Abstract
Taiwan experienced a series of outbreaks of nosocomial severe acute respiratory syndrome (SARS) infections in 2003. Two months after the final outbreak, we recruited 658 employees from the hospital that suffered the first and most severe SARS infections to help us investigate epidemiological and genetic factors associated with the SARS coronavirus (SARS-CoV). SARS-CoV infections were detected by using enzyme immunoassays and confirmed by a combination of Western blot assays, neutralizing antibody tests, and commercial SARS tests. Risk factors were analyzed via questionnaire responses and sequence-specific oligonucleotide probes of human leukocyte antigen (HLA) alleles. Our results indicate that 3% (20/658) of the study participants were seropositive, with one female nurse identified as a subclinical case. Identified SARS-CoV infection risk factors include working in the same building as the hospital's emergency room and infection ward, providing direct care to SARS patients, and carrying a Cw*0801 HLA allele. The odds ratio for contracting a SARS-CoV infection among persons with either a homozygous or a heterozygous Cw*0801 genotype was 4.4 (95% confidence interval, 1.5 to 12.9; P = 0.007).
Severe acute respiratory syndrome (SARS) is caused by SARS coronavirus (SARS-CoV) (10, 11, 17, 29), with transmission primarily thought to occur via respiratory droplets or direct contact. Contaminated sewage is believed to be responsible for the first major SARS outbreak in Hong Kong's Amoy Garden housing complex, with over 300 residents affected (28). The incubation period was measured at 2 to 10 days (mean, 6.4) (28). Manifesting as atypical pneumonia, SARS severity ranged from nonspecific symptoms (e.g., fever, chills, headache, and dry cough) to adult respiratory distress syndrome (28). Approximately 20% of all SARS patients became critically ill and required admission to intensive care units (3, 6). Very few subclinical cases have been identified to date (21, 34).
In 2003, Taiwan experienced a series of SARS outbreaks, and Municipal Hoping Hospital (referred to hereafter as HP) in Taipei City suffered the first and the most serious outbreak of SARS-CoV nosocomial infections: 137 probable cases and 26 deaths (19, 20). According to the Center for Disease Control (CDC) in Taiwan, 364 of the 664 probable Taiwanese SARS cases reported to the World Health Organization were confirmed by reverse transcriptase (RT) PCR and/or neutralizing antibody tests (8).
The human leukocyte antigen (HLA) complex plays an important role in determining susceptibility to infectious diseases. HLA class I gene products present antigenic peptides to T cells, initiating an immune response and the removal of foreign material (30). Genes that encode HLA class I molecules are highly polymorphic, apparently as the result of natural selection processes that enable mammals to resist a wide variety of pathogens (13). Researchers have demonstrated that specific HLA alleles are associated with susceptibility to and outcomes from such viral infections as human immunodeficiency virus type 1, human T-cell leukemia virus type 1, and hepatitis C virus (5, 15, 16). Previous studies have shown that people with the HLA-B*-4601 and HLA-B*-0703 alleles are susceptible to SARS-CoV infection (23, 27). We performed an epidemiological study of SARS-CoV infections among HP Hospital employees and used the data to conduct a case-control study to find a potential association between the HLA-Cw*0801 allele and SARS-CoV infections.
MATERIALS AND METHODS
Participants and questionnaires.
Invitations were sent to 938 HP Hospital employees between 9 June and 13 June 2003, which was the first week of their return to work following a 7-week hospital closure. Informed consent forms were signed by all study participants. Between 14 June and 19 June, we collected 658 blood samples and 647 completed questionnaires. Questionnaire items were designed to elicit information on demographics, residence, travel history, job description, work unit and location (building and floor), SARS-related work behaviors (i.e., contact with SARS patients), personal medical history (especially SARS diagnoses and symptoms), and quarantine history. In early December of the same year, 20 seropositive individuals were requested to donate blood samples to investigate changes in their serological status; 12 agreed to do so.
Serological assays.
Serum samples from the 12 seropositive participants were stored at −80°C prior to performing serological assays. An enzyme immunoassay (EIA) with a SARS-CoV nucleocapsid (N) recombinant protein (RP) was used as a screening test. Sera showing repeat positive results were confirmed by Western blot (WB) assay with SARS-CoV spike (S) and N RPs. Four commercially available serological tests for SARS infection were used for confirmation purposes: EUR-enzyme-linked immunosorbent assay (ELISA) (EUROIMMUN), GLD-ELISA (Genelabs Diagnostic Pte., Ltd.), EUR-immunofluorescence assay (EUROIMMUN), and a GLD quick test (Genelabs Diagnostic Pte. Ltd.).
pGST-N, pGST-SIa, and pGST-SIb plasmid construction.
To amplify the N and S genes of SARS-CoV, viral RNA from a local SARS-CoV isolate (TWC) (18) was used as a template in a single-tube RT-PCR (Stratagene, La Jolla, CA). The following primer pairs were used for gene amplification: for the N gene, NF, 5′-GCCGAATTCATGTCTGATAATGGACCCCA-3′, and NR, 5′-GCGCGTCGACGTTATGCCTGAGTTGAATCA-3′; for the SIa gene (amino acid residues 1 to 460 of the S protein), S1F, 5′-GCGGAATTCATGTTTATTTTCTTATTATTTCTTAC-3′, and S1R, 5′-GCGCTCGAGGAAAGGCACATTAGATATGTCT-3′; and for the SIb gene (amino acid residues 416 to 846 of the S protein), S2F, 5′-GCGGAATTCATGGGTTGTGTCCTTGCTTGGA-3′, and S2R, 5′-GCGCTCGAGCAGAGGTGGCAACACTGTAAGT-3′. The RT-PCR mixtures contained 40 mM deoxynucleoside triphosphate, 5 μl 10× high-fidelity RT-PCR buffer, 1.5 μl mix enzyme, and 30 μM of each primer. PCR products were subcloned into a pGEX-4T-1 vector (Amersham Biosciences, Piscataway, NJ). The resulting plasmids (labeled pGEX-N, pGEX-SIa, and pGEX-SIb) were sequenced using a DNA sequencer equipped with a dye terminator cycle core kit (model 373A, version 1.0.2; Applied Biosystems, Foster City, CA).
N, SIa, and SIb recombinant protein expression and purification.
RPs were induced into BL21 cells using isopropyl-β-d-thiogalactopyranoside (IPTG) (Sigma- Aldrich). The optic densities of BL21 cells containing plasmid pGEX-N, pGEX-SIa, or pGEX-SIb for induction were 0.6 to 0.7. After IPTG induction at 37°C for 2.5 h, bacterial cells were collected by centrifugation at 6,000 rpm at 4°C for 15 min. RP-N, RP-SIa, and RP-SIb quantities were purified using glutathione-Sepharose 4B beads (Pharmacia, Uppsala, Sweden) as described previously by Guan and Dixon (12). RP concentrations were measured using Pierce bicinchoninic acid protein assay reagent (Pierce, Rockford, IL); purity was analyzed by running samples on a 12.5% sodium dodecyl sulfate-polyacrylamide mini gel (Bio-Rad Laboratories, Richmond, CA).
Enzyme immunoassay.
We coated 96-well plates with RP-N protein at a concentration of 5 μg per ml (100 μl per well) in 50 mM carbonate buffer (pH 9.6) for 1 h at 37°C and blocked with 2% bovine serum albumin (BSA) in phosphate-buffered saline (PBS)-0.05% Tween (PBST) (pH 7.4). To each well, we added negative control and heat-inactivated sera from SARS patients (1:200 dilution in PBST-0.1% BSA at 100 μl/well) prior to incubating the plates for 60 min at 37°C. After five washes with PBST, we added 100 μl of horseradish peroxidase-conjugated goat anti-human immunoglobulin G (1:2,000 dilution in PBST-0.1% BSA; Santa Cruz Biotechnology, Santa Cruz, CA) to each well and reincubated the plates for 60 min at 37°C. Optical densities were determined at a wavelength of 450 nm using an EIA reader (BioTek Elx808). Our cutoff value was determined by using the mean optic density of six normal human sera plus 3 standard deviations.
Mouse anti-N, SIa, and SIb RP antisera preparation.
To raise mouse antibodies against nucleocapsid and spike proteins, we mixed purified RP-N, RP-SIa, or RP-SIb with Freund complete (for initial immunization) or incomplete adjuvant (for booster injections) (Sigma Co., St. Louis, MO) at 10-day intervals via intraperitoneal injection at a dosage of approximately 25 μg RP per inoculum. Serum samples were collected from tail veins prior to immunization and 1 week following each injection. All sera were heat inactivated at 56°C for 30 min and stored at −20°C.
Western blotting.
Recombinant proteins purified from glutathione-agarose bead proteins were used for WB detection of EIA-positive and borderline cases from Hoping Hospital. Polyclonal antisera generated by immunizing rabbits with RP-N or RP-SIa combined with RP-SIb were used as positive controls. Preimmunized rabbit serum and sera collected from healthy employees at Kong-Ning Hospital prior to the SARS epidemics were used as negative controls. Serum samples from various patients were WB assayed for their anti-N and anti-S antibody reactivity at a 1:200 dilution.
Neutralization antibody test.
Neutralization antibody (NA) titers from anti-SARS-CoV seropositive samples were evaluated using NA assays. Serum samples were prepared via a series of twofold dilutions from 1:8 to 1:1,024 using a phosphate-buffered saline solution prior to heat inactivation at 56°C for 30 min. Next, 50 μl of each diluted sample was mixed with an equal volume of 43 50% tissue culture infective doses of the SARS-CoV TWC strain at 37°C for 1 h. This mixture was added to 2.5 × 104 Vero-E6 cells in 100 μl Dulbecco's modified Eagle's medium (GIBCO BRL, Grand Island, NY) with 10% heat-inactivated fetal bovine serum (HyClone, Logan, UT), penicillin (100 IU/ml), streptomycin (100 μg/ml), nonessential amino acids (0.1 mM), fungizone (2.5 mg/ml), and l-glutamine (2 mM) in a 96-well tissue-culture plate and incubated in a humidified incubator with 5% CO2 for 5 days. The presence of NA titer was determined by the reciprocal of the highest serum dilution which showed no cytopathic effect in Vero-E6 cells multiplied by two for mixing with an equal volume of virus; the lowest titer value was measured as 16. A positive control serum was used in each plate to ensure consistency. To compare changes in NA titer, paired serum samples taken from each participant in June and December were tested at the same time using the same plate. For cases in which NA titer was <16, the SARS-CoV TWC virus was mixed with undiluted and heat-inactivated serum for the NA test.
HLA allele determination.
A DNA extraction kit (QIAGEN, Valencia, CA) was used to prepare genomic DNA from the participants' peripheral blood mononuclear cells. Allele typing for HLA-A, -B, -Cw, -DQB1, and -DRB1 loci was performed via PCR followed by sequence-specific oligonucleotide probing (Dynal Biotech, Ltd., Wirral, United Kingdom).
Statistical analysis.
Fisher's exact and χ2 tests were performed as part of univariate analyses to determine the statistical significance for all comparisons between seropositive and seronegative participants. Data were analyzed using SAS software (version 8.2). To determine associations between different HLA alleles and SARS-CoV infection, we designed a case-control study in which 20 seropositive cases were matched with 40 control employees according to age, gender, job description, work unit, and location. A conditional logistic regression was used to analyze the odds ratio of different HLA alleles and susceptibility to SARS infection. To confirm the statistical significance of the HLA allele in the case-control study, another 40 control employees (for a total of 80) were added to the analysis based on age, gender, job description, work unit, and location.
RESULTS
Serological tests for SARS-CoV infections.
The number of participating HP employees (658) represents more than 70 percent of all employees who were quarantined in the hospital during the April and May 2003 SARS outbreaks. Following an EIA screening test, serum samples with repeat positive results were confirmed using a WB assay with recombinant SARS-CoV N and S proteins. In all, 20 of the 658 participants (3%, 1 male and 19 female) were confirmed as having been infected with the SARS-CoV. The same 20 cases tested positive using two EIAs (EUR-ELISA and GLD-ELISA), one immunofluorescent antibody test (EUR-immunofluorescence assay), and one quick test (GLD quick test).
SARS-CoV infection risk factor analysis.
As shown in Table 1, the average ages of seropositive and seronegative participants were 34.5 and 37.6 years, respectively. In terms of job description, the seropositive breakdown was 15 of 304 participating nurses (4.9%), 3 of 16 nursing aides (18.7%), 1 of 2 laundry workers, and 1 of 2 occupational therapists. More than half of the 647 questionnaire respondents worked in HP building A, but 65% of the seropositive employees worked in building B (where the emergency room and the internal medicine ward are located) (P < 0.01). Of the 257 participants who reported treating or providing care for SARS patients, 15 (5.8%) had anti-SARS antibodies; 4 of the 375 (1.1%) who reported having no contact with SARS patients were seropositive (P < 0.01). Of the 25 employees who stated that they were diagnosed with SARS, 19 (76%) were confirmed serologically; of the 622 participants who denied having SARS, only 1 (0.16%) was found to have anti-SARS antibodies (P < 0.01). In terms of SARS-related symptoms, 19 of the 20 seropositive participants had body temperatures above 38°C, 15 had myalgia, 13 had rigor, 13 had diarrhea, and 8 had shortness of breath.
TABLE 1.
An analysis of risk factors and symptoms for SARS-CoV infection in Taipei Municipal Hoping Hospital in Taipei City
| Risk factor or symptoma | % of subjects
|
Totale (n = 647) | |
|---|---|---|---|
| Seropositiveb | Seronegativec | ||
| Sex (P = 0.15) | |||
| Male (n = 121) | 0.8 (1/121) | 99.2 (120/121) | 18.7 (121/647) |
| Female (n = 526) | 3.6 (19/526) | 96.4 (507/526) | 81.3 (526/647) |
| Aged (P = 0.66) | |||
| 20-29 (n = 182) | 4.4 (8/182) | 95.6 (174/182) | 28.1 (182/647) |
| 30-39 (n = 192) | 2.6 (5/192) | 97.4 (187/192) | 29.7 (192/647) |
| 40-49 (n = 169) | 3.0 (5/169) | 97.0 (164/169) | 26.1 (169/647) |
| >50 (n = 101) | 1.9 (2/101) | 98.1 (99/101) | 16.1 (101/647) |
| Occupation/status (P < 0.01) | |||
| Doctor (n = 68) | 0 (0/68) | 100 (68/68) | 10.7 (68/638) |
| Nurse (n = 304) | 4.9 (15/304) | 95.1 (289/304) | 47.7 (304/638) |
| Laboratory technician (n = 22) | 0 (0/22) | 100 (22/22) | 3.5 (22/638) |
| Pharmacist (n = 26) | 0 (0/26) | 100 (26/26) | 4.1 (26/638) |
| Nursing aide (n = 16) | 18.7 (3/16) | 81.3 (13/16) | 2.5 (16/638) |
| Laundry worker (n = 2) | 50 (1/2) | 50 (1/2) | 0.3 (2/638) |
| Housekeeper (n = 16) | 0 (0/16) | 100 (16/16) | 2.5 (16/638) |
| Patient's personal attendant (n = 12) | 0 (0/10) | 100 (12/12) | 1.9 (12/638) |
| Occupational therapist (n = 2) | 50 (1/2) | 50 (1/2) | 0.3 (2/638) |
| Physical therapist (n = 6) | 0 (0/6) | 100 (6/6) | 0.9 (6/638) |
| Administrative staff (n = 39) | 0 (0/39) | 100 (39/39) | 6.1 (39/638) |
| Other (n = 125) | 0 (0/125) | 100 (125/125) | 20.7 (125/638) |
| Work location (P < 0.01) | |||
| Building A (n = 369) | 0.5 (2/369) | 99.5 (367/369) | 57.7 (369/640) |
| Building B (n = 179) | 7.3 (13/179) | 92.7 (166/179) | 28.0 (179/640) |
| Buildings A and B (n = 92) | 5.4 (5/92) | 94.6 (87/92) | 14.4 (92/640) |
| Contact with SARS patients? (P < 0.01) | |||
| Yes (n = 275) | 5.8 (15/257) | 94.2 (242/257) | 40.7 (257/632) |
| No (n = 375) | 1.1 (4/375) | 98.9 (371/375) | 59.3 (375/632) |
| Diagnosed with SARS (P < 0.01) | |||
| Yes | 76 (19/25) | 24 (6/25) | 3.9 (25/647) |
| No | 0.2 (1/622) | 99.8 (621/622) | 96.1 (622/647) |
| Symptoms | |||
| Fever (>38°C) | 95 (19/20) | 2.1 (13/624) | 5.0 (32/644) |
| Myalgia (P < 0.01) | 75 (15/20) | 1.1 (7/624) | 3.4 (22/644) |
| Sore throat (P < 0.01) | 15 (3/20) | 1.8 (11/624) | 2.2 (14/644) |
| Cough (P < 0.01) | 60 (12/20) | 2.6 (16/624) | 4.3 (28/644) |
| Rigor (P < 0.01) | 65 (13/20) | 1.0 (6/624) | 3.0 (19/644) |
| Diarrhea (P < 0.01) | 65 (13/20) | 2.2 (14/624) | 4.2 (27/644) |
| Rhinorrhea (P < 0.04) | 10 (2/20) | 1.4 (9/624) | 1.7 (11/644) |
| Vomiting (P < 0.01) | 20 (4/20) | 0 (0/624) | 0.6 (4/644) |
| Shortness of breath (P < 0.01) | 40 (8/20) | 1.6 (10/624) | 2.8 (18/644) |
The Fisher's exact test P value is given for each risk factor or symptom.
In parentheses is the number of subjects seropositive/the number of subjects with risk factor or symptom. There were 20 subjects in the seropositive group.
In parentheses is the number of subjects seronegative/the number of subjects with risk factor or symptom. There were 627 subjects in the seronegative group.
Mean ages ± standard deviations were as follows: 34.5 ± 9.9 years for the seropositive group, 37.6 ± 10.6 years for the seronegative group, and 37.5 ± 10.5 years for both groups.
In parentheses is the total number of subjects with risk factor or symptom/total number of subjects either with any risk factor in the category or showing any symptom(s).
Data on demographics and neutralizing antibody titers for the 20 seropositive cases are shown in Table 2. Of the 19 participants with SARS-related symptoms, 8 (44%) suffered from acute respiratory distress syndrome. A 22-year-old nurse (case no. HP613) was the only seropositive participant who claimed that she had no SARS-related symptoms. Her first NA titer was measured at 32 in June of 2003 and 2 in December. As an employee of the internal medicine ward, she had frequent contact with SARS patients. She claimed that she took her own body temperature twice a day for more than 1 month during the nosocomial infection outbreak and that it was slightly above normal (37.4°C) only once.
TABLE 2.
Demographic data for 20 anti-SARS-CoV antibody-positive employees in the HP Hospital in Taipei City
| Case no. | Age (Yr) | Sex | Occupation | Workplace | SARS-related symptoms/ ARDSa | Neutralizing Ab titerb
|
HLA-Cw* genotype | |
|---|---|---|---|---|---|---|---|---|
| First | Second | |||||||
| HP11 | 29 | F | Nurse | Building A, 8th floor ward, orthopedics | +/− | 16 | NAc | 0302/0304 |
| HP84 | 34 | F | Nurse | Building A, 5th floor ward, kidney dialysis center | +/− | 16 | NA | 0401/0702 |
| HP580 | 28 | F | Nurse | Building B, 6th floor ward, internal medicine | +/+ | 128 | NA | 0303/1203 |
| HP583 | 32 | F | Nurse | Building B, 6th floor ward, internal medicine | +/− | 64 | 16 | 0801/1402 |
| HP604 | 35 | F | Nurse | Building B, 8th floor ward, internal medicine | +/+ | 128 | 32 | 0102/0304 |
| HP607 | 28 | F | Nurse | Building B, 8th floor ward, internal medicine | +/+ | 32 | NA | 0102/0801 |
| HP613 | 22 | F | Nurse | Building B, 8th floor ward, internal medicine | −/− | 32 | 2 | 0602/0801 |
| HP619 | 53 | F | Nurse | Building B, 9th floor ward, nursing home | +/+ | 128 | 128 | 0602/0801 |
| HP631 | 30 | F | Nurse | Building B, 5th floor, intensive care unit | +/− | 128 | 64 | 0702/1202 |
| HP681 | 21 | F | Nurse | Building B, 1st floor, emergency room | +/− | NA | NA | 0702/0801 |
| HP682 | 40 | F | Nurse | Building B, 1st floor, emergency room | +/− | 64 | NA | 0102/0702 |
| HP688 | 25 | F | Nurse | Building B, 1st floor, emergency room | +/− | 16 | NA | 0304/0801 |
| HP689 | 24 | F | Nurse | Building B, 1st floor, emergency room | +/+ | 64 | 64 | 0801/1402 |
| HP982 | 29 | F | Nurse | Building B, 1st floor, emergency room | +/+ | 128 | 16 | 0702/1202 |
| HP1001 | 44 | F | Nurse | Building B, B1 floor, family medicine center | +/− | 32 | NA | 0102/0702 |
| HP164 | 41 | F | Nursing aide | Building A, B1 floor, supply center | +/− | 128 | 32 | 0304/1202 |
| HP165 | 40 | F | Nursing aide | Building A, B1 floor, supply center | +/− | 128 | 64 | 0304/0702 |
| HP206 | 54 | F | Nursing aide | Building A, B1 floor, supply center | +/+ | 64 | 64 | 0302/0304 |
| HP225 | 49 | M | Laundry worker | Building A, B3 floor | +/− | 64 | 32 | 0801/0801 |
| HP769 | 31 | F | Occupational therapist | Building B, 4th floor, psychiatric department | +/+ | 128 | 128 | 0304/0802 |
ARDS, acute respiratory distress syndrome.
The first and second serum samples were drawn in June and December 2003, respectively. The nosocomial infection at HP Hospital was originated from a female patient who arrived at the emergency room on 6 April 2003. Ab, antibody.
NA, serum samples were not available for the assay.
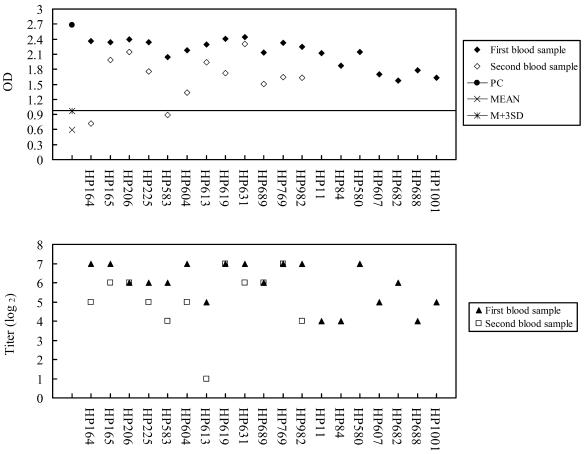
Our June 2003 NA test results indicate that 19 of the 20 seropositive participants had neutralizing antibodies (we had insufficient serum for the other participant). These antibodies were also detected in serum from the 12 participants who donated samples in December. Of the second group, the lowest titer level was 2; we noted that the mean decrease in NA titers for the 12 participants was fourfold between June and December (Fig. 1, lower panel). Of the 12 December samples, 10 were seropositive for anti-N antibodies according to EIA test results (Fig. 1, upper panel).
FIG. 1.
Results from EIA (upper panel) and NA (lower panel) tests for 19 anti-SARS-CoV antibody-positive cases. The EIA cutoff value equaled the mean optic density of normal controls plus 3 standard deviations. The lowest NA test titer was 2. First and second blood samples were collected 2 and 8 months after the outbreak, respectively. HP613 was a subclinical case. OD, optical density.
SARS-CoV infection genetic correlates.
We used PCR and sequence-specific oligonucleotide probing to determine HLA-A, -B, -Cw, -DQB1, and -DRB1 alleles in samples from the 20 seropositive cases and 40 control employees matched by age, gender, job description, and work location. Our results indicate that among all tested alleles, the only statistically significant difference was for Cw*0801 allele rates between the case and control groups (22.5% or 9/40 of the seropositive cases versus 6.3% or 5/80 for controls; P = 0.05) (Table 3, first control group). As explained in an earlier section, we increased the number of control individuals from 40 to 80 (also matched by age, gender, and occupation). When we retested our hypothesis of an association between the Cw*0801 allele and SARS-CoV infection susceptibility, our results indicated that the P for the difference between case and control group rates decreased to 0.003 (Table 3, second control group). Results from a conditional logistic regression analysis of our results by genotype show that the odds ratio for contracting a SARS-CoV infection among persons with either a homozygous or a heterozygous Cw*0801 genotype was 4.4 (95% confidence interval, 1.5 to 12.9; P = 0.007) (Table 4).
TABLE 3.
Comparison of HLA-Cw*0801 and HLA-DRB1*0301 allelic frequencies between seropositive and seronegative anti-SARS-CoV antibody groups
| Control groupa | Allele | Anti-SARS-CoV antibodies
|
Odds ratio | 95% CI | Pb | |
|---|---|---|---|---|---|---|
| % Seropositive | % Seronegative | |||||
| First | Cw*0801 | 22.5 (9/40) | 6.3 (5/80) | 2.37 | 1.00-5.61 | 0.05 |
| B*4601 | 5.0 (2/40) | 13.8 (11/80) | 0.44 | 0.10-1.97 | 0.28 | |
| DRB1*0301 | 2.5 (1/40) | 11.3 (9/80) | 0.28 | 0.04-2.15 | 0.22 | |
| Second | Cw*0801 | 22.5 (9/40) | 5.6 (9/160) | 3.40 | 1.50-7.58 | 0.003 |
| DRB1*0301 | 2.5 (1/40) | 8.1 (13/160) | 0.32 | 0.04-2.41 | 0.27 | |
In the first control group, there were 20 seropositive participants and 40 seronegative participants; in the second control group, there were 20 seropositive participants and 80 seronegative participants.
Results shown were determined by conditional logistic regression analysis.
TABLE 4.
A conditional logistic regression analysis on the odds ratios of contracting SARS-CoV infection among persons with heterozygous or homozygous HLA-Cw*0801 genotypea
| HLA-Cw*0801 genotype | Anti-SARS-CoV antibodies for the HP Hospital cohortb
|
Control populationc | |
|---|---|---|---|
| % Seropositive | % Seronegative | ||
| −/− | 60 (12/20) | 88.7 (71/80) | 86 (80/93) |
| +/− or +/+ | 40 (8/20) | 11.3 (9/80) | 14 (13/93) |
For the HP hospital cohort compared to the group with the HLA-Cw*0801 −/− genotype, the odds ratio (relative risk) of contracting SARS-CoV infection for individuals with the +/− or +/+ genotype was 4.4, with a 95% CI between 1.5 and 12.9 (P = 0.007). If we replace the seronegative group with another control population (7), then the odds ratio for individuals with the +/− or +/+ genotype was 4.1, with a 95% CI between 1.4 and 12.0 (P = 0.01).
In parentheses is the number of subjects with the indicated genotype/the number of subjects in the indicated group.
Control subjects were Han Chinese residing in Taiwan (72). In parentheses is the number of subjects with the indicated genotype/the number of control subjects.
DISCUSSION
The HP hospital that we included in this study was the first hospital reported to have cases of nosocomial infection of SARS-CoV in Taiwan (19). Before this outbreak, there were only several sporadic imported cases of SARS in Taiwan (19). In addition, judging by the dates of onset of the symptoms of the staff members at HP Hospital, we concluded that all of them contracted the infection in the hospital. Previous reports indicate that SARS patients develop antinucleocapsid antibodies 3 weeks after symptom onset (28). We assumed that all SARS-CoV-infected participants in our study would have detectable antibodies, since we used serum samples collected 2 months after the nosocomial outbreaks. In addition, we reduced the EIA cutoff value to mean optical density plus 2 standard deviations to include as many cases as possible for confirmation using WB and four commercial assay kits that became available after we finished our serological study.
In this study, 280 of 938 (29.9%) of the staff members of HP Hospital did not participate. According to Taiwan CDC records, there were 51 confirmed SARS cases, including 7 mortalities among staff members of HP Hospital. Among 20 cases that we detected in this study, 2 cases have never been identified and reported to Taiwan CDC. Therefore, 26 staff members who had SARS and were alive did not participate in this study. According to the medical records, most of them were still hospitalized in other hospitals due to the illness during our study period. These nonparticipants may introduce bias to our analysis.
The self-reported symptoms of the 19 symptomatic and serologically confirmed cases were similar to those described in other clinical SARS studies (22, 32): significantly higher incidences of fever (>38°C), myalgia, and rigor compared to symptomatic seronegative individuals (Table 1). Four of the six seronegative participants who claimed that they had been diagnosed with SARS also reported that they suffered from shortness of breath. Optical density readings for these employees were far below the EIA cutoff value; WB assay results showed that none of them had SARS-CoV infections.
The anti-SARS-CoV antibody reactivity of the asymptomatic nurse (case no. HP613) was verified by all available serological assays. Her antibody reactivity remained positive 6 months after her first blood sample was drawn (Fig. 1, upper panel), although her NA titer fell to 2 in her second sample (lower panel). We tried to isolate the SARS-CoV from a nasopharyngeal swab in order to refute the possibility that she was an asymptomatic SARS-CoV carrier, but the results were negative.
Regarding risk factors for contracting SARS, the data indicate that emergency room or internal medicine ward nurses were at highest risk. According to the Center for Disease Control in Taiwan, the index case of nosocomial infection for HP was a laundry worker who had access to both the emergency room and the building B internal medicine ward. Three SARS-CoV-infected nursing aides (case nos. HP164, HP165, and HP206) and one infected occupational therapist (HP769) claimed that they had no direct contact with SARS patients; they may have come into contact with patient specimens or infected bedclothes.
We found evidence of a link between HLA-Cw*0801 and SARS-CoV infection susceptibility. One of the cases was HLA-Cw*0801 homozygous; we observed that the relative risk of infection increased from 3.3 for heterozygous individuals to 6 for homozygous individuals (95% confidence intervals of 0.9 to 11.6 and 0.2 to 188.7, respectively). We also compared our data with data from a normal group provided by a separate Taiwanese research team (7) and obtained similar results (odds ratio, 4.1; 95% confidence interval [CI], 1.4 to 12.0; P, 0.01) (Table 4). We therefore conclude that Cw*0801 is a susceptibility marker for SARS-CoV infection.
During their study of potential SARS patients and high-risk health care workers, Lin et al. observed an association between HLA-B*4601 and SARS-CoV infections (23). However, their definition of a SARS patient was based on clinical diagnosis rather than serological evidence and health care workers may not have been a suitable control group. In contrast, 14.6% of our 80-member control group and 5% of the 20 seropositive participants carried the HLA-B*4601 allele (no statistical significance) (Table 3). In a Hong Kong study that used bone marrow donors as a control, Ng et al. described HLA-B*0703 and HLA-DRB1*0301 as susceptible and resistant alleles for SARS-CoV infection, respectively. However, they did not collect or analyze HLA-Cw allelic frequencies among their participants (27). In the present study, the seropositive group had a lower HLA-DRB1*0301 frequency than the seronegative group (Table 3), but not at a statistically significant level (P = 0.22). None of our participants carried HLA-B*0703.
Previous reports have stated that individuals carrying HLA-Cw*0801 are at significantly higher risk of contracting adult periodontitis (odds ratio, 6.2) (25) and that a link exists between HLA-Cw*04 and persistent hepatitis C viral infection (31). A research team in Beijing found that the total numbers of natural killer (NK) and CD158b+ NK cells were significantly lower in SARS patients compared to those in healthy patients (26). NK cells play a central role in innate antiviral immune response. In vivo, their activity is controlled via inhibitory and activation receptors for major histocompatibility complex class I molecules (1, 2, 24, 33). Individuals who are homozygous for the HLA-B/-C region of conserved MHC-extended haplotypes have lower NK cell activity and significantly lower numbers of CD16+ and CD158b+ NK cells compared to those for their heterozygous counterparts (14). This may help explain the higher susceptibility of homozygous individuals to viral infections. Khakoo et al. recently reported that genes that encode the inhibitory NK-cell receptor KIR2DL3 and its HLA C1 ligand exert a direct influence on the resolution of hepatitis C viral infections (16). Accordingly, HLA-Cw*0801 may affect SARS-CoV susceptibility via its interaction with the killer cell immunoglobulin-like receptors of NK cells.
Of the 242 asymptomatic participants in our study who had close contact with SARS patients, only 1 (0.41%) had a subclinical infection. The single asymptomatic case was unusual in several respects. In addition to carrying the heterozygous HLA-Cw*0801, she also carried an HLA haplotype (HLA-A*0101/2402, -B*4006/5701, -Cw*0801/0602, -DRB1*0701/0803, and -DQB1*0303/0601) that is considered rare among Chinese. Her HLA allelic frequencies were relatively low compared to the majority of Taiwanese: 1, 2, and 2.9% for HLA-A*0101, -B*4006, and -Cw*0602, respectively (http://www.allelefrequencies.net/). On the other hand, all of her HLA class I alleles were heterozygous, which is consistent with the hypothesis of a heterozygote advantage against infectious disease (4, 9, 35). We therefore suggest that HLA-Cw*0801 confers higher susceptibility to SARS-CoV infection but is not necessarily linked with disease severity.
Acknowledgments
We thank the administrative staff of Hoping Hospital for assistance in recruiting study participants. We obtained prior approval from thehospital's institutional review board. We also thank Jen-Hsiang Chuang and Yi-Fen Lin for their helpful discussions.
This study was supported in part by the following grants: no. 93004-62-007 from the Department of Health, Taipei City Government, NSC 92-2751-B-010-001-Y from the R.O.C. National Science Council, and VGHUST93-G7-07-1 from the Veterans General Hospital, University System of Taiwan.
Footnotes
This paper is dedicated to the physicians and nurses who died of SARS in 2003.
REFERENCES
- 1.Ahmad, R., S. T. Sindhu, P. Tran, E. Toma, R. Morisset, J. Menezes, and A. Ahmad. 2001. Modulation of expression of the MHC class I-binding natural killer cell receptors, and NK activity in relation to viral load in HIV-infected/AIDS patients. J. Med. Virol. 65:431-440. [PubMed] [Google Scholar]
- 2.Bauer, S., V. Groh, J. Wu, A. Steinle, J. H. Phillips, L. L. Lanier, and T. Spies. 1999. Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285:727-729. [DOI] [PubMed] [Google Scholar]
- 3.Booth, C. M., and T. E. Stewart. 2005. Severe acute respiratory syndrome and critical care medicine: the Toronto experience. Crit. Care Med. 33(Suppl):S53-S60. [DOI] [PubMed] [Google Scholar]
- 4.Carrington, M., G. W. Nelson, M. P. Martin, T. Kissner, D. Vlahov, J. J. Goedert, R. Kaslow, S. Buchbinder, K. Hoots, and S. J. O'Brien. 1999. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science 283:1748-1752. [DOI] [PubMed] [Google Scholar]
- 5.Carrington, M., and S. J. O'Brien. 2003. The influence of HLA genotype on AIDS. Annu. Rev. Med. 54:535-551. [DOI] [PubMed] [Google Scholar]
- 6.Chan-Yeung, M., and R. H. Xu. 2003. SARS: epidemiology. Respirology. 8(Suppl):S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chung, W. H., S. I. Hung, H. S. Hong, M. S. Hsih, L. C. Yang, H. C. Ho, J. Y. Wu, and Y. T. Chen. 2004. Medical genetics: a marker for Stevens-Johnson syndrome. Nature 428:486. [DOI] [PubMed] [Google Scholar]
- 8.Division of Surveillance and Investigation, Center for Disease Control, Taiwan. 2004. SARS probable cases in Taiwan-reclassification on September, 2003, p. 6-10. In I. J. Su (ed.), Memoir of severe acute respiratory syndrome control in Taiwan. Republic of China Center for Disease Control, Taipei, Taiwan.
- 9.Doherty, P. C., and R. M. Zinkernagel. 1975. Enhanced immunological surveillance in mice heterozygous at the H-2 gene complex. Nature 256:50-52. [DOI] [PubMed] [Google Scholar]
- 10.Drosten, C., S. Gunther, W. Preiser, S. van der Werf, H. R. Brodt, S. Becker, H. Rabenau, M. Panning, L. Kolesnikova, R. A. Fouchier, A. Berger, A. M. Burguiere, J. Cinatl, M. Eickmann, N. Escriou, K. Grywna, S. Kramme, J. C. Manuguerra, S. Muller, V. Rickerts, M. Sturmer, S. Vieth, H. D. Klenk, A. D. Osterhaus, H. Schmitz, and H. W. Doerr. 2003. Identification of a novel coronavirus in patients with severe acute respiratory syndrome. N. Engl. J. Med. 348:1967-1976. [DOI] [PubMed] [Google Scholar]
- 11.Fouchier, R. A., T. Kuiken, M. Schutten, G. van Amerongen, G. J. van Doornum, B. G. van den Hoogen, M. Peiris, W. Lim, K. Stohr, and A. D. Osterhaus. 2003. Aetiology: Koch's postulates fulfilled for SARS virus. Nature 423:240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guan, K. L., and J. E. Dixon. 1991. Eukaryotic proteins expressed in Escherichia coli: an improved thrombin cleavage and purification procedure of fusion proteins with glutathione S-transferase. Anal. Biochem. 192:262-267. [DOI] [PubMed] [Google Scholar]
- 13.Hughes, A. L., and M. Yeager. 1998. Natural selection at major histocompatibility complex loci of vertebrates. Annu. Rev. Genet. 32:415-435. [DOI] [PubMed] [Google Scholar]
- 14.Husain, Z., E. Levitan, C. E. Larsen, N. M. Mirza, S. Younes, E. J. Yunis, C. A. Alper, and D. P. Dubey. 2002. HLA-Cw7 zygosity affects the size of a subset of CD158b+ natural killer cells. J. Clin. Immunol. 22:28-36. [DOI] [PubMed] [Google Scholar]
- 15.Jeffery, K. J., A. A. Siddiqui, M. Bunce, A. L. Lloyd, A. M. Vine, A. D. Witkover, S. Izumo, K. Usuku, K. I. Welsh, M. Osame, and C. R. Bangham. 2000. The influence of HLA class I alleles and heterozygosity on the outcome of human T cell lymphotropic virus type I infection. J. Immunol. 165:7278-7284. [DOI] [PubMed] [Google Scholar]
- 16.Khakoo, S. I., C. L. Thio, M. P. Martin, C. R. Brooks, X. Gao, J. Astemborski, J. Cheng, J. J. Goedert, D. Vlahov, M. Hilgartner, S. Cox, A. M. Little, G. J. Alexander, M. E. Cramp, S. J. O'Brien, W. M. Rosenberg, D. L. Thomas, and M. Carrington. 2004. HLA and NK cell inhibitory receptor genes in resolving hepatitis C virus infection. Science 305:872-874. [DOI] [PubMed] [Google Scholar]
- 17.Ksiazek, T. G., D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret, S. Emery, S. Tong, C. Urbani, J. A. Comer, W. Lim, P. E. Rollin, S. F. Dowell, A. E. Ling, C. D. Humphrey, W. J. Shieh, J. Guarner, C. D. Paddock, P. Rota, B. Fields, J. DeRisi, J. Y. Yang, N. Cox, J. M. Hughes, J. W. LeDuc, W. J. Bellini, and L. J. Anderson. 2003. A novel coronavirus associated with severe acute respiratory syndrome. N. Engl. J. Med. 348:1953-1966. [DOI] [PubMed] [Google Scholar]
- 18.Lan, Y. C., H. F. Liu, Y. P. Shih, J. Y. Yang, H. Y. Chen, and Y. M. Chen. 2005. Phylogenetic analysis and sequence comparisons of structural and non-structural SARS coronavirus proteins in Taiwan. Infect. Genet. Evol. 5:261-269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lan, Y. C., T. T. Liu, J. Y. Yang, C. M. Lee, Y. J. Chen, Y. J. Chan, J. J. Lu, H. F. Liu, C. A. Hsuing, M. S. Ho, K. J. Hsiao, H. Y. Chen, and Y. M. A. Chen. 2005. Molecular epidemiology of severe acute respiratory syndrome-associated coronavirus infections in Taiwan. J. Infect. Dis. 191:1478-1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee, M. L., C. J. Chen, and I. J. Su. 2003. Severe acute respiratory syndrome—Taiwan, 2003. Morb. Mortal. Wkly. Rep. 52:461-466. [PubMed] [Google Scholar]
- 21.Leung, G. M., P. H. Chung, T. Tsang, W. Lim, S. K. Chan, P. Chau, C. A. Donnelly, A. C. Ghani, C. Fraser, S. Riley, N. M. Ferguson, R. M. Anderson, Y. L. Law, T. Mok, T. Ng, A. Fu, P. Y. Leung, J. S. Peiris, T. H. Lam, and A. J. Hedley. 2004. SARS-CoV antibody prevalence in all Hong Kong patient contacts. Emerg. Infect. Dis. 10:1653-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li, G., X. Chen, and A. Xu. 2003. Profile of specific antibodies to the SARS-associated coronavirus. N. Engl. J. Med. 349:508-509. [DOI] [PubMed] [Google Scholar]
- 23.Lin, M., H. K. Tseng, J. A. Trejaut, H. L. Lee, J. H. Loo, C. C. Chu, P. J. Chen, Y. W. Su, K. H. Lim, Z. U. Tsai, R. Y. Lin, R. S. Lin, and C. H. Huang. 2003. Association of HLA class I with severe acute respiratory syndrome coronavirus infection. BMC Med. Genet. 4:9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ljunggren, H. G., and K. Karre. 1990. In search of the ‘missing self’: MHC molecules and NK cell recognition. Immunol. Today 11:237-244. [DOI] [PubMed] [Google Scholar]
- 25.Machulla, H. K., J. Stein, A. Gautsch, J. Langner, H. G. Schaller, and S. Reichert. 2002. HLA-A, B, Cw, DRB1, DRB3/4/5, DQB1 in German patients suffering from rapidly progressive periodontitis (RPP) and adult periodontitis (AP). J. Clin. Periodontol. 29:573-579. [DOI] [PubMed] [Google Scholar]
- 26.National Research Project for SARS, Beijing Group. 2004. The involvement of natural killer cells in the pathogenesis of severe acute respiratory syndrome. Am. J. Clin. Pathol. 121:507-511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ng, M. H., K. M. Lau, L. Li, S. H. Cheng, W. Y. Chan, P. K. Hui, B. Zee, C. B. Leung, and J. J. Sung. 2004. Association of human-leukocyte-antigen class I (B*0703) and class II (DRB1*0301) genotypes with susceptibility and resistance to the development of severe acute respiratory syndrome. J. Infect. Dis. 190:515-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Peiris, J. S., C. M. Chu, V. C. Cheng, K. S. Chan, I. F. Hung, L. L. Poon, K. I. Law, B. S. Tang, T. Y. Hon, C. S. Chan, K. H. Chan, J. S. Ng, B. J. Zheng, W. L. Ng, R. W. Lai, Y. Guan, and K. Y. Yuen. 2003. Clinical progression and viral load in a community outbreak of coronavirus-associated SARS pneumonia: a prospective study. Lancet 361:1767-1772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peiris, J. S., S. T. Lai, L. L. Poon, Y. Guan, L. Y. Yam, W. Lim, J. Nicholls, W. K. Yee, W. W. Yan, M. T. Cheung, V. C. Cheng, K. H. Chan, D. N. Tsang, R. W. Yung, T. K. Ng, and K. Y. Yuen. 2003. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet 361:1319-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Segal, S., and A. V. Hill. 2003. Genetic susceptibility to infectious disease. Trends Microbiol. 11:445-448. [DOI] [PubMed] [Google Scholar]
- 31.Thio, C. L., X. Gao, J. J. Goedert, D. Vlahov, K. E. Nelson, M. W. Hilgartner, S. J. O'Brien, P. Karacki, J. Astemborski, M. Carrington, and D. L. Thomas. 2002. HLA-Cw*04 and hepatitis C virus persistence. J. Virol. 76:4792-4797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsang, K. W., P. L. Ho, G. C. Ooi, W. K. Yee, T. Wang, M. Chan-Yeung, W. K. Lam, W. H. Seto, L. Y. Yam, T. M. Cheung, P. C. Wong, B. Lam, M. S. Ip, J. Chan, K. Y. Yuen, and K. N. Lai. 2003. A cluster of cases of severe acute respiratory syndrome in Hong Kong. N. Engl. J. Med. 348:1977-1985. [DOI] [PubMed] [Google Scholar]
- 33.Valiante, N. M., M. Uhrberg, H. G. Shilling, K. Lienert-Weidenbach, K. L. Arnett, A. D'Andrea, J. H. Phillips, L. L. Lanier, and P. Parham. 1997. Functionally and structurally distinct NK cell receptor repertoires in the peripheral blood of two human donors. Immunity 7:739-751. [DOI] [PubMed] [Google Scholar]
- 34.Woo, P. C., S. K. Lau, H. W. Tsoi, K. H. Chan, B. H. Wong, X. Y. Che, V. K. Tam, S. C. Tam, V. C. Cheng, I. F. Hung, S. S. Wong, B. J. Zheng, Y. Guan, and K. Y. Yuen. 2004. Relative rates of non-pneumonic SARS coronavirus infection and SARS coronavirus pneumonia. Lancet 363:841-845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zinkernagel, R. M. 1996. Immunology taught by viruses. Science 271:173-178. [DOI] [PubMed] [Google Scholar]