Abstract
Little information is available about the genetic variability of Leishmania populations and the possible correlations with ecoepidemiological features of leishmaniases. The present study was carried out in French Guiana, a country where cutaneous leishmaniases (CL) are endemic over the whole territory. The genetic polymorphism of a nuclear sequence encompassing the end of the ribosomal small subunit and the internal transcribed spacer 1 of 265 isolates from patients with CL was examined by restriction fragment length polymorphism analysis. Genotypes based on the fingerprinting phenetic integration were compared to epidemiological, clinical, and geographical data. In agreement with previous reports, five different Leishmania species were identified, but Leishmania (Viannia) guyanensis represented 95.8% of the samples. Two distinct L. (V.) guyanensis populations were found to originate in two ecologically characterized regions. Higher lesional parasite densities and the need for additional treatments were significantly linked to genotype group I. Parasites of genotype group II were more likely to cause chronic and disseminated cutaneous forms in patients. L. (V.) guyanensis was previously said not to be very polymorphic; however, the present analysis resulted in a significant degree of discrimination among L. (V.) guyanensis isolates from diverse ecological areas and with different clinical implications.
In the highly biodiversified region of the New World (NW) that encompasses the northern Amazon Basin and the Guiana Shield, at least seven named species belonging to the Leishmania genus coexist with often overlapping hosts and vectors (39). American visceral leishmaniases due to Leishmania (Leishmania) infantum are found there, but cases of cutaneous leishmaniases (CL) are far more numerous. Human cutaneous infections may be unapparent; however, many of the Leishmania parasites are able to produce a spectrum of diseases rather than a single clinical form. Cases reported in that region range from localized and diffuse cutaneous forms to mucocutaneous leishmaniases. They are now being reported in areas where leishmaniases had previously not been endemic (2, 17): increasing risk factors related to natural and especially man-made environmental changes are making leishmaniases a growing public health concern in this particular region. However, little information is available about the genetic variability of parasite populations and the possible correlation with ecoepidemiological features of the diseases. The present study was carried out in French Guiana, a country where CL are endemic over the whole territory (incidence of 0.2% between 1979 and 2000 [6, 14]). The genetic polymorphism of a nuclear sequence encompassing the end of the ribosomal small subunit (SSU) and the internal transcribed spacer 1 (ITS1) of 265 samples from patients with CL was examined by restriction fragment length polymorphism (RFLP) analysis. Genotypes based on the fingerprinting phenetic integration were then compared to epidemiological, clinical, and geographical data.
MATERIALS AND METHODS
Patients.
A total of 265 patients positive for CL that have consulted the Dermatology Department of Cayenne General Hospital (Cayenne, French Guiana) and/or the health centers of French Guiana in 2003 and 2004 were included in this study. The inclusion of a confirmed case of CL was based on the following: (i) the presence of cutaneous lesions with 2 or more weeks of evolution and a compatible epidemiological history, (ii) no specific treatment before consultation, and (iii) a positive result by at least one of the two methods used for routine diagnosis, microscopic examination of dermal scraping smears and/or in vitro cultivation. All individuals enrolled in this study provided informed consent. The supposed geographical origin of infection was assessed for each patient both by the health center where the sample was taken and by the patient interview. Strict biopsy protocol and treatment schemes were applied for all patients included in this study. Data were centralized, and cases were monitored in the Parasitology Department of Cayenne General Hospital (Cayenne, French Guiana). Table 1 summarizes the clinical data of the 265 patients enrolled in this study.
TABLE 1.
Clinical characteristics of 265 patients with cutaneous leishmaniasis
| Characteristic | Value |
|---|---|
| Patients | |
| Sex (% male) | 79.5 |
| Mean age (yr) (SD) | 32.9 (13.5) |
| Median duration of the disease (mo) (range) | 1 (0.25-96) |
| Atypical clinical forms (%)a | 3 |
| Lesions | |
| Median no. (range) | 1 (1-61) |
| Median parasite density score (range)b | 4 (1-5) |
| Ulcerated lesion (%) | 92.9 |
| Nodular lesion (%) | 15.8 |
| Satellite papules (%) | 5.6 |
| Lesion location | |
| Arms (%) | 49.3 |
| Legs (%) | 45.4 |
| Trunk (%) | 18 |
| Head (%) | 18.9 |
| Treatment | |
| 1 pentamidine injection (%) | 66.3 |
| 2 pentamidine injections (%) | 30.3 |
| 3 pentamidine injections (%) | 2.2 |
| 4 pentamidine injections (%) | 0 |
| 5 pentamidine injections (%) | 1.1 |
| Additional pentavalent antimonial courses (%) | 16.3 |
| Additional fluconazole oral cures (%) | 2.2 |
Atypical clinical forms are disseminated cutaneous leishmaniases and chronic presentations.
Parasite density scores were estimated by direct microscopic examination and scored as follows. Parasite density scores were 1 (rare [≤1 parasite every 20 fields]), 2 (few [≥1 parasite every 20 fields]), 3 (quite numerous [≥1 parasite every 5 fields]), 4 (numerous [1 to 5 parasites/field]), and 5 (very numerous [>5 parasites/field]).
Clinical samples and standard diagnostic procedure.
Tissue scrapings for microscopic smear examination were collected by slitting the internal border of skin lesions with a surgical blade, and the tissue scraping was smeared onto a clean glass slide, fixed with methanol, and stained in Giemsa. For each patient, two slides were read by trained parasitologists; if the slides were positive, they were given a parasite density score from 1 (rare [less than one parasite every 20 fields]) to 5 (very numerous [more than five parasites/field]). Punch skin biopsy specimens of 4 mm were taken in the internal border of lesions under sterile conditions and local anesthesia (lidocaine). In vitro cultivations were performed by inoculation of sterilely crushed biopsy specimens in 3 ml of RPMI 1640 (Sigma) supplemented with 20% fetal calf serum, 1% nonessential amino acids, and 50 IU/ml penicillin. Culture flasks were incubated at 24°C in the dark and microscopically observed every 3 days for 4 weeks.
DNA extraction and PCR amplification.
Only a single round of cultivation separated isolation from DNA extraction. For each isolate, DNA extraction was conducted from cultured biopsy medium by a simple salting-out procedure as described elsewhere (40). PCR control DNAs were extracted from Leishmania (Viannia) guyanensis MHOM/GF/2003/LBC40, L. (V.) braziliensis MHOM/BR/75/M2903, L. (V.) naiffi MHOM/GF/97/CRE88, L. (V.) lainsoni IUBI/BR/00/M12025, and Leishmania (Leishmania) amazonensis MHOM/BR/73/M2269. Negative controls were also used in each run, and all samples were tested at least twice. The 1,200-bp sequences located between the end of the ribosomal SSU and the 5.8S region were amplified as described elsewhere (41). The primers SSU-12103-D (5′-GGGAATATCCTCAGCACGT-3′) and 5.8S-13333-R (5′-CGACACTGAGAATATGGCATG-3′) were used.
Restriction analysis and control sequencing.
Constant amounts of amplified DNA (approximately 100 ng) were then loaded and digested for 2 h with restriction enzymes according to the manufacturer's recommendations. Seven restriction enzymes were tested: TaqI, HaeIII, DdeI, PstI, and HpaII (Sigma) and RsaI and BfaI (New England BioLabs). Digestion products were separated for 2 h by simple electrophoresis in 2% agarose gels stained with ethidium bromide. Sequencing of the end of the RNA polymerase II large-subunit gene (330 bp) (8) was also performed to confirm the PCR-RFLP identification of 42 samples characteristic of each restriction pattern.
Numerical analyses.
PCR-RFLP gel scans were analyzed with the LabImage 2.7.1 software (Kapelan GmbH). A character matrix was then created by reporting all possible fragments obtained with each enzyme in the samples studied. Then, for each sample, the presence or absence of bands was given a score of 1 or 0, respectively. In a given RFLP profile, differences of intensity among fragments were not taken into account. These differences are common in tandemly repeated genes consisting of sequence variants present in different copy numbers (24, 53). The binary matrix was then processed for phenetic analyses, with the following programs of the PHYLIP version 3.63 package (Joseph Felsenstein, University of Washington, Seattle, Wash.) (July 2004 version): RESTDIST (restriction fragment distance method modified by the Nei and Li distance method [32]) followed by UPGMA (unweighted pair group method with arithmetic averages). A condensed tree was drawn using the Molecular Evolutionary Genetics Analysis version 2 (MEGA2) software (27). Statistical analyses were performed by the Intercooled Stata 8.2 version software (StataCorp LP). Qualitative binary data (genotype, gender, atypical clinical form, presence of ulcer, presence of nodule, presence of scab, presence of papule, presence of lymphangitis, lesion on the inferior parts, lesion on the superior parts, lesion on the head, lesion on the trunk, additional fluconazole cure, additional pentavalent antimonial cure, treatment different from one pentamidine injection), qualitative ordinal data (parasite density score), qualitative categorical information (supposed origin of infection), and quantitative values (number of lesions, number of pentamidine injections, age, and lesion duration) were successively analyzed by Pearson chi-square tests.
RESULTS
PCR on cultured biopsy specimens gave negative results in 49 (18.5%) of the 265 samples tested, most of these samples were positive by direct microscopic examination but negative by cultivation. Moreover, 32 samples (12.1%) were identified but not genotyped due to their low DNA levels. Species identification was based on comparison to the control strain profiles by checking the presence or absence of specific bands (41). In total, five different Leishmania species were identified from 216 samples: Leishmania (Viannia) guyanensis represented 95.8% of the samples, four L. Leishmania (Leishmania) amazonensis isolates (1.9%) were also found, as well as three L. (V.) braziliensis isolates (1.4%), one L. (V.) lainsoni isolate (0.5%), and one L. (V.) naiffi isolate (0.5%). Control sequencing of the RNA polymerase II gene (8) confirmed the PCR-RFLP identification of 42 samples characteristic of each restriction pattern. Finally, 175 L. (V.) guyanensis samples were genotyped with seven restriction enzymes.
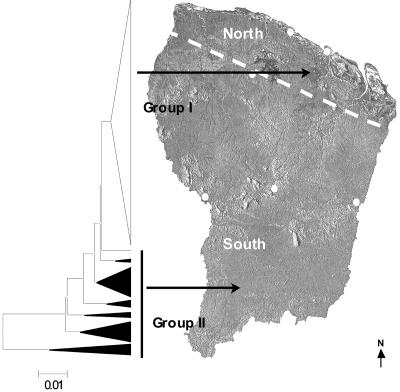
As currently observed, PCR-RFLP profiles were relatively complex and polymorphic. All amplification patterns were reproducible. Each of the seven enzymes tested showed different levels of variability depending on the sample tested. The unrooted UPGMA molecular tree presented in Fig. 1 depicts relationships between the 175 L. (V.) guyanensis samples. The Nei genetic distances between L. (V.) guyanensis samples ranged from 0.0005 to 1.2600. This molecular epidemiological tree distinguished two genotype groups forming more or less tight clusters. Genotype group I included 133 homogeneous samples (76.0%) that were closely related (with branch lengths no longer than 0.0074), whereas genotype group II, which was much more heterogeneous, was characterized by a smaller population of 42 polymorphic samples (24.0%) which were sometimes highly distant (branch length > 0.01).
FIG. 1.
Correlation between the UPGMA tree based on the PCR-RFLP genotype analyses of 175 Leishmania (Viannia) guyanensis samples and the supposed geographical origin of patient infection. A condensed unrooted UPGMA tree built with the PCR-RFLP data (seven restriction enzymes tested) of 175 samples is presented in the left part of the figure. The triangle width and height are commensurate with the divergence of the isolates and the number of samples, respectively. Samples were grouped according to their genotype: 133 closely related strains (branch length < 0.01) were included in genotype group I (white triangle), and 42 samples showing polymorphic heterogeneous patterns (branch length > 0.01) were included in genotype group II (black triangles). Sample origins are indicated by arrows pointing to the map of French Guiana. These links were supported by a Pearson chi-square test (P < 0.01).
When examining possible associations of the genotypic groups with epidemiological, clinical, and geographical data, several links could be observed (Table 2). All the L. (V.) braziliensis, L. (V.) lainsoni, and L. (V.) naiffi samples originated from the northern coastal part of French Guiana, whereas the four L. (L.) amazonensis isolates came from the southern part of the territory. Overall, L. (V.) guyanensis samples of genotype group I seemed to have originated in the northern part of French Guiana, whereas the samples of genotype group II seemed to have originated in the southern part of the country, from Suriname, and from the neighboring Brazilian states (Fig. 1). This link was supported by a Pearson chi-square test [P (>χ2) = 0.007]. Patients belonging to genotype group I were likely to present higher parasite density [P (>χ2) = 0.008] than patients belonging to genotype group II. Moreover, there were more patients from genotype group I who needed additional treatments (i.e., more than one intramuscular pentamidine injection) before they were cured (36.7%) compared to patients from genotype group II (8.3%) [P (>χ2) = 0.055]. Atypical clinical forms, such as disseminated presentations and chronic diseases, were mostly found in genotype group II [P (>χ2) = 0.056]. However, no clear correlation was observed between the genotype and number of lesions, duration of the disease, gender, age, localization and aspect of lesions, and associated clinical features, such as satellite papules or lymphangitis.
TABLE 2.
L. (V.) guyanensis genotype correlations with geographical, clinical, and epidemiological dataa
| Genotype | Population (%) | Supposed geographical zone of infection (%) | Parasite density score of >2 (%)b | Need for additional treatments (%)c | Atypical clinical forms (%)d |
|---|---|---|---|---|---|
| Group I | 76.0 | North (62.6) | 74.7 | 36.7 | 1.5 |
| Group II | 24.0 | South (60) | 48.1 | 8.3 | 7.1 |
| χ2 test probability | P = 0.007 | P = 0.008 | P = 0.055 | P = 0.056 |
The genotype groups of the 175 L. (V.) guyanensis samples studied and the geographical zones are defined in Results and in Fig. 1.
Parasite density scores were estimated by direct microscopic examination and scored as follows. Parasite density scores were 1 (rare [<1 parasite every 20 fields]), 2 (few [≥1 parasite every 20 fields]), 3 (quite numerous [≥1 parasite every 5 fields]), 4 (numerous [1 to 5 parasites/field]), and 5 (very numerous [>5 parasites/field]).
In patients with nonchronic typical localized forms, treatments used in addition to one pentamidine intramuscular injection are other pentamidine intramuscular injections, intralesional pentavalent antimonial courses, and/or fluconazole oral cures.
Atypical clinical forms are disseminated cutaneous leishmaniases and chronic presentations.
DISCUSSION
The different Leishmania species prevalence were in agreement with previous studies (4, 14, 18; reviewed in reference 39), showing a relative stability of the different pathogenic complex in French Guiana during the last 20 years. Nevertheless, considering the important man-made environmental changes and associated vector population adaptations (B. Rotureau et al., submitted), the incidence of the most clinically relevant species, Leishmania (Leishmania) amazonensis and Leishmania (Viannia) braziliensis, should be checked regularly in the future.
Linking the polymorphism of Leishmania samples with clinical outcomes has not always been evident in the past (19, 37, 44, 47, 52). As highlighted by Schriefer et al. (48), the wide geographic distribution and multiple sources (e.g., vectors, reservoirs, and human immunodeficiency virus-positive human hosts) of the isolates tested may be among the reasons for this lack of association. However, several studies using different methods have demonstrated at least a geographic structuring of Leishmania populations. Geographic structuring was demonstrated in L. (V.) braziliensis by randomly amplified polymorphic DNA (RAPD) analysis (48), RFLP analysis (21), multilocus enzyme electrophoresis (MLEE) (42), and serology (43); in L. (L.) infantum by MLEE (35), RAPD analysis (52, 54), and analysis with other molecular markers (22); in L. (V.) peruviana by RAPD analysis (3); in L. (L.) aethiopica by RFLP (44); and in L. (L.) donovani by sequencing (26, 28) and RFLP (51). Among Viannia parasites, reports have indicated that genetic variations were extensive, with some clones widely distributed and others localized to a particular endemic focus (13). Other results have suggested that their distribution was related to the origin of the gene pool as well as to present vector and reservoir movements (25). The present RFLP analysis resulted in a significant degree of discrimination among isolates from diverse regions, confirming the ability of this technique to discriminate between closely related parasites. Two main L. (V.) guyanensis genotype pools were distinguished by phenetic analysis and associated with two distinct geographical regions. L. (V.) guyanensis genotype group II originated in southern French Guiana and neighboring areas in Brazil and Suriname. These regions are characterized by a continuous dense primary rain forest spotted with gold-digging sites. Most of the patients were infected during occupational activities as reported by Dedet et al. (15). L. (V.) guyanensis genotype group I originated in the northern part of the country. As along the whole northern part of the Guiana Shield, this costal region is typically divided in distinct ecotopes: beaches, mangroves, coastal swamps, and rain forests. This area is heavily settled by humans, and most of the sylvatic ecotopes are secondary. Infections in this geographic area likely occurred principally during occupational or leisure activities (15). These two distinct L. (V.) guyanensis populations observed by ribosomal fingerprinting may have originated in the two different ecologically characterized regions. Thus, they may represent at least two nonsympatric lineages of this parasite. As it was proposed by Cupolillo (9, 11, 13), the polymorphism observed could be related to the great number of sand fly vectors and animal reservoir hosts of the rain forests, especially in the southern part of the country. This particular Leishmania species is said not to be very polymorphic; however, to our knowledge, no large-scale study on L. (V.) guyanensis populations has been conducted before. Cupolillo et al. (12, 13) did not find much intraspecific polymorphism in L. (V.) guyanensis isolates compared to L. (V.) braziliensis samples. This was probably due to the smaller number of samples studied. As suggested by Cupolillo for L. (V.) braziliensis (10), the distinct genotypic groups described herein within 175 L. (V.) guyanensis samples could explain the plasticity of these parasites and their ability to adapt to changing ecological conditions, especially to the man-made environmental changes happening in French Guiana. Entomological and zoological data are required to confirm this hypothesis.
The possibility of mixed infections has been previously reported (23, 29, 31, 49). However, in the present study, we did not observe any hybrid profiles and control genotyping did not detect any multiple infections. Biopsy and treatment protocols were identical at all the health centers, and dermal smear parasite density scores were all estimated by the same trained parasitologists at the Cayenne General Hospital. A higher parasite density in lesions and the need for additional treatments were significantly linked to genotype group I. This suggests the presence of two distinct nonsympatric L. (V.) guyanensis populations with different clinical implications. Patients with atypical clinical presentations, i.e., chronic and disseminated cutaneous forms, were mostly found in genotype group II. The immunogenetic susceptibility of the host is likely to be partially responsible for these particular presentations, but atypical forms could also be related to the higher biological diversity of the virgin rain forest in the southern part of French Guiana, and thus, to a potentially higher parasitic cycle diversity. This result reinforces the existence of significant genetic diversity within L. (V.) guyanensis samples and the possible association of genotypes with specific transmission cycles. The identification of specific patterns for L. (V.) guyanensis subpopulations also enables detection of unfavorable outcomes and proper adjustment of therapy.
The degree of polymorphism within each species varied according to the genetic markers studied. Because of the high level of polymorphism in the composition of the ITS locus, several molecular epidemiological studies have been conducted by RFLP analysis in New World taxa (5, 10, 21, 38), but most studies have been performed on Old World Leishmania species (7, 19, 34, 44-47). Analyses of the ribosomal ITS regions of different parasites have demonstrated the utility of this marker in detecting genetic diversity and any association of this diversity with several epidemiological aspects (16, 33). The mechanisms that control the expression of genes in the Leishmania genus remain unclear. In the absence of the characteristic eukaryotic modulation of primary transcription via individual promoter activity, Leishmania organisms rely on posttranscriptional (mostly trans-splicing and polyadenylation) or posttranslational mechanisms to generate the appropriate levels of gene products within the cell (reviewed in reference 50). Multicopy tandemly repeated genes are characterized by a high degree of conservation in peptide sequence, although there is considerable divergence in the 5′ and 3′ untranslated regions (UTRs) of each gene transcript, a feature perhaps related to differences in their regulation (1). Posttranscriptional regulation of transcript levels appeared to be dependent on 3′ UTR sequences (via their effects on mRNA processing or by binding of stability/degradation factors) (20, 30, 36). Although UTR and ITS regions present functional differences, such as transcription by distinct polymerases, one could hypothesize that the rRNA ITS region may also have a role in ribosomal gene expression itself. It is unlikely that the ITS sequences are directly genetically linked to the pathogenic mechanisms, due to the continuum of variation observed in this locus. These variations are undoubtedly linked to the divergence among the isolates. Nevertheless, the important genetic polymorphism, statistically linked to various geographical, epidemiological, and clinical parameters evokes a much more complex potential functional role of this particular locus. However, this is conjecture, and further studies will be required to clarify this point.
L. (V.) guyanensis was previously said not to be very polymorphic; however, the present RFLP analysis on a ribosomal sequence resulted in a significant degree of discrimination among L. (V.) guyanensis isolates from diverse ecological areas and with different clinical implications.
Acknowledgments
This work was supported by the University of the French West Indies and French Guiana, by the Contrat Plan Etat-Region no. 2365, by the Institut National de la Sante et de la Recherche Medicale (INSERM, Paris, France), the Centre National pour la Recherche Scientifique (CNRS, Paris, France), and the University of Montpellier 1 (Montpellier, France).
REFERENCES
- 1.Aly, R., M. Argaman, S. Halman, and M. Shapira. 1994. A regulatory role for the 5′ and 3′ untranslated regions in differential expression of hsp83 in Leishmania. Nucleic Acids Res. 22:2922-2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ashford, R. W. 2000. The leishmaniases as emerging and reemerging zoonoses. Int. J. Parasitol. 30:1269-1281. [DOI] [PubMed] [Google Scholar]
- 3.Banuls, A. L., J. C. Dujardin, F. Guerrini, S. De Doncker, D. Jacquet, J. Arevalo, S. Noel, D. Le Ray, and M. Tibayrenc. 2000. Is Leishmania (Viannia) peruviana a distinct species? A MLEE/RAPD evolutionary genetics answer. J. Eukaryot. Microbiol. 47:197-207. [DOI] [PubMed] [Google Scholar]
- 4.Basset, D., F. Pratlong, C. Ravel, J. Puechberty, J. Dereure, and J. Dedet. 2001. Les leishmanioses déclarées en France en 1999. Bull. Épidémiol. Hebd. 5:19-20. [Google Scholar]
- 5.Berzunza-Cruz, M., N. Cabrera, M. Crippa-Rossi, T. Sosa Cabrera, R. Perez-Montfort, and I. Becker. 2002. Polymorphism analysis of the internal transcribed spacer and small subunit of ribosomal RNA genes of Leishmania mexicana. Parasitol. Res. 88:918-925. [DOI] [PubMed] [Google Scholar]
- 6.Carme, B., C. Aznar, and R. Pradinaud. 2001. Absence of a proven resurgence of Chagas disease or cutaneous leishmaniasis in French Guiana over the last two decades. Ann. Trop. Med. Parasitol. 95:623-625. [DOI] [PubMed] [Google Scholar]
- 7.Chicharro, C., M. A. Morales, T. Serra, M. Ares, A. Salas, and J. Alvar. 2002. Molecular epidemiology of Leishmania infantum on the island of Majorca: a comparison of phenotypic and genotypic tools. Trans. R. Soc. Trop. Med. Hyg. 96(Suppl. 1):S93-S99. [DOI] [PubMed] [Google Scholar]
- 8.Croan, D. G., D. A. Morrison, and J. T. Ellis. 1997. Evolution of the genus Leishmania revealed by comparison of DNA and RNA polymerase gene sequences. Mol. Biochem. Parasitol. 89:149-159. [DOI] [PubMed] [Google Scholar]
- 9.Cupolillo, E., F. Aguiar Alves, L. R. Brahim, M. F. Naiff, L. O. Pereira, M. P. Oliveira-Neto, A. Falqueto, and G. Grimaldi, Jr. 2001. Recent advances in the taxonomy of the New World leishmanial parasites. Med. Microbiol. Immunol. (Berlin) 190:57-60. [DOI] [PubMed] [Google Scholar]
- 10.Cupolillo, E., L. R. Brahim, C. B. Toaldo, M. P. de Oliveira-Neto, M. E. de Brito, A. Falqueto, M. de Farias Naiff, and G. Grimaldi, Jr. 2003. Genetic polymorphism and molecular epidemiology of Leishmania (Viannia) braziliensis from different hosts and geographic areas in Brazil. J. Clin. Microbiol. 41:3126-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cupolillo, E., G. Grimaldi, Jr., and H. Momen. 1997. Genetic diversity among Leishmania (Viannia) parasites. Ann. Trop. Med. Parasitol. 91:617-626. [DOI] [PubMed] [Google Scholar]
- 12.Cupolillo, E., G. Grimaldi, Jr., H. Momen, and S. M. Beverley. 1995. Intergenic region typing (IRT): a rapid molecular approach to the characterization and evolution of Leishmania. Mol. Biochem. Parasitol. 73:145-155. [DOI] [PubMed] [Google Scholar]
- 13.Cupolillo, E., H. Momen, and G. Grimaldi, Jr. 1998. Genetic diversity in natural populations of New World Leishmania. Mem. Inst. Oswaldo Cruz 93:663-668. [DOI] [PubMed] [Google Scholar]
- 14.Dedet, J. P. 1990. Cutaneous leishmaniasis in French Guiana: a review. Am. J. Trop. Med. Hyg. 43:25-28. [DOI] [PubMed] [Google Scholar]
- 15.Dedet, J. P., R. Pradinaud, and F. Gay. 1989. Epidemiological aspects of human cutaneous leishmaniasis in French Guiana. Trans. R. Soc. Trop. Med. Hyg. 83:616-620. [DOI] [PubMed] [Google Scholar]
- 16.Dengjel, B., M. Zahler, W. Hermanns, K. Heinritzi, T. Spillmann, A. Thomschke, T. Loscher, R. Gothe, and H. Rinder. 2001. Zoonotic potential of Enterocytozoon bieneusi. J. Clin. Microbiol. 39:4495-4499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Desjeux, P. 2004. Leishmaniasis: current situation and new perspectives. Comp. Immunol. Microbiol. Infect. Dis. 27:305-318. [DOI] [PubMed] [Google Scholar]
- 18.Desjeux, P., and J. P. Dedet. 1989. Isoenzyme characterization of 112 Leishmania isolates from French Guiana. Trans. R. Soc. Trop. Med. Hyg. 83:610-612. [DOI] [PubMed] [Google Scholar]
- 19.El Tai, N. O., M. El Fari, I. Mauricio, M. A. Miles, L. Oskam, S. H. El Safi, W. H. Presber, and G. Schonian. 2001. Leishmania donovani: intraspecific polymorphisms of Sudanese isolates revealed by PCR-based analyses and DNA sequencing. Exp. Parasitol. 97:35-44. [DOI] [PubMed] [Google Scholar]
- 20.Folgueira, C., L. Quijada, M. Soto, D. R. Abanades, C. Alonso, and J. M. Requena. 2005. The translational efficiencies of the two Leishmania infantum HSP70 mRNAs, differing in their 3′-untranslated regions, are affected by shifts in the temperature of growth through different mechanisms. J. Biol. Chem. 280:35172-35183. [DOI] [PubMed] [Google Scholar]
- 21.Garcia, A. L., A. Kindt, K. W. Quispe-Tintaya, H. Bermudez, A. Llanos, J. Arevalo, A. L. Banuls, S. De Doncker, D. Le Ray, and J. C. Dujardin. 2005. American tegumentary leishmaniasis: antigen-gene polymorphism, taxonomy and clinical pleomorphism. Infect. Genet. Evol. 5:109-116. [DOI] [PubMed] [Google Scholar]
- 22.Guerbouj, S., I. Guizani, N. Speybroeck, D. Le Ray, and J. C. Dujardin. 2001. Genomic polymorphism of Leishmania infantum: a relationship with clinical pleomorphism? Infect. Genet. Evol. 1:49-59. [DOI] [PubMed] [Google Scholar]
- 23.Ibrahim, M. E., A. J. Smyth, M. H. Ali, D. C. Barker, and A. Kharazmi. 1994. The polymerase chain reaction can reveal the occurrence of naturally mixed infections with Leishmania parasites. Acta Trop. 57:327-332. [DOI] [PubMed] [Google Scholar]
- 24.Inga, R., S. De Doncker, J. Gomez, M. Lopez, R. Garcia, D. Le Ray, J. Arevalo, and J. C. Dujardin. 1998. Relation between variation in copy number of ribosomal RNA encoding genes and size of harbouring chromosomes in Leishmania of subgenus Viannia. Mol. Biochem. Parasitol. 92:219-228. [DOI] [PubMed] [Google Scholar]
- 25.Ishikawa, E. A., F. T. Silveira, A. L. Magalhaes, R. B. Guerra, Jr., M. N. Melo, R. Gomes, T. G. Silveira, and J. J. Shaw. 2002. Genetic variation in populations of Leishmania species in Brazil. Trans. R. Soc. Trop. Med. Hyg. 96(Suppl. 1):S111-S121. [DOI] [PubMed] [Google Scholar]
- 26.Kuhls, K., I. L. Mauricio, F. Pratlong, W. Presber, and G. Schonian. 2005. Analysis of ribosomal DNA internal transcribed spacer sequences of the Leishmania donovani complex. Microbes Infect. 7:1224-1234. [DOI] [PubMed] [Google Scholar]
- 27.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 28.Lewin, S., G. Schonian, N. El Tai, L. Oskam, P. Bastien, and W. Presber. 2002. Strain typing in Leishmania donovani by using sequence-confirmed amplified region analysis. Int. J. Parasitol. 32:1267-1276. [DOI] [PubMed] [Google Scholar]
- 29.Martinez, E., S. Mollinedo, M. Torrez, M. Munoz, A. L. Banuls, and F. Le Pont. 2002. Co-infection by Leishmania amazonensis and L. infantum/L. chagasi in a case of diffuse cutaneous leishmaniasis in Bolivia. Trans. R. Soc. Trop. Med. Hyg. 96:529-532. [DOI] [PubMed] [Google Scholar]
- 30.McNicoll, F., M. Muller, S. Cloutier, N. Boilard, A. Rochette, M. Dube, and B. Papadopoulou. 2005. Distinct 3′-untranslated region elements regulate stage-specific mRNA accumulation and translation in Leishmania. J. Biol. Chem. 280:35238-35246. [DOI] [PubMed] [Google Scholar]
- 31.Mebrahtu, Y. B., P. G. Lawyer, L. D. Hendricks, R. Muigai, C. N. Oster, P. V. Perkins, D. K. Koech, H. Pamba, and C. R. Roberts. 1991. Concurrent infection with Leishmania donovani and Leishmania major in a Kenyan patient: clinical description and parasite characterization. Am. J. Trop. Med. Hyg. 45:290-296. [DOI] [PubMed] [Google Scholar]
- 32.Nei, M., and W. H. Li. 1979. Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc. Natl. Acad. Sci. USA 76:5269-5273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nimri, L. F., I. N. Moura, L. Huang, C. del Rio, D. Rimland, J. S. Duchin, E. M. Dotson, and C. B. Beard. 2002. Genetic diversity of Pneumocystis carinii f. sp. hominis based on variations in nucleotide sequences of internal transcribed spacers of rRNA genes. J. Clin. Microbiol. 40:1146-1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nimri, L. F., and H. D. Schallig. 2003. Application of riboprinting for the identification of isolates of cutaneous Leishmania spp. Parasitology 127:201-205. [DOI] [PubMed] [Google Scholar]
- 35.Pratlong, F., J. P. Dedet, P. Marty, M. Portus, M. Deniau, J. Dereure, P. Abranches, J. Reynes, A. Martini, M. Lefebvre, et al. 1995. Leishmania-human immunodeficiency virus coinfection in the Mediterranean basin: isoenzymatic characterization of 100 isolates of the Leishmania infantum complex. J. Infect. Dis. 172:323-326. [DOI] [PubMed] [Google Scholar]
- 36.Ramamoorthy, R., K. G. Swihart, J. J. McCoy, M. E. Wilson, and J. E. Donelson. 1995. Intergenic regions between tandem gp63 genes influence the differential expression of gp63 RNAs in Leishmania chagasi promastigotes. J. Biol. Chem. 270:12133-12139. [DOI] [PubMed] [Google Scholar]
- 37.Reiner, N. E., R. Lo, A. Llanos-Cuentas, H. Guerra, L. L. Button, and W. R. McMaster. 1989. Genetic heterogeneity in Peruvian Leishmania isolates. Am. J. Trop. Med. Hyg. 41:416-421. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez-Bonfante, C., R. Bonfante-Garrido, G. Grimaldi, Jr., H. Momen, and E. Cupolillo. 2003. Genotypically distinct Leishmania colombiensis isolates from Venezuela cause both cutaneous and visceral leishmaniasis in humans. Infect. Genet. Evol. 3:119-124. [DOI] [PubMed] [Google Scholar]
- 39.Rotureau, B. 2006. Ecology of the Leishmania species in the Guianan ecoregion complex. Am. J. Trop. Med. Hyg. 74:87-96. [PubMed] [Google Scholar]
- 40.Rotureau, B., A. Gego, and B. Carme. 2005. Trypanosomatid protozoa: a simplified DNA isolation procedure. Exp. Parasitol. 111:207-209. [DOI] [PubMed] [Google Scholar]
- 41.Rotureau, B., C. Ravel, P. Couppié, F. Pratlong, M. Nacher, J.-P. Dedet, and B. Carme. 2006. Use of PCR-restriction fragment length polymorphism analysis to identify the main New World Leishmania species and analyze their taxonomic properties and polymorphism by application of the assay to clinical samples. J. Clin. Microbiol. 44:459-467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saravia, N. G., I. Segura, A. F. Holguin, C. Santrich, L. Valderrama, and C. Ocampo. 1998. Epidemiologic, genetic, and clinical associations among phenotypically distinct populations of Leishmania (Viannia) in Colombia. Am. J. Trop. Med. Hyg. 59:86-94. [DOI] [PubMed] [Google Scholar]
- 43.Saravia, N. G., K. Weigle, C. Navas, I. Segura, L. Valderrama, A. Z. Valencia, B. Escorcia, and D. McMahon-Pratt. 2002. Heterogeneity, geographic distribution, and pathogenicity of serodemes of Leishmania viannia in Colombia. Am. J. Trop. Med. Hyg. 66:738-744. [DOI] [PubMed] [Google Scholar]
- 44.Schonian, G., H. Akuffo, S. Lewin, K. Maasho, S. Nylen, F. Pratlong, C. L. Eisenberger, L. F. Schnur, and W. Presber. 2000. Genetic variability within the species Leishmania aethiopica does not correlate with clinical variations of cutaneous leishmaniasis. Mol. Biochem. Parasitol. 106:239-248. [DOI] [PubMed] [Google Scholar]
- 45.Schonian, G., M. El Fari, S. Lewin, C. Schweynoch, and W. Presber. 2001. Molecular epidemiology and population genetics in Leishmania. Med. Microbiol. Immunol. (Berlin) 190:61-63. [DOI] [PubMed] [Google Scholar]
- 46.Schonian, G., A. Nasereddin, N. Dinse, C. Schweynoch, H. D. Schallig, W. Presber, and C. L. Jaffe. 2003. PCR diagnosis and characterization of Leishmania in local and imported clinical samples. Diagn. Microbiol. Infect. Dis. 47:349-358. [DOI] [PubMed] [Google Scholar]
- 47.Schonian, G., L. Schnur, M. el Fari, L. Oskam, A. A. Kolesnikov, W. Sokolowska-Kohler, and W. Presber. 2001. Genetic heterogeneity in the species Leishmania tropica revealed by different PCR-based methods. Trans. R. Soc. Trop. Med. Hyg. 95:217-224. [DOI] [PubMed] [Google Scholar]
- 48.Schriefer, A., A. L. Schriefer, A. Goes-Neto, L. H. Guimaraes, L. P. Carvalho, R. P. Almeida, P. R. Machado, H. A. Lessa, A. R. de Jesus, L. W. Riley, and E. M. Carvalho. 2004. Multiclonal Leishmania braziliensis population structure and its clinical implication in a region of endemicity for American tegumentary leishmaniasis. Infect. Immun. 72:508-514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Silveira, F. T., R. Lainson, J. J. Shaw, and R. S. Ribeiro. 1984. Cutaneous leishmaniasis in Amazonia. Report of the 1st human case of mixed infection, determined by 2 different Leishmania species: Leishmania brasiliensis and Leishmania mexicana amazonensis. Rev. Inst. Med. Trop. Sao Paulo 26:272-275. (In Portuguese.) [DOI] [PubMed] [Google Scholar]
- 50.Stiles, J. K., P. I. Hicock, P. H. Shah, and J. C. Meade. 1999. Genomic organization, transcription, splicing and gene regulation in Leishmania. Ann. Trop. Med. Parasitol. 93:781-807. [DOI] [PubMed] [Google Scholar]
- 51.Tintaya, K. W., X. Ying, J. P. Dedet, S. Rijal, X. De Bolle, and J. C. Dujardin. 2004. Antigen genes for molecular epidemiology of leishmaniasis: polymorphism of cysteine proteinase B and surface metalloprotease glycoprotein 63 in the Leishmania donovani complex. J. Infect. Dis. 189:1035-1043. [DOI] [PubMed] [Google Scholar]
- 52.Toledo, A., J. Martin-Sanchez, B. Pesson, C. Sanchiz-Marin, and F. Morillas-Marquez. 2002. Genetic variability within the species Leishmania infantum by RAPD. A lack of correlation with zymodeme structure. Mol. Biochem. Parasitol. 119:257-264. [DOI] [PubMed] [Google Scholar]
- 53.Victoir, K., A. L. Banuls, J. Arevalo, A. Llanos-Cuentas, R. Hamers, S. Noel, S. De Doncker, D. Le Ray, M. Tibayrenc, and J. C. Dujardin. 1998. The gp63 gene locus, a target for genetic characterization of Leishmania belonging to subgenus Viannia. Parasitology 117:1-13. [PubMed] [Google Scholar]
- 54.Zemanova, E., M. Jirku, I. L. Mauricio, M. A. Miles, and J. Lukes. 2004. Genetic polymorphism within the Leishmania donovani complex: correlation with geographic origin. Am. J. Trop. Med. Hyg. 70:613-617. [PubMed] [Google Scholar]