Abstract
At least 13 characterized Leishmania species are known to infect humans in South America. Five of these parasites are transmitted in the sylvatic ecotopes of the whole French Guianan territory and responsible for cutaneous leishmaniasis. For the diagnosis of cutaneous leishmaniasis, restriction fragment length polymorphism (RFLP) analyses have shown promising results. Thus, the end of the small subunit and internal transcribed spacer 1 of the rRNA genes were sequenced and targeted by PCR-RFLP analysis in the 10 main New World (NW) Leishmania species from the two subgenera. Then, the procedure was tested on 40 samples from patients with cutaneous leishmaniasis, and its results were compared with those of conventional methods. (i) The results of this simple genus-specific method were in agreement with those of previous isoenzyme analyses. (ii) This method distinguished the most medically relevant Leishmania species with only one enzyme (RsaI). (iii) This method could be performed directly on human biopsy specimens (sensitivity of 85.7%). Performing NW Leishmania species typing rapidly and easily in the field constitutes a very valuable improvement for detection of Leishmania spp. Revealing great diversity with several enzymes, this method could also be useful for taxonomic, ecological, and epidemiological studies in space and time.
Among the 13 characterized Leishmania species known to infect humans in South America (22), at least 7 Leishmania parasites coexist in the Guiana Shield ecotopes (41). Cutaneous leishmaniases (CL) are endemic in French Guiana, and the annual incidence was estimated at 2 cases per 1,000 inhabitants between 1979 and 2000 (6, 14). At least five Leishmania species are known to be sympatrically transmitted in the sylvatic ecotopes of the whole French Guianan territory (41): Leishmania (Viannia) guyanensis (in more than 90% of the cases), Leishmania (Leishmania) amazonensis, L. (V.) braziliensis, L. (V.) naiffi, and L. (V.) lainsoni (2, 14). They are mainly responsible for localized cutaneous forms, but diffuse and mucocutaneous presentations have been reported (41).
Characterization of Leishmania species in clinical infections in geographic areas where different species are sympatrically transmitted is important for both clinical and epidemiologic reasons as follows. (i) Symptoms may be caused by other pathogens (streptococcal pyoderma, Buruli ulcer, etc.). (ii) Different Leishmania species can cause the same clinical presentation. (iii) Different species may require different treatments [e.g., Glucantime for L. (V.) braziliensis infections and pentamidine in CL cases due to L. (V.) guyanensis (reviewed in reference 9)]. (iv) These treatments are expensive and significantly toxic. (v) Therapeutic failures have to be monitored to discriminate between relapse of a latent infection and reinfection by another species. (vi) Increased human mobility and long-distance travel due to socioeconomic factors may contribute to the spread of leishmaniasis to areas where leishmaniasis had previously not been endemic.
Since neither microscopic examination nor cultivation permits species identification and to bypass the laborious and time-consuming isoenzyme analysis (multilocus enzyme electrophoresis [MLEE]), many methods have been developed to detect Leishmania species and to study the molecular diversity and relationships within Leishmania species. To detect Leishmania species, these methods often have to discriminate among closely related species. However, it is often difficult for a single method to cope with the problems of sensitivity and discrimination. Swiftness, cost-effectiveness, and feasibility in the field also govern the routine use of these methods. Many different PCR-based methods targeting microsatellites, kinetoplastic DNA, telomeric sequences, or gp63, hsp70, miniexon, β-tubulin, or rRNA genes have been proposed (reviewed in references 46, 48, and 51). Among these methods, restriction fragment length polymorphism (RFLP) analyses of PCR-amplified sequences of multicopy genes have shown promising results (1, 12, 23, 28, 30, 33, 52). Since the work of Cupolillo etal. (12), PCR-RFLP methods on the internal transcribed spacers (ITS) of the ribosomal genes have been performed on New World (NW) Leishmania species (4, 10, 19, 39), but these methods have been conducted mostly on Old World Leishmania species (7, 17, 35, 42-45). However, up to now, no study has integrated complete analyses of specificity, sensitivity, taxonomic reliability, intraspecies polymorphism, and trial on clinical samples for diagnosis of leishmaniasis in the field for the two subgenera of Leishmania.
A simple PCR-RFLP analysis targeting the end of the ribosomal small subunit (SSU) and the whole ITS1 (PRSI) is here proposed and compared to other results (12, 44) for the specific diagnosis of NW leishmaniases. The taxonomic robustness of PRSI for NW Leishmania identification was first investigated. The 10 main Leishmania species isolated from humans were studied. We especially focused our search on the most common Amazonian taxa. Then, the fingerprinting properties of PRSI were applied to the diagnosis of leishmaniasis on clinical samples from 40 patients with CL.
MATERIALS AND METHODS
Parasite strains.
Thirty-three protozoan strains of 13 different species representative of the main parasites responsible for human diseases in the NW were selected (Table 1). All the strains studied were supplied by the French National Reference Centre for Leishmania (Montpellier, France). Thirteen enzymatic complexes of the strains were first analyzed by MLEE as described elsewhere for controls (38). Lutzomyia umbratilis females were supplied by the Pasteur Institute of French Guiana (Cayenne, French Guiana).
TABLE 1.
Strains examined in this study
| Genus | Subgenus | Species | CNRLNa | International code | CNRLZb | GenBank ANc |
|---|---|---|---|---|---|---|
| Leishmania | Leishmania | infantum | 75 | MHOM/FR/78/LEM75 | MON-1 | DQ182535 |
| infantum | 307 | MHOM/ES/81/BCN1 | MON-29 | |||
| infantum | 417 | MHOM/DZ/82/LIPA59 | MON-24 | |||
| infantum | 1208 | MHOM/EG/87/RTC2 | MON-98 | |||
| infantum | 2205 | MHOM/ES/90/BCN61 | MON-28 | |||
| mexicana | 695 | MHOM/BZ/82/BEL21 | MON-156 | |||
| mexicana | 279 | MHOM/VE/57/LL1 | MON-40 | |||
| aristidesi | 693 | MORY/PA/69/GML3 | MON-133 | |||
| amazonensis | 395 | IFLA/BR/67/PH8 | MON-41 | |||
| amazonensis | 690 | MHOM/BR/73/M2269 | MON-132 | DQ182536 | ||
| amazonensis | 2236 | MHOM/PA/87/GML417 | MON-41 | |||
| amazonensis | 2238 | MPRO/BR/72/M1845 | MON-41 | |||
| amazonensis | 2246 | IFLA/TT/71/71-110 | MON-157 | |||
| hertigi | 694 | MCOE/PA/65/C8 | MON-135 | |||
| deanei | 718 | MCOE/BR/75/M2808 | MON-134 | |||
| Viannia | braziliensis | 2252 | MHOM/BR/84/LTB300 | MON-166 | DQ182537 | |
| braziliensis | 396 | MHOM/BR/75/M2903 | MON-43 | |||
| braziliensis | 469 | MHOM/CO/83/LEM469 | MON-44 | |||
| braziliensis | 2222 | MCAN/PE/91/LEM2222 | MON-141 | |||
| braziliensis | 2242 | MHOM/BZ/81/BEL13 | MON-176 | |||
| peruviana | 1534 | MHOM/PE/87/LC106 | MON-128 | |||
| guyanensis | 4745 | MHOM/GF/2004/LBC41 | MON-131 | DQ182538 | ||
| guyanensis | 4519 | MHOM/GF/2003/LBC40 | MON-131 | DQ182539 | ||
| guyanensis | 4769 | MHOM/GF/2004/LBC43 | MON-131 | DQ182540 | ||
| guyanensis | 4773 | MHOM/GF/2004/LBC45 | MON-45 | DQ182541 | ||
| naiffi | 2204 | MDAS/BR/79/M5533 | MON-148 | DQ182542 | ||
| naiffi | 2807 | MHOM/00/94/CRE54 | MON-193 | |||
| naiffi | 3426 | MHOM/GF/97/CRE88 | MON-254 | |||
| lainsoni | 2208 | MHOM/BR/81/M6426 | MON-149 | DQ182543 | ||
| lainsoni | 2227 | IUBI/BR/00/M12025 | MON-150 | |||
| lainsoni | 2229 | MCUN/BR/85/M9342 | MON-151 | |||
| Crithidia | fasciculata | 2744 | IOOO/00/00/LV116 | DQ182544 | ||
| Trypanosoma | cruzi | 578 | MHOM/AR/00/000 | MON-91 |
Centre National de Référence des Leishmania (National Reference Center for Leishmania [previously known as Medical Ecology Laboratory]) strain numbers.
Centre National de Référence des Leishmania (National Reference Center for Leishmania) zymodemes obtained from 13 isoenzymes.
GenBank accession numbers for ribosomal internal transcribed spacer sequences.
Patients.
A total of 40 consecutive patients with confirmed cases of CL who had consulted the Department of Dermatology of Cayenne General Hospital (Cayenne, French Guiana) and three health centers in the southern part of French Guiana (Camopi, Maripasoula, and Saul) during 1 month in mid-dry season when CL cases are the most abundant (15 April to 15 May 2004) were included in this study. The inclusion of a confirmed case of CL was based upon (i) the presence of typical lesions (ulcerative, nodulous, or papulous cutaneous lesions) of 2 or more weeks of evolution and a compatible epidemiological history, (ii) the absence of specific treatment before consultation, and (iii) two positive results by at least one of the five methods used in this study: microscopic examination of dermal smears; isolation by in vitro cultivation; and PCRs on fresh biopsy specimens, dermal smears, and cultured biopsy medium. All individuals enrolled in this study provided informed consent.
Clinical samples and standard diagnostic procedure.
Tissue scrapings for microscopic smear examination were collected by scraping the internal border of skin lesions with a surgical blade; the tissue scraping was then smeared onto a glass slide, fixed with methanol, and stained with Giemsa. For each patient, two slides were read by two different trained parasitologists. Punch skin biopsy specimens of 4 mm were taken from the internal border of lesions under sterile conditions and local anesthesia (Xylocaine). The cutaneous biopsy specimens were divided into two samples. One was processed by PRSI and the second by in vitro cultivation. In vitro cultures were performed by inoculation of the sterilely crushed biopsy specimen in 3 ml of RPMI 1640 (Sigma), 20% fetal calf serum, 1% nonessential amino acids, and 50 IU/ml penicillin. Culture flasks (25-cm2 flasks) were incubated at 24°C and microscopically observed every 3 days for 6 weeks before they were considered negative.
DNA extractions.
For each patient, DNA was extracted from fresh biopsy specimens, cultured biopsy media, and dermal smear materials. All DNA extractions were conducted using the DNeasy tissue kit (QIAGEN) as recommended by the manufacturer. DNA from Mycobacterium ulcerans was supplied by the Pasteur Institute of French Guiana (Cayenne, French Guiana).
Sequencing.
Sequencing of the end of the ribosomal SSU and the whole ITS region from 10 strains listed in Table 1 was performed without cloning to find conserved regions for primer hybridization and to improve the restriction enzyme choice. Two primer pairs were designed complementary to the consensus sequences from the 3′ end of the SSU and the 5′ end of the ribosomal large subunit unit (LSU) of nine Leishmania species, L. major Friedlin (29), L. mexicana, L. amazonensis, L. venezuelensis, L. gerbilli, L. tropica, L. braziliensis, L. adleri, and L. turanica (GenBank accession numbers AC005806, AF466383, AF466382, AF466381, AF466380, AF339753, AF339752, AJ300486, AJ300485, AJ300484, AJ300483, AJ300482, AJ300481, AJ300480, J272383, AJ272382, AJ272381, AJ272380, AJ272379, and AJ272378). The two primer pairs used wereprimers SSU-12733-D (5′-TACGTCCCTGCCATTTGTAC-3′) and 5.8S-13333-R (5′-CGACACTGAGAATATGGCATG-3′) for the SSU-ITS1 region and primers 5.8S-13277-D (5′-GCATGGGAGAAGCTCTATTG-3′) and LSU-14237-R1 (5′-TAACACCTTCTTTGGAATAC-3′) for the ITS2 sequence. Part of the DNAs was sequenced in an automatic ABI-Prism 310 sequencer (Applied Biosystems) at the CIRAD campus (Kourou, French Guiana) following the BigDye Terminator procedure of the manufacturer. The other DNAs were sequenced by the MWG service team (New York, N.Y.) by a Comfort Read procedure. The SSU-ITS sequences of the 10 strains were determined (Table 1). For the trial on clinical samples, sequencing of the RNA polymerase II gene (8) was also performed to confirm the PRSI identification of the Leishmania (Viannia) guyanensis samples characteristic of each genotype.
PCR amplification.
Primers were designed to amplify a 1.2-kb region including ITS1 from the 5′ end of the SSU to the 5′ end of the 5.8S subunit. The primers SSU-12103-D (5′-GGGAATATCCTCAGCACGT-3′) and 5.8S-13333-R (5′-CGACACTGAGAATATGGCATG-3′) were used. For each sample, PRSI with each enzyme was performed two or three times in order to guarantee the procedure reproducibility and to confirm the total digestion. Amplification reactions were performed in 50 μl containing the following: 100 mM Tris-HCl, pH 8.3; 500 mM KCl; 1.5 mM MgCl2; 150 μM each of dATP, dCTP, dTTP, and dGTP (Sigma); 0.1 μM each primer (MWG); 10 ng of genomic DNA (approximately 2.5 × 106 cells); and 2.5 units REDTaq DNA polymerase (Sigma). Amplification was performed by a GenAmp PCR System 9700 (Applied Biosystems) with an initial step of denaturation of 3 min at 96°C; followed by 40 cycles, with each cycle consisting of 30 s at 94°C, 1 min at 51°C, and 2 min at 72°C; and finally, a polymerization step of 5 min at 72°C. PCR efficiency was estimated with the SmartLadder (EUROGENTEC) molecular marker for weight and quantity by electrophoresis in 1.5% agarose gel stained with ethidium bromide.
The PCR protocol was improved on DNA extracted from L. (V.) guyanensis MHOM/GF/2003/LBC40. A good sensitivity and time yield issued from a 15 thermocycler program test set. The optimal result was achieved by beginning the PCR with a hot start to ensure complete denaturation prior to the first prolonged annealing cycle, increasing the number of cycles from 35 to 40, and prolonging the last cycle with a 5-min extension step. These modified PCR conditions were employed in subsequent routine amplifications.
Restriction analysis.
Enzymes were selected by in silico restriction of the alignment of the 10 new sequences. Constant amounts of DNA for each sample were loaded (approximately 100 ng DNA) and digested for 2 h with restriction enzymes according to the manufacturer's recommendations. Eleven restriction enzymes were tested: AluI, TaqI, HaeIII, and HpaII (Sigma), RsaI, BfaI, ApoI, and AflIII (New England BioLabs), SphI and PstI (GibcoBRL), and DdeI (Promega). Digestion products were separated for 2 h by simple electrophoresis at 130 V in 2% agarose gel stained with ethidium bromide.
Numerical analysis.
RFLP gel scans were analyzed with the LabImage 2.7.1 software (Kapelan GmbH). Character matrices were then created manually by reporting all possible PCR-RFLP fragments in the sample studied. Then, for each sample, the presence or absence of bands was given a score of 1 or 0, respectively. In a given RFLP profile, differences of intensity among fragments were not taken into account. These differences are common in tandemly repeated genes consisting of sequence variants present in different copy numbers (24, 52). The binary PRSI matrices were then processed for phenetic analyses with the following programs of PHYLIP version 3.63 package (Joseph Felsenstein, University of Washington, Seattle, Wash.) (July 2004 version): (i) SEQBOOT (bootstrap analysis); (ii) RESTDIST (restriction fragment distance method modified by the Nei and Li distance method [34]), followed by NEIGHBOR (neighbor-joining method); (iii) CONSENSE (majority rule consensus); and finally (iv) DRAWGRAM (dendrogram tree drawing). The bootstrap analyses were performed for 1,000 replications to estimate the robustness of the nodes. The tree built up with the reference strain data was rooted with Crithidia fasciculata (Cf2744). The percentage of genetic polymorphism within each species (n > 2) was calculated by dividing the number of different genotypes by the number of strains studied. Statistical analyses were performed by the Intercooled Stata 8.2 version software (StataCorp LP).
Nucleotide sequence accession numbers.
The SSU-ITS sequences of 10 Leishmania strains were deposited in GenBank and given accession numbers DQ182535 to DQ182544 (Table 1).
RESULTS
Reference strains.
As expected from Leishmania (Leishmania) major (1.2 kb) (29), amplicons ranged from 1.1 kb to 1.3 kb with no correlation between subgenera (Table 2). Little or no variability was observed within isolates of the same species. C. fasciculata (Cf2744) presented a smaller amplicon of 1.1 kb. DNA from Trypanosoma cruzi (Tc578) was not amplified despite a positive result in a genus-specific PCR control assay (5). Control DNA samples from humans, Lutzomyia umbratilis, and Mycobacterium ulcerans were also found negative.
TABLE 2.
Fragment diversity and polymorphism in the SSU-ITS1 region
| Species | Unit size | AluI
|
ApoI
|
BfaI
|
DdeI
|
HaeIII
|
HpaII
|
PstI
|
RsaI
|
TaqI
|
Polymorphismc | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| NFa | SFb | NF | SF | NF | SF | NF | SF | NF | SF | NF | SF | NF | SF | NF | SF | NF | SF | |||
| Leishmania (Leishmania) species | ||||||||||||||||||||
| L. (L.) infantum | ||||||||||||||||||||
| Genotype group I (Li75, Li307) | 1.3 | 8 | 3.1 (0.6) | 6 | 2.7 (1.2) | 0 | 0.0 | 0 | 0.0 | 7 | 2.7 | 4 | 1.9 (0.2) | 3 | 1.4 (0.4) | 7 | 3.2 (1.8) | 6 | 1.5 (0.5) | 40 |
| Genotype group II (Li417, Li1208, Li2205) | 1.3 | 7 | 2.7 (0.2) | 10 | 4.5 (0.7) | 0 | 0.0 | 0 | 0.0 | 7 | 2.7 | 4 | 1.8 | 2 | 1.5 (0.5) | 5 | 2.0 | 7 | 2.4 | |
| L. (L.) mexicana | ||||||||||||||||||||
| Lm695 | 1.3 | 7 | 2.9 | 5 | 2.4 | 0 | 0.0 | 0 | 0.0 | 7 | 3.2 | 3 | 1.3 | 3 | 2.1 | 6 | 2.6 | 4 | 0.9 | |
| Lm279 | 1.3 | 10 | 4.4 | 4 | 1.6 | 0 | 0.0 | 0 | 0.0 | 5 | 1.2 | 3 | 1.2 | 4 | 1.5 | 5 | 2.0 | 5 | 1.4 | |
| L. (L.) amazonensis | 1.3 | 9 | 4.2 (0.1) | 4 | 2.5 (0.4) | 0 | 0.0 | 0 | 0.0 | 7 | 3.0 (0.1) | 5 | 2.5 (0.4) | 3 | 1.9 (0.4) | 5 | 2.2 (0.4) | 7 | 2.2 (0.7) | 32 |
| L. (L.) aristidesi | 1.3 | 10 | 4.4 | 4 | 2.0 | 0 | 0.0 | 0 | 0.0 | 6 | 2.9 | 3 | 1.4 | 3 | 1.7 | 6 | 2.8 | 8 | 2.2 | |
| L. (L.) hertigi | 1.3 | 7 | 3.0 | 4 | 2.6 | 0 | 0.0 | 3 | 1.5 | 2 | 1.2 | 4 | 2.1 | 3 | 2.2 | 6 | 2.0 | 3 | 0.8 | |
| L. (L.) deanei | 1.3 | 6 | 2.0 | 4 | 1.7 | 0 | 0.0 | 2 | 1.3 | 2 | 1.4 | 4 | 1.9 | 2 | 1.3 | 7 | 3.4 | 8 | 2.2 | |
| Leishmania (Viannia) species | ||||||||||||||||||||
| L. (V.) braziliensis | ||||||||||||||||||||
| Genotype group I (Lb396, Lb469, Lb2222, Lb2242) | 1.2 | 11 | 4.3 (0.5) | 7 | 3.5 (0.9) | 2 | 1.3 | 3 | 1.5 (0.4) | 8 | 4.2 (0.6) | 5 | 2.3 (0.4) | 4 | 2.3 (0.2) | 6 | 3.3 (0.9) | 6 | 2.0 (0.8) | 52 |
| Lb2252 | 1.2 | 9 | 3.8 | 5 | 2.5 | 2 | 1.3 | 4 | 2.0 | 9 | 4.8 | 6 | 2.9 | 4 | 1.9 | 5 | 2.4 | 9 | 3.1 | |
| L. (V.) peruviana | 1.1 | 8 | 3.3 | 6 | 3.4 | 4 | 2.8 | 4 | 1.4 | 3 | 1.3 | 5 | 2.3 | 4 | 2.4 | 5 | 3.5 | 4 | 0.8 | |
| L. (V.) guyanensis | 1.2 | 9 | 3.3 (0.3) | 2 | 1.1 | 0 | 0.0 | 0 | 0.0 | 4 | 1.5 (0.7) | 6 | 2.4 (0.5) | 3 | 1.2 (0.1) | 2 | 0.8 (0.3) | 5 | 1.1 | 40 |
| L. (V.) naiffi | ||||||||||||||||||||
| Genotype group I (Ln2807, Ln3428) | 1.3 | 9 | 3.4 (0.6) | 7 | 3.6 (0.1) | 2 | 1.2 | 2 | 1.2 | 7 | 3.2 (1.6) | 6 | 2.7 (0.9) | 3 | 1.7 (0.8) | 7 | 3.5 (0.8) | 6 | 1.5 (0.3) | 67 |
| Ln2204 | 1.3 | 6 | 2.0 | 3 | 1.3 | 2 | 1.2 | 2 | 1.2 | 5 | 2.7 | 5 | 2.0 | 2 | 1.3 | 4 | 1.2 | 8 | 2.3 | |
| L. (V.) lainsoni | ||||||||||||||||||||
| Genotype group I (Ll2227, Ll2229) | 1.2 | 12 | 5.3 (0.1) | 2 | 1.1 | 0 | 0.0 | 0 | 0.0 | 7 | 2.4 | 5 | 2.0 (0.1) | 2 | 1.1 | 4 | 1.4 (0.3) | 6 | 1.6 (0.9) | 53 |
| Ll2208 | 1.2 | 11 | 4.5 | 3 | 1.3 | 0 | 0.0 | 0 | 0.0 | 5 | 1.7 | 3 | 1.2 | 2 | 1.3 | 3 | 1.3 | 7 | 2.2 | |
| Crithidia fasciculata | 1.0 | 8 | 3.1 | 2 | 1.2 | 0 | 0.0 | 5 | 1.7 | 6 | 3.2 | 3 | 1.2 | 5 | 2.5 | 5 | 2.8 | 3 | 0.6 | |
NF, average number of fragments differing from the unit size.
SF, sum of fragment sizes. The average and standard deviation (in parentheses) are shown for some values.
For each species (n > 2), polymorphism is calculated by dividing the sum of the different genotypes encountered for each enzyme by the number of strains analyzed and given as a percentage.
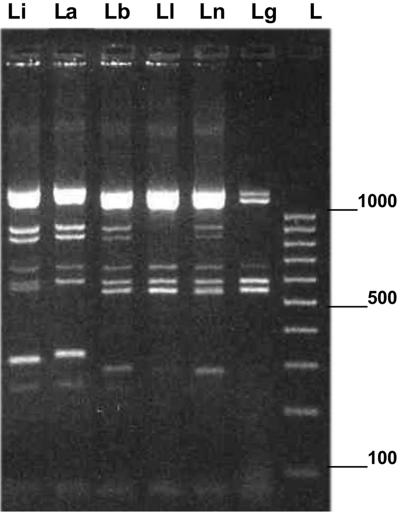
The PRSI profiles of the 33 strains digested with nine enzymes are shown in Table 2. Relatively complex and polymorphic patterns were obtained, a feature typical for a repetitive sequence. Most of the patterns evidenced specific bands that corresponded to particular subgenera and species. All amplification patterns were reproducible. Restriction patterns for species in the Viannia subgenus were relatively more complex than those in the Leishmania subgenus. At the intraspecific level, polymorphism was also observed in all taxa (more than two strains studied) up to 67% within Leishmania (Viannia) naiffi. Each of the nine enzymes tested showed different levels of variability depending on the taxon set. For a given enzyme, the sum of restriction fragment sizes often exceeded the amplicon unit size as it has been observed in previous studies (12, 23). RsaI produced different subgenus-specific and species-specific fragments and was the most powerful enzyme to distinguish each species (Fig. 1). However, Viannia parasites showed very closely related patterns, and the L. (V.) guyanensis and L. (V.) lainsoni strains tested were not distinguishable by this restriction enzyme alone.
FIG. 1.
PRSI profiles of six Leishmania strains digested with RsaI. PRSI restriction patterns of six strains were obtained after 2 h of digestion with RsaI. From left to right, the strains studied were Leishmania (Leishmania) infantum Li75, L. (L.) amazonensis La395, Leishmania (Viannia) braziliensis Lb2252, L. (V.) lainsoni Ll2208, L. (V.) naiffi Ln2204, and L. (V.) guyanensis Lg4745. A 100-bp low-molecular-size ladder (L) was used, and electrophoresis was performed for 4 h. The positions of molecular size markers (in base pairs) are shown to the right of the gel.
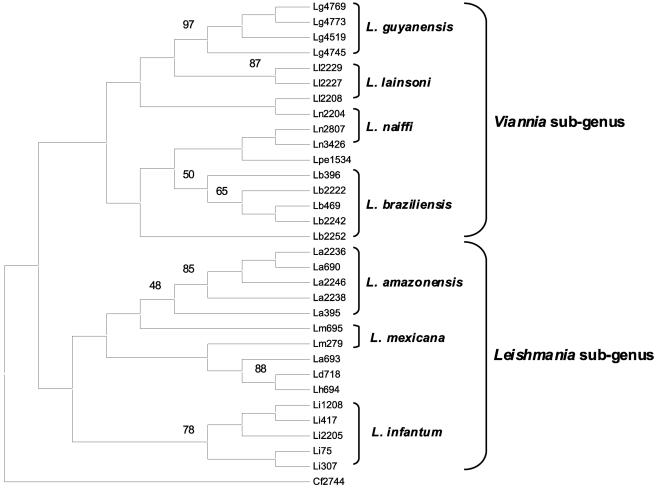
In order to obtain a comprehensive view of the genetic polymorphism within the samples studied, all the PRSI data were processed by phenetic analyses. The neighbor-joining molecular tree presented in Fig. 2 depicts relationships between strains. A similar topology with minor differences was obtained using the unweighted-pair group method using average linkages (tree not shown). PRSI evidenced an extensive polymorphism and revealed specific markers for subgenus and species in congruence with previous MLEE reports (36, 38, 47, 49, 50). This dendrogram distinguished the two subgenera of Leishmania, Leishmania and Viannia (26). L. (L.) amazonensis, L. (L.) infantum, and L. (V.) guyanensis formed independent clusters, whereas L. (L.) mexicana, L. (V.) braziliensis, L. (V.) lainsoni, and L. (V.) naiffi could be clustered into subgroups. L. (V.) guyanensis, L. (L.) infantum, L. (L.) amazonensis, L. (L.) deanei, and L. (L.) hertigi showed higher bootstrap values (>75%). However, L. (V.) braziliensis, L. (V.) lainsoni, L. (V.) naiffi, and L. (L.) mexicana branching patterns were not as strong. Strains Ln2204, Ll2208, Lb2252, and Lm695 were not found in the same cluster as the other strains of the same species.
FIG. 2.
PRSI phenetic tree. This bootstrapped neighbor-joining tree was built up with the PRSI data given in Table 2 (nine restriction enzymes tested) by the Nei and Li restriction distance-modified method in PHYLIP RESTDIST and NEIGHBOR programs. One thousand bootstrap replicates were applied, and values were given for each informative node clustering at the species level. All the strains studied are given in Table 1. This tree was rooted on Crithidia fasciculata (Cf2744). The Leishmania species and subgenera are indicated to the right of the tree.
Clinical samples.
Results of the trial of different methods on clinical samples were summarized in Table 3. PCR on dermal smear materials and cultured biopsy media appeared to have lower sensitivity, whereas PCR on fresh biopsy specimens had a higher sensitivity for detection of Leishmania parasites than each of the other diagnostic tests did. However, PCR on fresh biopsy specimens was as sensitive as the two conventional reference methods combined (87.5% of positivity).
TABLE 3.
Results of conventional methods and PCR on different sample types for the diagnosis of cutaneous leishmaniases
| Diagnostic test | Frequency of positivitya (%) | Sensitivity comparison with PCR on fresh biopsy specimens (%) |
|---|---|---|
| Conventional methods | ||
| Microscopic smear examination | 29/40 (72.5) | 89.6 |
| In vitro culture | 31/40 (77.5) | 87.1 |
| Microscopy and cultivation combined | 35/40 (87.5) | 85.7 |
| PCR on: | ||
| Material from dermal smear | 13/29 (44.8)b | 84.6 |
| Cultured biopsy medium | 20/38 (52.6)b | 90 |
| Fresh biopsy specimens | 35/40 (87.5) |
Number of positive samples/total number of samples tested among 40 patients.
These values are significantly different (P < 0.001) from the value for PCR on fresh biopsy specimens.
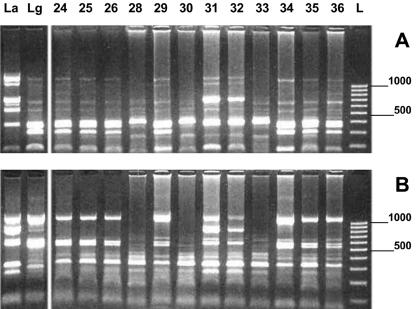
PRSI profiles obtained with seven enzymes were relatively complex and polymorphic. All amplification patterns were reproducible. All amplified DNAs were identified as L. (V.) guyanensis, except for two samples (samples 31 and 32) that were similar to the L. (L.) amazonensis control strain (Fig. 3). The Nei genetic distances between the genotyped L. (V.) guyanensis samples ranged from 0.0000 to 1.2600. Sequencing of the RNA polymerase II gene (8) was also performed and confirmed the PRSI identification.
FIG. 3.
PRSI profiles of 14 samples digested with two restriction enzymes. PRSI restriction patterns of 14 samples were obtained after 2 h of digestion by HpaII (A) and RsaI (B). L. (L.) amazonensis (La395) (La) and L. (V.) guyanensis (Lg4745) (Lg) were used as references. A 100-bp low-molecular-size ladder (L) was loaded, and electrophoresis ran for 2 h. The positions of molecular size markers (in base pairs) are shown to the right of the gel.
DISCUSSION
Taxonomic robustness of PRSI.
Good agreement was obtained with previous trees based upon MLEE data (36, 38, 47, 49, 50). Hierarchical relationships among taxa at the subgenus and species levels were found to be similar. PRSI results were congruent with the classification proposed by Rioux et al., based on an analysis of isoenzyme complexes (38, 47, 49), and validating the previous Lainson and Shaw phylogenetic analysis of phenotypic characters (26), which divides the Leishmania genus Ross 1903 into two subgenera: Leishmania (Leishmania) Saf'janova 1982 and Leishmania (Viannia) Lainson and Shaw 1987. Up to now, all the L. (L.) chagasi strains from the NW analyzed at the French National Reference Center for Leishmania were identical to Li75 and were attributed to the MON-1 zymodeme. Thus, L. (L.) chagasi and L. (L.) infantum are likely to be considered the same taxon (11, 21, 31, 32, 38). In the PRSI tree, Li75 grouped with all the other strains of the same species.
The higher intraspecies polymorphism resolution in PRSI compared with MLEE was likely to be responsible for the weaker branching patterns observed especially in the L. (V.) naiffi, L. (V.) braziliensis, L. (V.) lainsoni, and L. (L.) mexicana groups. Probably for the same reason, dendrogram built from PRSI data did not allow L. (V.) braziliensis to be distinguished from L. (V.) peruviana, unlike the dendrogram built by PCR-RFLP of the gp63 locus (52). In the PRSI tree, L. (L.) mexicana and L. (L.) amazonensis were distinguishable but closely related, similar to previous observations (12). As reported before (12, 13), L. (V.) lainsoni was shown to be distinct from the other Viannia species but closely related to L. (V.) naiffi. The two L. (L.) mexicana strains studied were found to be different (Nei distance index of 0.028564), whereas a previous study showed that ITS patterns in this species were invariant (4). However, the number of enzymes used and the size of the region studied were larger in the present study. A larger number of characters, states, and strains studied will be required to accurately resolve relationships and improve the clustering strength at supraspecies levels. Unclear points in clustering may have also been provoked by the great intraspecies polymorphism detail given by this method, which could become an advantage in ecoepidemiological studies. Nevertheless, good agreement was also obtained with species groupings in previous PCR-RFLP studies of the gp63 locus (19, 52) and ribosomal genes (10, 12, 13) and by the random-amplified polymorphic deoxyribonucleic acid technique (25). As evidenced in this study, but unlike Garcia et al. (19), Cupolillo et al. (10, 12) and Ishikawa et al. (25) showed that isolates of L. (V.) naiffi and L. (V.) braziliensis were highly polymorphic. In contrast, when studying polymorphism of clinical samples from L. (L.) infantum, very low or no intraspecific variation was observed by the Schonian (18, 43) and Cupolillo (13) teams. In the present study, L. (L.) infantum isolates showed the lowest heterogeneity rates but were not identical.
Sensitivity of PCR compared to other methods.
Detection of Leishmania parasites in a clinical sample is necessary to confirm a suspected case of leishmaniasis. Most commonly used methods for the direct detection of the parasite (microscopic examination of stained smears and in vitro cultivation) lack sensitivity because of the paucity of parasites in some specimens or are hampered by the problem of infection. As underlined by Ramirez et al. (37) and Weigle et al. (54), the lack of an ideal “gold standard” for diagnosing CL poses problems for estimating the sensitivities of PCR methods. No single test could serve as a “gold standard” against which the different assays could be evaluated. Therefore, assessment of test performance was based on the assumptions that, with sufficient coherence of clinical information, a patient was positive for CL when the infection was detected by at least one of the five diagnostic assays. One positive case in cultivation was negative by direct examination and by all the duplicate PCR assays. Two samples were positive only by duplicate PCR on biopsy specimens. They were subsequently identified as L. (V.) guyanensis and genotyped. All other cases were positive by at least two methods.
The present trial on different clinical samples from patients with CL gave several useful results for diagnosis applications. Compared with conventional reference methods for diagnosis, i.e., microscopic examination and in vitro cultures, PCR was the most sensitive. However, unlike some previous reports (3, 44), PCRs on cultured biopsy media and materials from smear slides were less sensitive. These results were probably due to (i) the presence of PCR-inhibitory compounds in the medium and in the chemicals used for slide staining, even when a commercial DNA purification kit was used, and/or more likely due to (ii) a reduced quantity of parasitic material compared with fresh punch biopsy specimens. The sensitivity of PCR on fresh cutaneous biopsy specimens was comparable to those obtained by other molecular protocols targeting the same locus (95.4% in reference 44) or different multicopy genes, such as kinetoplastic DNA (75.7% to 100% in references 3, 16, 40, and 54), miniexon sequences (89.7% in reference 27), the gp63 gene (85% in reference 53), and the hsp70 locus (100% in reference 20). However, we obtained better sensitivity results than those reported by Marfurt et al. (only 51.7%) (27) with a PCR-RFLP method targeting the ITS region (44). Thus, use of this method for the diagnosis of the NW CL seemed to be appropriate, since parasites are said to be scarce in clinical forms caused byViannia parasites, especially in mucosal and chronic cases (16,54).
PRSI for clinical diagnosis.
The identification of two L. (L.) amazonensis strains among the 40 parasites studied here was in agreement with a previous study in the same area [6.25% of L. (L.) amazonensis in reference 15]. It was confirmed that only one digestion with RsaI is required to identify parasites to the species level [except L. (V.) lainsoni and L. (V.) guyanensis]. Unlike the results of reference 44, HaeIII was not sufficient to clearly distinguish all species in the Viannia subgenus, especially L. (V.) braziliensis/L. (V.) naiffi and L. (V.) lainsoni/L. (V.) guyanensis. Control patterns from HaeIII, HpaII, and/or PstI restrictions could yet be envisaged to ensure the results at least to the subgenus level. Interestingly, clinical confusion between commonly found leishmanial cutaneous ulcers and Buruli ulcers caused by Mycobacterium ulcerans could easily be avoided. When simple commercial kits for DNA extraction were used and following our optimized thermocycler program, PRSI diagnosis was shown to be rapid: it could be performed within 12 h from fresh biopsy specimens (DNA extraction, PCR, RFLP, and analyses). PRSI was also cost-effective: depending on the number of samples simultaneously treated, its cost (about $3 to $10 per sample, according to the number of samples and including product, reagent, and working time charges in French Guiana) was comparable to that of cultivation. This method proved to be reproducible and, compared to culture/MLEE, requires only minimal laboratory equipment. Thus, PRSI seemed to fit with the routine use requirements. It is noteworthy that some other tests on different clinical samples, such as cutaneous swabs, bone marrow aspirates, and blood samples, and studying samples from other areas where leishmaniasis is endemic would be interesting to complete this study. Further development also eventually includes the use of nested primers to control negative PCR results and comparison with results obtained from other gene targets, such as gp63, hsp70, and miniexon genes.
However, because the detection sensitivities of the two combined conventional parasitological methods in this study are quite good and comparable to that in reference 40, specific guidelines for the use of PRSI for diagnosis of leishmaniasis are needed. Since more than 90% of infections are known to be caused by L. (V.) guyanensis (15), we recommend the use of this method mainly (i) for detection (PCR) in patients suspected to have leishmaniasis with negative microscopic examination and/or culture results, (ii) for identification (PRSI) of chronic and/or atypical clinical forms, (iii) for regular randomized prospective screening in epidemiological surveillance studies on a limited set of patients.
Conclusion: PRSI as a multipurpose tool.
On one hand, the possibility of performing Leishmania species typing rapidly and easily in the field constitutes a valuable improvement for diagnosis. PRSI performed with one enzyme and directly on human biopsy specimens without parasite isolation appeared to be a good detection and identification method for clinical diagnosis. Our results demonstrated the taxonomic value of PRSI as well as its clinical usefulness for the diagnosis of CL, at least in French Guiana. On the other hand, because PRSI revealed considerable molecular diversity when more endonucleases were used, it could also be useful for taxonomic, ecological, and epidemiological studies over time and in different geographic areas.
Acknowledgments
We thank the CIRAD team (Kourou, French Guiana) for their help in sequencing.
This work was supported by the University of the French West Indies and French Guiana, by the French Contrat Plan Etat-Region no. 2365, the Institut National de la Santé et de la Recherche Médicale (INSERM, Paris, France), the Centre National pour la Recherche Scientifique (CNRS, Paris, France), and the University of Montpellier 1 (Montpellier, France).
REFERENCES
- 1.Abdeen, Z. A., S. S. Sawalha, C. L. Eisenberger, H. M. Khanfar, C. L. Greenblatt, O. Yousef, L. F. Schnur, K. Azmi, A. Warburg, K. A. Bader, C. L. Jaffe, and G. Baneth. 2002. Epidemiology of visceral leishmaniasis in the Jenin District, West Bank: 1989-1998. Am. J. Trop. Med. Hyg. 66:329-333. [DOI] [PubMed] [Google Scholar]
- 2.Basset, D., F. Pratlong, C. Ravel, J. Puechberty, J. Dereure, and J. Dedet. 2001. Les leishmanioses déclarées en France en 1999. Bull. Épidémiol. Hebd. 5:19-20. [Google Scholar]
- 3.Belli, A., B. Rodriguez, H. Aviles, and E. Harris. 1998. Simplified polymerase chain reaction detection of New World Leishmania in clinical specimens of cutaneous leishmaniasis. Am. J. Trop. Med. Hyg. 58:102-109. [DOI] [PubMed] [Google Scholar]
- 4.Berzunza-Cruz, M., N. Cabrera, M. Crippa-Rossi, T. Sosa Cabrera, R. Perez-Montfort, and I. Becker. 2002. Polymorphism analysis of the internal transcribed spacer and small subunit of ribosomal RNA genes of Leishmania mexicana. Parasitol. Res. 88:918-925. [DOI] [PubMed] [Google Scholar]
- 5.Breniere, S. F., M. F. Bosseno, S. Revollo, M. T. Rivera, Y. Carlier, and M. Tibayrenc. 1992. Direct identification of Trypanosoma cruzi natural clones in vectors and mammalian hosts by polymerase chain reaction amplification. Am. J. Trop. Med. Hyg. 46:335-341. [DOI] [PubMed] [Google Scholar]
- 6.Carme, B., C. Aznar, and R. Pradinaud. 2001. Absence of a proven resurgence of Chagas disease or cutaneous leishmaniasis in French Guiana over the last two decades. Ann. Trop. Med. Parasitol. 95:623-625. [DOI] [PubMed] [Google Scholar]
- 7.Chicharro, C., M. A. Morales, T. Serra, M. Ares, A. Salas, and J. Alvar. 2002. Molecular epidemiology of Leishmania infantum on the island of Majorca: a comparison of phenotypic and genotypic tools. Trans. R. Soc. Trop. Med. Hyg. 96(Suppl. 1):S93-S99. [DOI] [PubMed] [Google Scholar]
- 8.Croan, D. G., D. A. Morrison, and J. T. Ellis. 1997. Evolution of the genus Leishmania revealed by comparison of DNA and RNA polymerase gene sequences. Mol. Biochem. Parasitol. 89:149-159. [DOI] [PubMed] [Google Scholar]
- 9.Croft, S. L., V. Yardley, and H. Kendrick. 2002. Drug sensitivity of Leishmania species: some unresolved problems. Trans. R. Soc. Trop. Med. Hyg. 96(Suppl. 1):S127-S129. [DOI] [PubMed] [Google Scholar]
- 10.Cupolillo, E., L. R. Brahim, C. B. Toaldo, M. P. de Oliveira-Neto, M. E. de Brito, A. Falqueto, M. de Farias Naiff, and G. Grimaldi, Jr. 2003. Genetic polymorphism and molecular epidemiology of Leishmania (Viannia) braziliensis from different hosts and geographic areas in Brazil. J. Clin. Microbiol. 41:3126-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cupolillo, E., G. Grimaldi, Jr., and H. Momen. 1994. A general classification of New World Leishmania using numerical zymotaxonomy. Am. J. Trop. Med. Hyg. 50:296-311. [DOI] [PubMed] [Google Scholar]
- 12.Cupolillo, E., G. Grimaldi, Jr., H. Momen, and S. M. Beverley. 1995. Intergenic region typing (IRT): a rapid molecular approach to the characterization and evolution of Leishmania. Mol. Biochem. Parasitol. 73:145-155. [DOI] [PubMed] [Google Scholar]
- 13.Cupolillo, E., H. Momen, and G. Grimaldi, Jr. 1998. Genetic diversity in natural populations of New World Leishmania. Mem. Inst. Oswaldo Cruz 93:663-668. [DOI] [PubMed] [Google Scholar]
- 14.Dedet, J. P. 1990. Cutaneous leishmaniasis in French Guiana: a review. Am. J. Trop. Med. Hyg. 43:25-28. [DOI] [PubMed] [Google Scholar]
- 15.Desjeux, P., and J. P. Dedet. 1989. Isoenzyme characterization of 112 Leishmania isolates from French Guiana. Trans. R. Soc. Trop. Med. Hyg. 83:610-612. [DOI] [PubMed] [Google Scholar]
- 16.Disch, J., M. J. Pedras, M. Orsini, C. Pirmez, M. C. de Oliveira, M. Castro, and A. Rabello. 2005. Leishmania (Viannia) subgenus kDNA amplification for the diagnosis of mucosal leishmaniasis. Diagn. Microbiol. Infect. Dis. 51:185-190. [DOI] [PubMed] [Google Scholar]
- 17.El Tai, N. O., M. El Fari, I. Mauricio, M. A. Miles, L. Oskam, S. H. El Safi, W. H. Presber, and G. Schonian. 2001. Leishmania donovani: intraspecific polymorphisms of Sudanese isolates revealed by PCR-based analyses and DNA sequencing. Exp. Parasitol. 97:35-44. [DOI] [PubMed] [Google Scholar]
- 18.el Tai, N. O., O. F. Osman, M. el Fari, W. Presber, and G. Schonian. 2000. Genetic heterogeneity of ribosomal internal transcribed spacer in clinical samples of Leishmania donovani spotted on filter paper as revealed by single-strand conformation polymorphisms and sequencing. Trans. R. Soc. Trop. Med. Hyg. 94:575-579. [DOI] [PubMed] [Google Scholar]
- 19.Garcia, A. L., A. Kindt, K. W. Quispe-Tintaya, H. Bermudez, A. Llanos, J. Arevalo, A. L. Banuls, S. De Doncker, D. Le Ray, and J. C. Dujardin. 2005. American tegumentary leishmaniasis: antigen-gene polymorphism, taxonomy and clinical pleomorphism. Infect. Genet. Evol. 5:109-116. [DOI] [PubMed] [Google Scholar]
- 20.Garcia, L., A. Kindt, H. Bermudez, A. Llanos-Cuentas, S. De Doncker, J. Arevalo, K. Wilber Quispe Tintaya, and J. C. Dujardin. 2004. Culture-independent species typing of neotropical Leishmania for clinical validation of a PCR-based assay targeting heat shock protein 70 genes. J. Clin. Microbiol. 42:2294-2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Grimaldi, G., Jr., and R. B. Tesh. 1993. Leishmaniases of the New World: current concepts and implications for future research. Clin. Microbiol. Rev. 6:230-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Grimaldi, G., Jr., R. B. Tesh, and D. McMahon-Pratt. 1989. A review of the geographic distribution and epidemiology of leishmaniasis in the New World. Am. J. Trop. Med. Hyg. 41:687-725. [DOI] [PubMed] [Google Scholar]
- 23.Guevara, P., G. Alonso, J. F. da Silveira, M. de Mello, J. V. Scorza, N. Anez, and J. L. Ramirez. 1992. Identification of New World Leishmania using ribosomal gene spacer probes. Mol. Biochem. Parasitol. 56:15-26. [DOI] [PubMed] [Google Scholar]
- 24.Inga, R., S. De Doncker, J. Gomez, M. Lopez, R. Garcia, D. Le Ray, J. Arevalo, and J. C. Dujardin. 1998. Relation between variation in copy number of ribosomal RNA encoding genes and size of harbouring chromosomes in Leishmania of subgenus Viannia. Mol. Biochem. Parasitol. 92:219-228. [DOI] [PubMed] [Google Scholar]
- 25.Ishikawa, E. A., F. T. Silveira, A. L. Magalhaes, R. B. Guerra, Jr., M. N. Melo, R. Gomes, T. G. Silveira, and J. J. Shaw. 2002. Genetic variation in populations of Leishmania species in Brazil. Trans. R. Soc. Trop. Med. Hyg. 96(Suppl. 1):S111-S121. [DOI] [PubMed] [Google Scholar]
- 26.Lainson, R., and J. J. Shaw. 1987. Evolution, classification and geographical distribution, p. 1-120. In W. Peters and R. Killick-Kendrick (ed.), The leishmaniasis in biology and medicine, vol. 1. Academic Press, London, United Kingdom. [Google Scholar]
- 27.Marfurt, J., A. Nasereddin, I. Niederwieser, C. L. Jaffe, H. P. Beck, and I. Felger. 2003. Identification and differentiation of Leishmania species in clinical samples by PCR amplification of the miniexon sequence and subsequent restriction fragment length polymorphism analysis. J. Clin. Microbiol. 41:3147-3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Marfurt, J., I. Niederwieser, N. D. Makia, H. P. Beck, and I. Felger. 2003. Diagnostic genotyping of Old and New World Leishmania species by PCR-RFLP. Diagn. Microbiol. Infect. Dis. 46:115-124. [DOI] [PubMed] [Google Scholar]
- 29.Martinez-Calvillo, S., S. M. Sunkin, S. Yan, M. Fox, K. Stuart, and P. J. Myler. 2001. Genomic organization and functional characterization of the Leishmania major Friedlin ribosomal RNA gene locus. Mol. Biochem. Parasitol. 116:147-157. [DOI] [PubMed] [Google Scholar]
- 30.Mauricio, I. L., M. W. Gaunt, J. R. Stothard, and M. A. Miles. 2001. Genetic typing and phylogeny of the Leishmania donovani complex by restriction analysis of PCR amplified gp63 intergenic regions. Parasitology 122:393-403. [DOI] [PubMed] [Google Scholar]
- 31.Mauricio, I. L., M. K. Howard, J. R. Stothard, and M. A. Miles. 1999. Genomic diversity in the Leishmania donovani complex. Parasitology 119:237-246. [DOI] [PubMed] [Google Scholar]
- 32.Momen, H., G. Grimaldi, Jr., and L. M. Deane. 1987. Leishmania infantum, the aetiological agent of American visceral leishmaniasis (AVL)? Mem. Inst. Oswaldo Cruz 82:447-448. [DOI] [PubMed] [Google Scholar]
- 33.Morales, M. A., C. Chicharro, M. Ares, C. Canavate, D. C. Barker, and J. Alvar. 2001. Molecular tracking of infections by Leishmania infantum. Trans. R. Soc. Trop. Med. Hyg. 95:104-107. [DOI] [PubMed] [Google Scholar]
- 34.Nei, M., and W. H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nimri, L. F., and H. D. Schallig. 2003. Application of riboprinting for the identification of isolates of cutaneous Leishmania spp. Parasitology 127:201-205. [DOI] [PubMed] [Google Scholar]
- 36.Noyes, H., F. Pratlong, M. Chance, J. Ellis, G. Lanotte, and J. P. Dedet. 2002. A previously unclassified trypanosomatid responsible for human cutaneous lesions in Martinique (French West Indies) is the most divergent member of the genus Leishmania ss. Parasitology 124:17-24. [DOI] [PubMed] [Google Scholar]
- 37.Ramirez, J. R., S. Agudelo, C. Muskus, J. F. Alzate, C. Berberich, D. Barker, and I. D. Velez. 2000. Diagnosis of cutaneous leishmaniasis in Colombia: the sampling site within lesions influences the sensitivity of parasitologic diagnosis. J. Clin. Microbiol. 38:3768-3773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rioux, J. A., G. Lanotte, E. Serres, F. Pratlong, P. Bastien, and J. Perieres. 1990. Taxonomy of Leishmania. Use of isoenzymes. Suggestions for a new classification. Ann. Parasitol. Hum. Comp. 65:111-125. [DOI] [PubMed] [Google Scholar]
- 39.Rodriguez-Bonfante, C., R. Bonfante-Garrido, G. Grimaldi, Jr., H. Momen, and E. Cupolillo. 2003. Genotypically distinct Leishmania colombiensis isolates from Venezuela cause both cutaneous and visceral leishmaniasis in humans. Infect. Genet. Evol. 3:119-124. [DOI] [PubMed] [Google Scholar]
- 40.Romero, G. A., M. V. Guerra, M. G. Paes, E. Cupolillo, C. Bentin Toaldo, V. O. Macedo, and O. Fernandes. 2001. Sensitivity of the polymerase chain reaction for the diagnosis of cutaneous leishmaniasis due to Leishmania (Viannia) guyanensis. Acta Trop. 79:225-229. [DOI] [PubMed] [Google Scholar]
- 41.Rotureau, B. 2006. Ecology of the Leishmania species in the Guianan ecoregion complex. Am. J. Trop. Med. Hyg., 74(1):87-96. [PubMed]
- 42.Schonian, G., H. Akuffo, S. Lewin, K. Maasho, S. Nylen, F. Pratlong, C. L. Eisenberger, L. F. Schnur, and W. Presber. 2000. Genetic variability within the species Leishmania aethiopica does not correlate with clinical variations of cutaneous leishmaniasis. Mol. Biochem. Parasitol. 106:239-248. [DOI] [PubMed] [Google Scholar]
- 43.Schonian, G., M. El Fari, S. Lewin, C. Schweynoch, and W. Presber. 2001. Molecular epidemiology and population genetics in Leishmania. Med. Microbiol. Immunol. (Berlin) 190:61-63. [DOI] [PubMed] [Google Scholar]
- 44.Schonian, G., A. Nasereddin, N. Dinse, C. Schweynoch, H. D. Schallig, W. Presber, and C. L. Jaffe. 2003. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn. Microbiol. Infect. Dis. 47:349-358. [DOI] [PubMed] [Google Scholar]
- 45.Schonian, G., L. Schnur, M. el Fari, L. Oskam, A. A. Kolesnikov, W. Sokolowska-Kohler, and W. Presber. 2001. Genetic heterogeneity in the species Leishmania tropica revealed by different PCR-based methods. Trans. R. Soc. Trop. Med. Hyg. 95:217-224. [DOI] [PubMed] [Google Scholar]
- 46.Singh, S., and R. Sivakumar. 2003. Recent advances in the diagnosis of leishmaniasis. J. Postgrad. Med. 49:55-60. [DOI] [PubMed] [Google Scholar]
- 47.Soccol, V. 1993. Les Leishmania du Nouveau Monde. Analyse enzymatique, démarche progressive phénétique et cladistique. Relations phylogénétiques avec les Leishmania de l'ancien monde. Ph.D. thesis. University of Montpellier I, Montpellier, France.
- 48.Tavares, C. A., A. P. Fernandes, and M. N. Melo. 2003. Molecular diagnosis of leishmaniasis. Expert Rev. Mol. Diagn. 3:657-667. [DOI] [PubMed] [Google Scholar]
- 49.Thomaz-Soccol, V., G. Lanotte, J. A. Rioux, F. Pratlong, A. Martini-Dumas, and E. Serres. 1993. Phylogenetic taxonomy of New World Leishmania. Ann. Parasitol. Hum. Comp. 68:104-106. [PubMed] [Google Scholar]
- 50.Thomaz-Soccol, V., I. D. Velez, F. Pratlong, S. Agudelos, G. Lanotte, and J. A. Rioux. 2000. Enzymatic polymorphism and phylogenetic relationships in Leishmania Ross, 1903 (Sarcomastigophora: Kinetoplastida): a case study in Colombia. Syst. Parasitol. 46:59-68. [DOI] [PubMed] [Google Scholar]
- 51.Vega-Lopez, F. 2003. Diagnosis of cutaneous leishmaniasis. Curr. Opin. Infect. Dis. 16:97-101. [DOI] [PubMed] [Google Scholar]
- 52.Victoir, K., A. L. Banuls, J. Arevalo, A. Llanos-Cuentas, R. Hamers, S. Noel, S. De Doncker, D. Le Ray, M. Tibayrenc, and J. C. Dujardin. 1998. The gp63 gene locus, a target for genetic characterization of Leishmania belonging to subgenus Viannia. Parasitology 117:1-13. [PubMed] [Google Scholar]
- 53.Victoir, K., S. De Doncker, L. Cabrera, E. Alvarez, J. Arevalo, A. Llanos-Cuentas, D. Le Ray, and J. C. Dujardin. 2003. Direct identification of Leishmania species in biopsy specimens from patients with American tegumentary leishmaniasis. Trans. R. Soc. Trop. Med. Hyg. 97:80-87. [DOI] [PubMed] [Google Scholar]
- 54.Weigle, K. A., L. A. Labrada, C. Lozano, C. Santrich, and D. C. Barker. 2002. PCR-based diagnosis of acute and chronic cutaneous leishmaniasis caused by Leishmania (Viannia). J. Clin. Microbiol. 40:601-606. [DOI] [PMC free article] [PubMed] [Google Scholar]