Applied Biosystems, Mannheim, Germany), and 25 pmol each of the GP5+ (5′-TTT GTT ACT GTG GTA GAT ACT AC-3′) and 5′-biotinylated GP6+ (5′-GAA AAA TAA ACT GTA AAT CAT ATT C-3′) primers (MWG-Biotech AG, Ebersberg, Germany). A 4-min denaturation step at 94°C was followed by 40 cycles of amplification with a PCR thermocycler (Gene Amp PCR System 2400; Perkin-Elmer, Wellesley, MA). Each cycle included a denaturation step at 94°C for 20 s, an annealing step at 38°C for 30 s, and an elongation step at 71°C for 80 s. The final elongation step was prolonged for a further 4 min. Similar reaction and cycling conditions were used for GP5+/6+-PCR on 10 μl of crude extracts of cervical scrapings (4, 5).
Quantification of PCR products.
Serial 1:2 dilutions of HPV PCR products in TE (10 mM Tris-HCl, 1 mM EDTA, pH 8.0) ranging from 4.0 μl to 0.25 μl were separated on a 2.0% agarose gel together with 5.0 μl of Smart-Ladder DNA quantification marker (Eurogentec, Seraing, Belgium). After staining with ethidium bromide (1.0 mg/liter), digital photographs were taken and concentrations were determined using ImageQuant TL v2003.02 software (Amersham Biosciences) by comparing net intensities of nonsaturated PCR product bands with those of the quantification marker.
Coupling of oligonucleotide probes.
The sequences of 5′ amino-modifier C-12-linked oligonucleotide probes (MWG-Biotech) are shown in Table 1. Published (“old”) probes that generated insufficient signal-to-noise ratios or recognizing too few variants were redesigned (“novel”). Probes were coupled to carboxylated beads by a carbodiimide-based coupling procedure. For each combination of probe and bead set, 2.5 million carboxylated beads (xMAP; Luminex Corp., Austin, TX) were suspended in 25 μl of 0.1 M 2-(N-morpholino)ethanesulfonic acid, pH 4.5 (MES). Probe oligonucleotides (400 pmol) and 200 μg of N-(3-dimethylaminopropyl)-N-ethylcarbodiimide (EDC) were added and thoroughly mixed with the beads. Incubation was performed in the dark under agitation for 30 min and was interrupted by a thorough mixing step after 15 min. The addition of EDC and incubation steps were repeated, and the coupled beads were finally washed once with 0.5 ml of 0.2 g/liter Tween-20 and once with 0.5 ml of 1.0 g/liter sodium dodecyl sulfate before being stored in 100 μl of TE buffer at 4°C in the dark. Coupling efficiency of new bead batches compared to old ones was verified by hybridization to 1.0 to 4.0 μl of biotinylated PCR product of the respective HPV. New coupling batches showing a coefficient of variation (CV) below 15% were used for further analyses (data not shown).
Hybridization assay.
Following PCR amplification, 10 μl of each reaction mixture was transferred to 96-well plates (96-well PCR thermo polystyrene plates; Costar, Wiesbaden, Germany) containing 33 μl of tetramethylammonium chloride (TMAC) hybridization solution (0.15 M TMAC, 75 mM Tris-HCl, 6 mM EDTA, 1.5 g/liter Sarkosyl, pH 8.0) and a mixture of 2,000 probe-coupled beads of each set. TE buffer (7.0 μl) was added, followed by gentle mixing with a 12-channel pipette (Biohit PLC). The mixture was heated to 95°C for 10 min in a laboratory oven (Bachofer, Reutlingen, Germany), immediately placed on ice for 2 min, and then transferred to a thermomixer (Eppendorf, Hamburg, Germany). Hybridization was performed at 41°C for 30 min under agitation. Using a 12-channel pipette (Brand, Roskilde, Denmark), the samples were transferred to a 96-well wash plate (Millipore, Bedford, MA) preequilibrated with blocking buffer (phosphate-buffered saline, 1 mg/ml casein). Subsequently, the beads were washed once with 100 μl of blocking buffer on a vacuum wash station (Millipore). On a horizontal shaker at room temperature, beads were resuspended for 20 min in 75 μl of streptavidin-R-phycoerythrin (Strep-PE; Molecular Probes, Eugene, OR) diluted 1:1,600 in 2.0 M TMAC, 75 mM Tris-HCl, 6 mM EDTA, 1.5 g/liter Sarkosyl, 1.0 g/liter casein, pH 8.0. Beads were then washed three times with 100 μl blocking buffer and finally resuspended in 100 μl blocking buffer for 5 min on a shaker. Beads were analyzed for internal bead color and R-phycoerythrin reporter fluorescence on a Luminex 100 analyzer. The median reporter fluorescence intensity (MFI) of at least 100 beads was computed for each bead set in the sample.
Cutoff definition and statistics.
Reactions of probes with PCR products from other HPV types were considered background values. The cutoff was defined for each HPV probe individually according to the following criteria: (i) mean background below 5 MFI led to a cutoff of 10 MFI (HPV types 6, 11, 16, 31, 33, 35, 43, 56, 58, 59, 66, and 68), (ii) mean background of 6 to 10 MFI to a cutoff of 15 MFI (HPV types 18, 39 [new probe], 44, 45, 52, 70, 73, 82, and the universal probe), and (iii) mean background of 11 to 20 MFI to a cutoff of 25 MFI (HPV type 42 [new probe]). The cutoffs for the old HPV-39 and HPV-42 probes were 100 and 75 MFI, respectively. The HPV-51 probe background showed a certain degree of interexperimental variation (between 7 and 17 MFI) and required cutoff adjustment (15 or 25 MFI) for each experiment (Table 2 and Table 3). In all cases the defined cutoff value was above the mean background plus five times the standard deviations (data not shown).
TABLE 2.
Specificity and detection limits of 22 type-specific probes and the universal probe in multiplex HPV genotyping (each line represents a single well with the PCR product hybridized to a mixture of 23 distinct bead sets)
| HPV PCR product | HPV type-specific probe:
|
unia | mmb uni | DLc uni | |||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 11 | 16 | 18 | 31 | 33 | 35 | 39 | 42 | 43 | 44 | 45 | 51 | 52 | 56 | 58 | 59 | 66 | 68 | 70 | 73 | 82 | ||||
| 6 | 179 | 2d | 3 | 7 | 2 | 2 | 2 | 8 | 18 | 2 | 6 | 9 | 16 | 8 | 4 | 3 | 4 | 3 | 4 | 6 | 8 | 6 | 34 | 0 | 6.4 |
| 11 | 3 | 624 | 3 | 7 | 2 | 2 | 2 | 8 | 16 | 2 | 6 | 8 | 15 | 8 | 4 | 2 | 4 | 3 | 4 | 6 | 7 | 6 | 113 | 1 | 3.2 |
| 16 | 4 | 2 | 592 | 6 | 2 | 2 | 2 | 8 | 16 | 2 | 5 | 9 | 14 | 8 | 5 | 9e | 4 | 3 | 3 | 4 | 7 | 7 | 70 | 1 | 3.2 |
| 18 | 4 | 1 | 2 | 316 | 2 | 2 | 2 | 8 | 17 | 2 | 6 | 10 | 15 | 9 | 4 | 3 | 4 | 3 | 3 | 5 | 7 | 6 | 205 | 1 | 1.6 |
| 31 | 4 | 1 | 3 | 6 | 146 | 2 | 2 | 8 | 15 | 2 | 6 | 9 | 14 | 9 | 4 | 2 | 5 | 3 | 4 | 5 | 8 | 6 | 117 | 1 | 0.8 |
| 33 | 4 | 1 | 2 | 8 | 2 | 269 | 2 | 8 | 16 | 3 | 6 | 8 | 16 | 9 | 4 | 3 | 4 | 4 | 4 | 6 | 8 | 7 | 125 | 1 | 1.6 |
| 35 | 4 | 2 | 2 | 7 | 2 | 2 | 328 | 8 | 16 | 2 | 6 | 8 | 15 | 7 | 4 | 2 | 4 | 3 | 3 | 5 | 7 | 6 | 144 | 0 | 0.8 |
| 39 | 4 | 1 | 2 | 7 | 2 | 1 | 2 | 186 | 16 | 2 | 6 | 8 | 15 | 8 | 4 | 2 | 4 | 4 | 3 | 6 | 7 | 6 | 62 | 1 | 1.6 |
| 42 | 4 | 2 | 2 | 7 | 2 | 4 | 2 | 8 | 513 | 2 | 6 | 9 | 16 | 10 | 4 | 2 | 3 | 3 | 4 | 5 | 7 | 6 | 56 | 2 | 6.4 |
| 43 | 4 | 1 | 2 | 8 | 2 | 2 | 2 | 8 | 15 | 293 | 5 | 8 | 19 | 8 | 4 | 2 | 4 | 4 | 3 | 5 | 7 | 6 | 227 | 0 | 1.6 |
| 44 | 4 | 1 | 2 | 11 | 2 | 2 | 2 | 8 | 18 | 3 | 455 | 8 | 15 | 9 | 4 | 2 | 4 | 3 | 4 | 7 | 8 | 6 | 208 | 1 | 1.6 |
| 45 | 3 | 1 | 12e | 8 | 2 | 2 | 2 | 7 | 15 | 2 | 6 | 324 | 14 | 8 | 3 | 2 | 4 | 4 | 3 | 6 | 7 | 5 | 92 | 1 | 3.2 |
| 51 | 4 | 1 | 2 | 7 | 2 | 2 | 2 | 7 | 16 | 2 | 6 | 8 | 537 | 9 | 4 | 2 | 4 | 3 | 4 | 5 | 8 | 13f | 87 | 0 | 1.6 |
| 52 | 4 | 2 | 3 | 7 | 2 | 2 | 3 | 8 | 17 | 3 | 5 | 8 | 16 | 653 | 4 | 2 | 4 | 8f | 3 | 6 | 8 | 6 | 50 | 1 | 3.2 |
| 56 | 3 | 2 | 4 | 7 | 2 | 2 | 1 | 7 | 17 | 2 | 6 | 8 | 14 | 8 | 830 | 2 | 4 | 3 | 4 | 6 | 7 | 7 | 121 | 1 | 3.2 |
| 58 | 3 | 1 | 2 | 7 | 1 | 2 | 2 | 8 | 17 | 2 | 6 | 9 | 15 | 9 | 3 | 255 | 4 | 4 | 4 | 6 | 8 | 6 | 258 | 0 | 0.4 |
| 59 | 4 | 2 | 2 | 7 | 2 | 2 | 2 | 8 | 17 | 2 | 6 | 7 | 14 | 8 | 4 | 2 | 171 | 3 | 3 | 6 | 8 | 6 | 119 | 1 | 3.2 |
| 66 | 4 | 2 | 4 | 7 | 2 | 1 | 2 | 8 | 17 | 2 | 6 | 8 | 14 | 9 | 4 | 2 | 5 | 547 | 3 | 6 | 7 | 6 | 70 | 1 | 6.4 |
| 68 | 4 | 1 | 2 | 8 | 2 | 2 | 2 | 8 | 15 | 2 | 6 | 9 | 14 | 8 | 3 | 2 | 4 | 4 | 467 | 6 | 7 | 7 | 418 | 0 | 0.8 |
| 70 | 4 | 1 | 2 | 7 | 2 | 2 | 2 | 8 | 17 | 2 | 6 | 9 | 14 | 8 | 4 | 2 | 4 | 3 | 3 | 480 | 7 | 6 | 353 | 0 | 0.8 |
| 73 | 4 | 1 | 2 | 7 | 2 | 2 | 2 | 7 | 16 | 2 | 5 | 8 | 15 | 8 | 4 | 2 | 4 | 3 | 4 | 5 | 120 | 6 | 43 | 3 | 12.8 |
| 82 | 4 | 2 | 2 | 7 | 2 | 2 | 2 | 8 | 17 | 2 | 6 | 9 | 15 | 9 | 4 | 3 | 4 | 3 | 4 | 6 | 8 | 459 | 95 | 1 | 1.6 |
| Noneg | 4 | 2 | 2 | 8 | 2 | 3 | 2 | 7 | 16 | 3 | 7 | 9 | 14 | 9 | 5 | 3 | 4 | 4 | 5 | 7 | 7 | 6 | 7 | ||
| Cutoff | 10 | 10 | 10 | 15 | 10 | 10 | 10 | 15 | 25 | 10 | 15 | 15 | 25 | 15 | 10 | 10 | 10 | 10 | 10 | 15 | 15 | 15 | 15 | ||
| DLh | 0.4 | 0.2 | 0.1 | 0.1 | 0.4 | 0.2 | 0.2 | 0.2 | 0.1 | 0.4 | 0.8 | 0.2 | 0.2 | 0.2 | 0.2 | 0.4 | 0.8 | 0.2 | 0.2 | 0.2 | 0.8 | 0.2 | |||
uni, Universal probe.
mm uni, mismatch of universal probe to prototype sequence.
DL uni, detection limit of universal probe to PCR product, in nanograms.
Probe-dependent background generated from PCR products that did not hybridize to the respective probe.
Weak additional reactivity probably due to probe contamination during synthesis (see Results, section “Specificity”).
Weak possible cross-reactivity despite the presence of at least four mismatches.
Background generated without PCR product present.
Detection limit of specific probe (in nanograms) based on the amount of PCR products required to obtain a signal above cutoff.
TABLE 3.
Representative examples for multiplex HPV genotyping of GP5+/6+-PCR products from clinical specimens, including all 17 typing reactions (in specimens 1 to 14) discordant between MPG and RLBe
| Clinical specimen no. | HPV type-specific probe
|
unib | Typing result
|
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 | 11 | 16 | 18 | 31 | 33 | 35 | 39a | 42a | 43 | 44 | 45 | 51 | 52 | 56 | 58 | 59 | 66 | 68 | 70 | 73 | 82 | MPG | RLB | ||
| 1 | 4 | 1 | 3 | 8 | 2 | 2 | 3 | 70 | 52 | 2 | 6 | 9 | 144 | 9 | 193 | 3 | 27 | 3 | 5 | 7 | 7 | 5 | 91 | 51, 56, 59 | 51, 56 |
| 2 | 4 | 1 | 2 | 8 | 2 | 2 | 2 | 68 | 48 | 2 | 6 | 9 | 7 | 501 | 4 | 2 | 238 | 3 | 6 | 7 | 6 | 5 | 134 | 52, 59 | 52 |
| 3 | 4 | 1 | 3 | 8 | 2 | 32 | 3 | 66 | 45 | 2 | 6 | 8 | 8 | 10 | 4 | 3 | 187 | 4 | 6 | 7 | 7 | 5 | 164 | 33, 59 | 33 |
| 4 | 4 | 1 | 3 | 8 | 2 | 2 | 2 | 66 | 219 | 2 | 6 | 9 | 7 | 10 | 4 | 3 | 180 | 3 | 5 | 7 | 7 | 6 | 65 | 42, 59 | 42 |
| 5 | 5 | 2 | 20 | 8 | 2 | 457 | 4 | 66 | 48 | 3 | 6 | 10 | 7 | 9 | 5 | 3 | 5 | 4 | 5 | 8 | 8 | 5 | 67 | 16, 33 | 33 |
| 6 | 4 | 1 | 677 | 8 | 2 | 2 | 3 | 72 | 76 | 2 | 5 | 10 | 8 | 11 | 3 | 11c | 4 | 4 | 6 | 36 | 6 | 5 | 147 | 16, 42, 70 | 16 |
| 7 | 4 | 1 | 14c | 8 | 3 | 2 | 2 | 80 | 41 | 2 | 6 | 302 | 6 | 47 | 4 | 2 | 4 | 3 | 6 | 6 | 6 | 6 | 233 | 45, 52 | 45 |
| 8 | 3 | 2 | 2 | 8 | 2 | 2 | 3 | 86 | 76 | 2 | 6 | 5 | 17 | 869 | 3 | 2 | 5 | 3 | 5 | 7 | 8 | 6 | 254 | 42, 51, 52 | 52 |
| 9 | 3 | 1 | 28 | 8 | 2 | 2 | 3 | 71 | 49 | 2 | 5 | 9 | 18 | 1043 | 838 | 2 | 4 | 3 | 6 | 8 | 7 | 5 | 208 | 16, 51, 52, 56 | 52, 56 |
| 10 | 3 | 2 | 2 | 119 | 2 | 2 | 4 | 60 | 51 | 2 | 5 | 10 | 9 | 9 | 4 | 2 | 4 | 3 | 5 | 6 | 7 | 21 | 90 | 18, 82 | 18 |
| 11 | 4 | 2 | 12 | 345 | 2 | 2 | 2 | 68 | 42 | 2 | 5 | 9 | 6 | 7 | 4 | 2 | 3 | 4 | 5 | 6 | 6 | 5 | 130 | 16, 18 | 18 |
| 12 | 4 | 1 | 3 | 7 | 2 | 2 | 3 | 76 | 44 | 2 | 5 | 7 | 6 | 7 | 4 | 3 | 4 | 3 | 4 | 6 | 6 | 5 | 13 | 70 | |
| 13 | 4 | 1 | 3 | 7 | 2 | 2 | 2 | 68 | 79 | 2 | 6 | 9 | 6 | 1260 | 4 | 2 | 4 | 3 | 5 | 7 | 7 | 5 | 176 | 42, 52 | 52 |
| 14 | 4 | 1 | 2 | 8 | 1 | 2 | 3 | 65 | 671 | 2 | 6 | 8 | 7 | 9 | 4 | 2 | 4 | 3 | 5 | 6 | 6 | 5 | 56 | 42 | |
| 15 | 3 | 2 | 2 | 7 | 2 | 2 | 2 | 71 | 44 | 2 | 5 | 8 | 6 | 9 | 6 | 3 | 4 | 3 | 21 | 6 | 6 | 6 | 92 | 68 | 68 |
| 16 | 3 | 1 | 2 | 8 | 2 | 1 | 2 | 62 | 55 | 2 | 6 | 9 | 6 | 10 | 4 | 2 | 4 | 3 | 6 | 6 | 6 | 6 | 36 | uni | 30 |
| 17 | 4 | 2 | 2 | 7 | 3 | 3 | 3 | 83 | 38 | 3 | 7 | 7 | 5 | 7 | 4 | 2 | 5 | 3 | 6 | 7 | 6 | 6 | 45 | uni | |
| 18 | 4 | 2 | 2 | 7 | 2 | 2 | 2 | 71 | 45 | 3 | 6 | 9 | 7 | 9 | 4 | 3 | 4 | 4 | 5 | 7 | 6 | 7 | 11 | ||
| Noned | 4 | 1 | 2 | 7 | 2 | 2 | 2 | 69 | 48 | 2 | 6 | 6 | 7 | 9 | 3 | 2 | 4 | 3 | 5 | 5 | 7 | 6 | 6 | ||
| Cutoff | 10 | 10 | 10 | 15 | 10 | 10 | 10 | 100 | 75 | 10 | 15 | 15 | 15 | 15 | 10 | 10 | 10 | 10 | 15 | 15 | 15 | 15 | 15 | ||
Old HPV-39 and HPV-42 probes (later replaced due to high background; see Results, section “Type-specific probe design”).
uni, Universal probe.
Unspecific reactivities due to probe contamination (see Results, section “Specificity”).
Background generated without PCR product present.
Each row represents a single well with one PCR product hybridized to a mixture of 23 distinct bead sets.
The correlation between RLB and MPG was assessed using kappa statistics. The coefficient of correlation (R2), the slope of the regression line, and the coefficient of variation (CV) were computed to describe assay reproducibility.
RLB of GP5+/6+-PCR products.
RLB analyses were performed as previously described (20). Using a miniblotter, 37 different oligonucleotide probes modified with 5′-amino groups were spotted on a carboxylated nylon membrane and bound by carbodiimide coupling chemistry. Subsequently, up to 40 PCR products were pipetted into the parallel channels of the miniblotter in such a way that the channels were perpendicular to the rows of probes deposited previously (20). Hybridization of the biotinylated PCR products to the probes was followed by incubation of the membrane with antibiotin conjugate and enhanced chemiluminescence detection.
RESULTS
Development of multiplex HPV genotyping. (i) Principle of the assay.
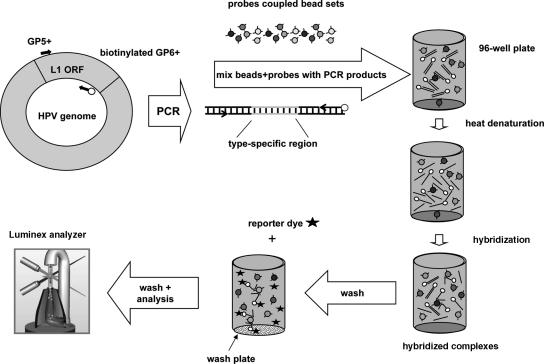
GP5+/bio6+-PCR products are generated, denatured, and hybridized to the bead-coupled probes in 96-well plates, allowing PCR products from 96 individual specimens to be processed in parallel. After transfer into wash plates with filter bottoms, unhybridized DNA is removed. Subsequently, biotinylated PCR products are stained by Strep-PE conjugate. After further washing steps, beads are analyzed in the Luminex reader, which contains two lasers to identify the bead set by the internal bead color and to quantify the reporter fluorescence on the bead. The result is expressed as the median fluorescence intensity (MFI) of at least 100 beads per set (Fig. 1).
FIG. 1.
Schematic overview of HPV genotyping of GP5+/6+-PCR products by bead-based multiplex HPV genotyping. ORF, open reading frame. (Picture of the Luminex analyzer reproduced from a tutorial on the Luminex Corp. website with permission.)
(ii) Assay optimization.
Final assay conditions used for this study were established after systematic variation of the following parameters: coupling procedure of probes to beads (EDC and probe input); temperature, salt concentration, and hybridization conditions; Strep-PE concentration; incubation time for staining; concentrations of different blocking substances in the washing buffer; and number of wash cycles and wash buffer composition (see Materials and Methods). For optimization of the protocol reproducibility, signal-to-noise ratios and detection limits were analyzed (data not shown).
(iii) Type-specific probe design.
HPV type-specific hybridization probes for GP5+/6+-PCR products analyzed here have been published previously (20) and were used in the initial experiments. However, some of these “old” probes exhibited different limitations (see below); therefore, alternative “novel” probe sequences were designed and evaluated (Table 1). All novel probes were designed to completely match their corresponding HPV prototype and all other sequences of this type (except HPV-16; see Materials and Methods) available in the National Center for Biotechnology Information nucleotide sequence database (GenBank) and to exhibit at least three mismatches with all other mucostropic HPV types. Published probes for HPV types 16, 39, 42, 56, 59, 66, and 70 generated insufficient signals and/or high probe-dependent background (between 20 and 90 MFI), resulting in poor detection limits (>800 pg of PCR product). In order to recognize additional published HPV type variants, novel HPV-73 and HPV-82 probes with one degenerated nucleotide were designed that target highly conserved regions shared by all variants of the respective type. Novel probes were designed to anneal at melting temperatures between 50 and 55°C, resulting in probe lengths of 18 to 22 nucleotides. When hybridized to 100 ng of plasmid-derived PCR product, novel probes showed reduced background (except for HPV-73) compared to old probes, 2- to 8-fold enhanced signal values, and 2- to 30-fold improved signal-to-noise ratios. Detection limit was four- to eightfold improved by using novel probes, except for HPV-73, for which it remained unchanged (data not shown). PCR products from five of eight HPV-42-positive clinical samples also were available for retesting with the novel probe and showed identical typing results.
(iv) Universal probe.
We developed a universal probe comprising a conserved region of 11 nucleotides upstream of the GP6+ sequence that detects all 22 mucosotropic HPV types included in this study. The universal probe overlapped in eight nucleotides with the GP6+ primer sequence. Nucleotide variations found with some of the 22 HPV types were partly overcome with the introduction of inosines (base pairing with adenine and cytosine) or nucleotide degenerations at five positions. The universal probe showed perfect matches with 7 of the 22 examined HPV types (32%), one mismatch with 13 HPV types (59%), and two and three mismatches, respectively, with HPV-42 and HPV-73. The universal probe recognized all 22 HPV types analyzed, but the hybridization signal was mostly dependent on the number of mismatches (Table 2).
(v) Specificity.
In order to determine the specificity of MPG, the 22 type-specific probes and the universal probe were coupled individually to defined bead sets (“23-plex”) and hybridized to 1 μl (∼20 to 30 ng DNA) of PCR products derived from saturated GP5+/6+-PCRs on 1 ng of cloned plasmid template. Typing of all HPV types was highly specific (Table 2); it was also highly specific when 100 ng DNA of PCR products was used (data not shown). Depending on the concentration of the PCR product and the HPV type, 1 μl of GP5+/6+-PCR product generated signals between 120 and 830 MFI. From the 22 examined HPV types, 18 probes reacted with absolute specificity. However, four probes showed very weak reactivity with one additional HPV type each (Table 2). The reactivity of the HPV-66 probe with HPV-52 and of the HPV-82 probe with HPV-51 could be due to cross-hybridization despite the presence of four mismatches in each pair. The reactivity of the HPV-16 probe with HPV-45 and of the HPV-58 probe with HPV-16 was probably due to probe contamination during synthesis in the same synthesis batch. This interpretation is based on high dissimilarity (at least six mismatches separate the pairs) and on the observation that resynthesized HPV-16 and HPV-58 probes lacked the additional reactivity. Taken together, DNA detection of 22 mucosal HPV types examined was highly specific and revealed a median signal-to-noise ratio (specific MFI/mean background) of 73 (range, 17 to 312) (Table 2). The table also demonstrates the simple read-out of results due to quantitative signal values and the stability of the background observed with all probes.
(vi) Detection limit.
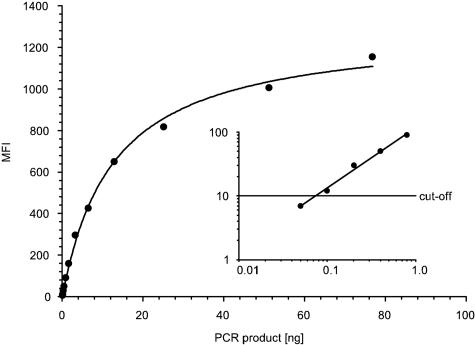
In general, the analytic sensitivity of RLB and MPG is a function of the sensitivity of the PCR amplification and the detection limit of probes. The objective of this work was the development and validation of a multiplex hybridization assay for HPV genotyping, taking advantage of the well-characterized system of the GP5+/6+-PCR. To determine detection limits of probes using cutoff values, twofold dilutions of PCR products (between 76.8 ng and 50 pg obtained from the individual HPV plasmids) were hybridized to the universal and HPV type-specific probes. For type-specific probes, detection limits varied between 800 and 100 pg of PCR products (Fig. 2, Table 2). For the universal probe, the detection limits varied between 12.8 ng and 400 pg of PCR products depending on the number of mismatches (Table 2).
FIG. 2.
Detection limit analysis of the HPV-16 probe on twofold dilutions of HPV-16 GP5+/6+-PCR products. The graph displays the fit of the experimental values to a hyperbola. The inset shows the linear fitting (R2 = 0.991) of the five smallest amounts of PCR products in double-logarithmic scale. The detection limit was determined using the cutoff of 10 MFI.
(vii) Reproducibility.
To test reproducibility of MPG, we used a set of 21 different plasmid-derived PCR products. These products were mixed in 16 samples to obtain double positives and were hybridized to 10 distinct probe-coupled bead sets. The experiment was repeated twice, once on the same plate and once on a second plate. Intraplate and interplate reproducibility of all 160 data points was very high, with neither individual (R2 = 0.996 and 0.993, respectively) nor systematic variation (slope of the regression line, 0.965 and 0.953, respectively). The median intraplate coefficient of variation (CV) for specific signals was 4.5% (range, 0.4 to 13.2%), and the median interplate CV for specific signals was 5.2% (range, 0.4 to 14.7%). However, signals obtained with the universal probe exhibited higher intraplate (9.9%; range, 5.2 to 23.6%) and interplate (11.8%; range, 4.0 to 52.1%) CVs.
We also determined interday assay reproducibility for genotyping of PCR products derived from 40 clinical specimens (4, 5). The two experiments were performed within 2 weeks using different mixes of 22 type-specific probe-coupled bead sets from the same coupling batch. Of all typing results, 52 were concordantly positive (5.9%), 824 were concordantly negative (93.6%), and 4 were discordant (0.5%), yielding a kappa value of 0.96. For concordantly positive-typed reactions, the linear correlation coefficient (R2) was 0.878 (data not shown). The slope of the trend line was 0.996, indicating low systematic variation between both experiments. The median interday CV for specific signals was 13.8% (range, 0.4 to 44.8%), but again the universal probe showed a more elevated median CV of 39.2% (range, 3.3 to 105.5%). The discordant reactions, two with HPV-16 and two with HPV-68, showed borderline signals differing by an MFI of 8 or less in three cases and one HPV-16 reaction exhibited 3 and 20 MFI, respectively. Among all multiple infections detected in one of the two experiments, 10 of 13 double and 2 of 3 triple infections were concordantly genotyped.
Analysis of clinical samples and comparison with reverse line blot (RLB).
We next aimed to test the suitability of MPG for the analysis of clinical samples. To this end, we analyzed GP5+/GP6+-PCR products of 94 cervical smears (88 HPV positive, 6 HPV negative) from a population-based screening trial (4, 5). Forty of these samples could be analyzed twice; the four discordant reactions (see above) were classified as negative. According to RLB typing, the 40 samples contained a total of 17 different HPV types, present in single or multiple infections. The PCR products were genotyped by MPG, and the previous RLB results were unblinded only after the analysis was completed. Typing was performed with 10 μl of product from the same PCR run both in the previous RLB analysis and in this study, excluding any PCR variation.
Of all typing results, 107 were concordantly positive (5%), 1,944 were concordantly negative (94%), and 17 were discordant (1%), yielding a kappa value of 0.922. This resulted in identical typing results for 80 of the 94 clinical samples (85%).
Compared to RLB, MPG failed to detect a single infection with HPV-70 in one sample but identified 16 additional infections (Tables 3 and 4). Fifteen of them were found in samples with multiple infections by MPG only. Eight of these reactions showed strong MFI signals, while weak MFI signals (<10 MFI above cutoff) were generated in eight of the remaining reactions, indicating low viral loads. Among the eight discordant samples with strong MPG MFI signals, HPV-59 was detected in four cases, whereas HPV-42, HPV-52, HPV-70, and HPV-82 were scored within the other four. From six samples HPV-negative by RLB, five were concordant but one was strongly HPV-42 positive by MPG.
TABLE 4.
Detection of HPV genotypes within 94 clinical samples by GP5+/6+-PCR followed by MPG or RLB
| Typing reaction | HPV typea
|
Total | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 11 | 16 | 18 | 31 | 33 | 35 | 39 | 42 | 45 | 51 | 52 | 56 | 58 | 59 | 66 | 68 | 70 | 82 | ||
| Both | 1b | 24 | 16 | 6 | 7 | 5 | 2 | 4 | 9 | 5 | 7 | 9 | 4 | 2 | 1 | 4 | 1 | 107 | |
| RLB only | 1 | 1 | |||||||||||||||||
| MPG only | 3 | 4 | 2 | 1 | 4 | 1 | 1 | 16 | |||||||||||
| Total | 1 | 27 | 16 | 6 | 7 | 5 | 2 | 8 | 9 | 7 | 8 | 9 | 4 | 4 | 2 | 1 | 6 | 2 | 124 |
For sequence accession numbers see Materials and Methods.
Number of positive reactions for given HPV type; from eight total HPV-42-positive samples, four were detected by both methods and four additional reactions were detected only by MPG. HPV-6, HPV-43, HPV-44, and HPV-73 had no positive reactions.
The increased number of MPG positives may reflect an overall higher sensitivity of the MPG assay, improved detection of variants by the novel probes, e.g., HPV-59, and/or higher sensitivity of the novel probes. The latter is further supported by a sample showing an HPV-16 signal that was weak in the RLB but almost 10-fold above cutoff in the MPG (data not shown). Alternatively, the detection of additional types by MPG may also arise from the use of novel probes (e.g., HPV probes 59, 70, and 82) that better recognize variants displaying sequence variations in the GP5+/6+ region.
A sample of MPG typing results including all discordant typing reactions is presented in Table 4. Triple and quadruple HPV infections were detected as easily as single infections (samples 1, 6, 8, and 9), and known additional unspecific reactivity did not interfere with final analysis (samples 6 and 7). Sample 16, typed HPV-30 positive by RLB, showed no MPG type-specific result but a positive universal probe result since HPV-30 was not included in the MPG setup. The result seen with sample 17, which was both RLB- and MPG-negative type specific but MPG positive with the universal probe, suggested the presence of an amplified HPV type that was not recognized by type-specific probes. The weak HPV-68 (21 MFI) but high universal probe (92 MFI) signal in sample 15 might also reflect the presence of additional HPV types. HPV-negative sample 18 exhibited neither type-specific nor universal probe signal.
Taken together, these data demonstrate an excellent agreement of our MPG assay with RLB. Furthermore, they indicate a higher analytic sensitivity of MPG, as demonstrated here for multiple infections.
DISCUSSION
Conventional membrane-based genotyping methods such as the reverse line blot (RLB) assay are based on PCR technology and type-specific hybridization with oligonucleotide probes. The GP5+/6+-PCR-RLB allows the detection of multiple infections and is very sensitive (0.1 to 1 ng of PCR product required) and well validated (20). However, RLB assays are limited with regard to the number of HPV types that can be detected simultaneously in one reaction and are relatively laborious (20). Genotyping of epidemiological studies and vaccination trials with thousands of specimens is time-consuming and costly. In addition, RLB assays provide only semiquantitative results as read-out is visual, which may result in reproducibility problems and risks of mistakes during data entry.
Here, we developed a high-throughput multiplex assay that allows fast and simple genotyping of all clinically relevant genital HPV types in a single reaction yielding quantitative results. Comparing MPG to an established RLB assay, we found excellent agreement between both methods. We obtained evidence for a higher sensitivity of MPG by the detection of 15 additional HPV types in multiple infections. Three factors probably contribute to higher sensitivity: (i) improved detection of variants by novel probes, (ii) improved detection limits of novel probes, and (iii) overall higher sensitivity of the MPG assay. The third aspect will be addressed in more detail in a larger collection of EIA-negative PCR products from clinical samples.
The high sensitivity of this detection system revealed very weak, possible cross-hybridization of the HPV-66 probe with high concentrations of HPV-52 PCR products and of the HPV-82 probe with HPV-51, despite the four mismatches within the pairs. Additionally, probe contamination during commercial oligonucleotide synthesis can occur (as observed in our first batch of HPV-16 and HPV-58 probes) and mimic cross-hybridization.
Specific MFI hybridization signals depend on many variables, including the amount, size, sequence, and secondary structure of the PCR product used for hybridization, as well as size and sequence of the probe. As shown in this study, MPG is compatible with GP5+/6+-PCR products that have a size of approximately 150 nucleotides. It remains to be established whether longer PCR products, such as those derived from the PGMY09/11-PCR (450 bp), may impair hybridization due to sterical hindrance on the bead surface (Luminex Corp. website) or whether smaller PCR products improve the hybridization signal. One may speculate, however, that longer PCR products allow the variation of probe sequences in order to increase specificity and/or signal-to-noise ratios, which is hardly possible with, e.g., the short SPF10 PCR products (65 nucleotides) that exhibit little freedom for probe variation within the 22-nucleotide-long interprimer region. Therefore, GP5+/6+-PCR products may represent a good compromise for both high hybridization signals and specificity.
Recently, xMAP technology has been employed for HPV genotyping of HPV types (“BARCODE HPV assay”) using PCR products generated with PGMY09/11-PCR (22). Although this PCR system allows amplification of 45 different HPV types (10), specificity of the BARCODE HPV assay was evaluated with PCR products of only six HPV types (6, 11, 16, 18, 45, and 51), and the analytic sensitivity of type-specific probes was not determined. The assay was evaluated by comparing the PCR-based method with a non-PCR method, the hybrid capture 2 (HC2) assay. The HC2 assay lacks the ability of type-specific genotyping and only defines samples as positive for high-risk or low-risk HPV types. The comparison proved a reduced analytic sensitivity of BARCODE HPV, since HC2 detected a higher rate of high-risk HPV types in cervical lesions (81.8% versus 60.9%).
In this study, we successfully developed MPG for 22 out of 22 attempted genital HPV types and validated it for each type using PCR products derived from defined genomic plasmids. Meanwhile, after plasmids for validation were available, this array was extended to also include the initially missing putative high-risk HPV types 26 and 53. Given the design of the Luminex array comprising 100 sets of beads, further types can be easily integrated into the assay. As demonstrated here, the design of novel type-specific but variant-independent probes, such as the HPV-59 probe, allows genotyping of an even broader spectrum of HPV isolates, provided they are amplified by the PCR primer employed. On the other hand, HPV subtype-specific probes may also recognize those variants that do not show sufficient homology in the inner primer region.
MPG facilitates simultaneous and accurate detection of at least six different HPV types in the same sample without skewing the results (data not shown). Analytic sensitivity reached the level of 100 pg of PCR product and thus is in agreement with conventional membrane-based HPV genotyping assays (20). Investigation of the reproducibility revealed a high degree of robustness even for weak signals. We designed and included a universal probe in the assay that recognized all 22 HPV types analyzed. Clinical samples positive with the universal probe but negative with the type-specific probes could indicate the presence of (i) a known low-risk HPV type for which no type-specific probe has been included, (ii) an as-yet unknown new type, or (iii) a variant missed by the type-specific probe. Alignment analyses revealed that at least 18 additional mucosotropic HPV types (7, 13, 26, 30, 32, 34, 40, 53, 54, 55, 64, 67, 69, 71, 74, 84, 86cand and 87cand) might also be recognized by the universal probe, since they exhibit a maximum of three mismatches. However, other genital HPV types (61, 62, 72, 81, 83, and 89cand) with at least four mismatches may be missed by this probe. Preliminary data suggest that use of a second universal probe may allow the detection of those HPV types poorly detected by the first.
The quantitative data output of multiplex HPV genotyping permits researchers to assess the amount of PCR product amplified. The 96-well format allows fast, simple, and highly reproducible analyses of up to 96 PCR products from clinical specimens in less than 2.5 h, hence at least 500 samples per day can be examined by one person. Large epidemiological and vaccination studies with thousands of samples can be analyzed in a small amount of time, probably also decreasing intrastudy variation. Another benefit of the technology is that it can be automated from preparation of DNA samples to PCR, hybridization, and final detection. Apart from the start-up costs for the analyzer, costs for consumables amount to $0.10 to $0.15 per sample and HPV type. In conclusion, multiplex HPV genotyping appears to be highly suitable for the conduction of large-scale epidemiological and vaccination studies as well as diagnostic screening.
Acknowledgments
M.S. is grateful to the “Studienstiftung des Deutschen Volkes” for a travel grant.
REFERENCES
- 1.Armstrong, B., M. Stewart, and A. Mazumder. 2000. Suspension arrays for high throughput, multiplexed single nucleotide polymorphism genotyping. Cytometry 40:102-108. [PubMed] [Google Scholar]
- 2.Bosch, F. X., and S. de Sanjose. 2003. Chapter 1: human papillomavirus and cervical cancer-burden and assessment of causality. J. Natl. Cancer Inst. Monogr. 31:3-13. [DOI] [PubMed] [Google Scholar]
- 3.Bosch, F. X., A. Lorincz, N. Munoz, C. J. Meijer, and K. V. Shah. 2002. The causal relation between human papillomavirus and cervical cancer. J. Clin. Pathol. 55:244-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bulkmans, N. W., M. C. Bleeker, J. Berkhof, F. J. Voorhorst, P. J. Snijders, and C. J. Meijer. 2005. Prevalence of types 16 and 33 is increased in high-risk human papillomavirus positive women with cervical intraepithelial neoplasia grade 2 or worse. Int. J. Cancer 117:177-181. [DOI] [PubMed] [Google Scholar]
- 5.Bulkmans, N. W., L. Rozendaal, P. J. Snijders, F. J. Voorhorst, A. J. Boeke, G. R. Zandwijken, F. J. van Kemenade, R. H. Verheijen, K. v Groningen, M. E. Boon, H. J. Keuning, M. van Ballegooijen, A. J. van den Brule, and C. J. Meijer. 2004. POBASCAM, a population-based randomized controlled trial for implementation of high-risk HPV testing in cervical screening: design, methods and baseline data of 44,102 women. Int. J. Cancer 110:94-101. [DOI] [PubMed] [Google Scholar]
- 6.Dannecker, C., U. Siebert, C. J. Thaler, D. Kiermeir, H. Hepp, and P. Hillemanns. 2004. Primary cervical cancer screening by self-sampling of human papillomavirus DNA in internal medicine outpatient clinics. Ann. Oncol. 15:863-869. [DOI] [PubMed] [Google Scholar]
- 7.de Roda Husman, A. M., J. M. Walboomers, A. J. van den Brule, C. J. Meijer, and P. J. Snijders. 1995. The use of general primers GP5 and GP6 elongated at their 3′ ends with adjacent highly conserved sequences improves human papillomavirus detection by PCR. J. Gen. Virol. 76(Part 4):1057-1062. [DOI] [PubMed] [Google Scholar]
- 8.Dunbar, S., R. Godbout, H. Newkirk, and J. Hetzel. 2003. Microsphere suspension array technology for SNP detection in cattle. IEEE Eng. Med. Biol. Mag. 22:158-162. [DOI] [PubMed] [Google Scholar]
- 9.Dunbar, S. A., C. A. Vander Zee, K. G. Oliver, K. L. Karem, and J. W. Jacobson. 2003. Quantitative, multiplexed detection of bacterial pathogens: DNA and protein applications of the Luminex LabMAP system. J. Microbiol. Methods 53:245-252. [DOI] [PubMed] [Google Scholar]
- 10.Gravitt, P. E., C. L. Peyton, T. Q. Alessi, C. M. Wheeler, F. Coutlee, A. Hildesheim, M. H. Schiffman, D. R. Scott, and R. J. Apple. 2000. Improved amplification of genital human papillomaviruses. J. Clin. Microbiol. 38:357-361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gravitt, P. E., C. L. Peyton, R. J. Apple, and C. M. Wheeler. 1998. Genotyping of 27 human papillomavirus types by using L1 consensus PCR products by a single-hybridization, reverse line blot detection method. J. Clin. Microbiol. 36:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobs, M. V., P. J. Snijders, A. J. van den Brule, T. J. Helmerhorst, C. J. Meijer, and J. M. Walboomers. 1997. A general primer GP5+/GP6(+)-mediated PCR-enzyme immunoassay method for rapid detection of 14 high-risk and 6 low-risk human papillomavirus genotypes in cervical scrapings. J. Clin. Microbiol. 35:791-795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin, X. W., K. Zanotti, and B. Yen-Lieberman. 2005. New cervical cancer screening strategy: combined Pap and HPV testing. Cleveland Clin. J. Med. 72:141-148. [DOI] [PubMed] [Google Scholar]
- 14.Kleter, B., L. J. van Doorn, L. Schrauwen, A. Molijn, S. Sastrowijoto, J. ter Schegget, J. Lindeman, B. ter Harmsel, M. Burger, and W. Quint. 1999. Development and clinical evaluation of a highly sensitive PCR-reverse hybridization line probe assay for detection and identification of anogenital human papillomavirus. J. Clin. Microbiol. 37:2508-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Munoz, N., F. X. Bosch, S. de Sanjose, R. Herrero, X. Castellsague, K. V. Shah, P. J. Snijders, and C. J. Meijer. 2003. Epidemiologic classification of human papillomavirus types associated with cervical cancer. N. Engl. J. Med. 348:518-527. [DOI] [PubMed] [Google Scholar]
- 16.Munoz, N., S. Franceschi, C. Bosetti, V. Moreno, R. Herrero, J. S. Smith, K. V. Shah, C. J. Meijer, and F. X. Bosch. 2002. Role of parity and human papillomavirus in cervical cancer: the IARC multicentric case-control study. Lancet 359:1093-1101. [DOI] [PubMed] [Google Scholar]
- 17.Nieminen, P., S. Vuorma, M. Viikki, M. Hakama, and A. Anttila. 2004. Comparison of HPV test versus conventional and automation-assisted Pap screening as potential screening tools for preventing cervical cancer. BJOG 111:842-848. [DOI] [PubMed] [Google Scholar]
- 18.Notredame, C., D. G. Higgins, and J. Heringa. 2000. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J. Mol. Biol. 302:205-217. [DOI] [PubMed] [Google Scholar]
- 19.Parkin, D. M., F. I. Bray, and S. S. Devesa. 2001. Cancer burden in the year 2000. The global picture. Eur. J. Cancer 37(Suppl. 8):S4-S66. [DOI] [PubMed] [Google Scholar]
- 20.van den Brule, A. J., R. Pol, N. Fransen-Daalmeijer, L. M. Schouls, C. J. Meijer, and P. J. Snijders. 2002. GP5+/6+ PCR followed by reverse line blot analysis enables rapid and high-throughput identification of human papillomavirus genotypes. J. Clin. Microbiol. 40:779-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Walboomers, J. M., M. V. Jacobs, M. M. Manos, F. X. Bosch, J. A. Kummer, K. V. Shah, P. J. Snijders, J. Peto, C. J. Meijer, and N. Munoz. 1999. Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J. Pathol. 189:12-19. [DOI] [PubMed] [Google Scholar]
- 22.Wallace, J., B. A. Woda, and G. Pihan. 2005. Facile, comprehensive, high-throughput genotyping of human genital papillomaviruses using spectrally addressable liquid bead microarrays. J. Mol. Diagn. 7:72-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waterboer, T., P. Sehr, K. M. Michael, S. Franceschi, J. D. Nieland, T. O. Joos, M. F. Templin, and M. Pawlita. 2005. Multiplex human papillomavirus serology based on in situ-purified glutathione S-transferase fusion proteins. Clin. Chem. 51:1845-1853. [DOI] [PubMed] [Google Scholar]
- 24.Ye, F., M. S. Li, J. D. Taylor, Q. Nguyen, H. M. Colton, W. M. Casey, M. Wagner, M. P. Weiner, and J. Chen. 2001. Fluorescent microsphere-based readout technology for multiplexed human single nucleotide polymorphism analysis and bacterial identification. Hum. Mutat. 17:305-316. [DOI] [PubMed] [Google Scholar]
- 25.zur Hausen, H. 2002. Papillomaviruses and cancer: from basic studies to clinical application. Nat. Rev. Cancer 2:342-350. [DOI] [PubMed] [Google Scholar]