Abstract
We developed two colorimetric methods for the detection of vancomycin- and oxacillin-resistant Staphylococcus aureus in ≤6 h: (i) a nitrate reductase assay and (ii) a resazurin microplate method. MICs agreed with results obtained by CLSI methods for oxacillin. However, detection of vancomycin resistance required a larger inoculum. These methods may be recommended for the detection of vancomycin- and oxacillin-resistant S. aureus.
In a number of clinical microbiology laboratories performing routine work, the detection of methicillin-resistant Staphylococcus aureus (MRSA) is based on phenotypic assays such as disk diffusion and broth microdilution. These methods require at least 24 h to perform (10). Detection of the mecA gene or of PBP 2a is the “gold standard,” but these methods are not yet always available (1).
Recently, vancomycin-resistant and intermediate S. aureus (VISA) strains have been detected in many countries (4-6, 13). For these reasons, a rapid and reliable antibiotic susceptibility testing method for detection of such strains is needed (11). In this paper, we describe two rapid and colorimetric methods for early and accurate detection of vancomycin and oxacillin resistance in S. aureus.
We tested three vancomycin-resistant S. aureus isolates from Detroit, Hershey, and New York City (VRS1, VRS2, and VRS3, respectively) (4-6). Three VISA strains, NRS1, NRS12, and NRS17, were obtained from the Network on Antimicrobial Resistance in Staphylococcus aureus through Focus Technologies, Herndon, Va. In addition, 90 recent clinical isolates (31 oxacillin susceptible and 59 resistant, recently isolated from Ondokuz Mayis University Medical School, Clinical Microbiology Laboratory), as well as S. aureus ATCC 25923 as a control strain, were also tested. Oxacillin resistance was confirmed using the latex slide agglutination MRSA-Screen test (Oxoid, Ltd., United Kingdom).
Oxacillin, vancomycin, resazurin, sulfanilamide, and N-(1-naphthyl)ethylenediamine dihydrochloride were purchased from Sigma, Inc., St. Louis, Mo. In the resazurin microplate method (RMM) and reference method, susceptibility testing for vancomycin was performed in cation-adjusted Mueller-Hinton broth (CAMHB; Oxoid), and the test for oxacillin was performed in CAMHB with 2% NaCl (10). In the nitrate reductase assay (NRA), potassium nitrate (KNO3; final concentrations 1,000 μg/ml) was added to both media. The reference broth microdilution method was performed according to CLSI recommendations (10).
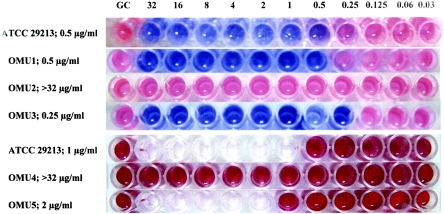
The RMM was performed in 96-well trays. Final drug concentrations were 32 to 0.03 μg/ml for both drugs. Aliquots of 100 μl of 1-McFarland-standard bacterial suspensions were inoculated in each well for vancomycin (final bacterial count, 0.5 × 108 CFU/ml), and 100 μl of 0.5-McFarland-standard bacterial suspensions were inoculated in each well for oxacillin (final bacterial count, 0.25 × 108 CFU/ml). Plates were incubated at 35°C. After 5 h of incubation, 30 μl of 0.02% resazurin solution was added to each well, and trays were reincubated for 1 h. The MIC was defined as the lowest concentration of the drug that prevented a change in color (Fig. 1).
FIG. 1.
MICs of oxacillin by the RMM (top four lines) and NRA (bottom three lines). OMU, Ondokus Magis University; GC, growth control.
In the NRA, CAMHB with 1,000 μg/ml potassium nitrate was used. All experiments were performed in the same way as the RMM. At the end of 5 h of incubation, 50 μl Griess reagent (one part 50% [vol/vol] concentrated hydrochloric acid, two parts 0.2% [wt/vol] sulfanilamide, and two parts 0.1% [wt/vol] N-(1-naphthyl)ethylenediamine dihydrochloride) was added in each well. After 5 min, a pink to red/purple color developed in the presence of growth (Fig. 1). The MIC was defined as the lowest drug concentration without color change. All tests were performed in duplicate. The results were compared by calculating essential agreement, absolute agreement, minor, major, and very major errors, as recommended by the FDA (8).
In this study, for oxacillin, essential agreement and absolute agreement were 86.5% and 100%, respectively, with RMM and 91.6% and 100%, respectively, with NRA (Tables 1 and 2). For vancomycin, essential agreement and absolute agreement were 81.2% and 98.9%, respectively, with RMM and 63.5% and 98.9%, respectively, with NRA. Minor errors were 1.04% in RMM and NRA, and there were no major or very major errors for vancomycin (Tables 1 and 2).
TABLE 1.
The errors and agreement between broth microdilution method and RMM and NRA for S. aureus
| Method and drug | No. (%) with errors
|
No. (%) with agreement
|
|||
|---|---|---|---|---|---|
| Minor | Major | Very major | Absolute | Essential | |
| RMM | |||||
| Oxacillin | 0 (0) | 0 (0) | 0 (0) | 100 (100) | 83 (86.5) |
| Vancomycin | 1 (1.04) | 0 (0) | 0 (0) | 95 (98.9) | 78 (81.2) |
| NRA | |||||
| Oxacillin | 0 (0) | 0 (0) | 0 (0) | 96 (100) | 88 (91.6) |
| Vancomycin | 1 (1.04) | 0 (0) | 0 (0) | 95 (98.9) | 61 (63.5) |
TABLE 2.
Results of agreement for oxacillin and vancomycin MIC results obtained by broth microdilution compared to RMM and NRAa
| Organism (no. tested) | Comparison | MIC variation in log 2 dilutions (%)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Oxacillin
|
Vancomycin
|
||||||||
| −1 | 0 | +1 | +2 | −1 | 0 | +1 | +2 | ||
| All S. aureus (96) | RMM | 1.0 | 72.9 | 12.6 | 13.5 | 1.0 | 22.9 | 57.3 | 18.8 |
| NRA | 0 | 79.1 | 12.5 | 8.4 | 1.0 | 9.4 | 53.2 | 36.4 | |
| MSSA (32) | RMM | 3.1 | 21.9 | 34.4 | 40.6 | 0 | 25 | 53.2 | 21.8 |
| NRA | 0 | 37.5 | 37.5 | 25 | 0 | 18.8 | 34.4 | 46.8 | |
| MRSA (64) | RMM | 0 | 98.4 | 1.6 | 0 | 1.5 | 21.9 | 59.4 | 17.2 |
| NRA | 0 | 100 | 0 | 0 | 1.5 | 4.7 | 62.6 | 31.2 | |
MSSA, methicillin-susceptible S. aureus.
Vancomycin MICs for VRS1 and VRS3 were >32 μg/ml and the MIC of VRS2 was 32 μg/ml, while in two colorimetric methods, the MICs obtained for VRS1, VRS2, and VRS3 were>32, 32, and 16, respectively. The MICs of three VISA strains were 8 μg/ml by the reference method and 16 μg/ml by the two colorimetric methods (Table 3).
TABLE 3.
Comparison of the MICs of VRSA and VISA strains by the reference methoda and two colorimetric methods
| Strain | MIC for vancomycin (μg/ml)
|
MIC for oxacillin (μg/ml)
|
||||
|---|---|---|---|---|---|---|
| Referencea | RMM | NRA | Reference | RMM | NRA | |
| ATCC 29213 | 1 | 2 | 2 | 0.25 | 1 | 1 |
| VRS1 | >32 | >32 | >32 | >32 | >32 | >32 |
| VRS2 | 32 | 16 | 16 | >32 | >32 | >32 |
| VRS3 | >32 | 32 | 32 | >32 | >32 | >32 |
| NRS1 | 8 | 16 | 16 | >32 | >32 | >32 |
| NRS12 | 8 | 16 | 16 | 0.25 | 1 | 0.25 |
| NRS17 | 8 | 16 | 16 | 4 | 8 | 4 |
Reference is broth microdilution method.
Early detection of methicillin resistance is an important parameter for choosing correct antibiotic therapy for infections caused by S. aureus. For this reason, in antibiotic susceptibility testing of these microorganisms, key antimicrobial agents should be oxacillin, penicillin, and vancomycin (2). Baker and Tenover (2) suggested that the Alamar colorimetric MIC method might be acceptable in antibiotic susceptibility testing for staphylococci and enterococci. They also commented on the difficulty of performing susceptibility testing, especially for some oxacillin-resistant coagulase-negative staphylococcal isolates, unless the inocula of all the agents and their incubation period were increased. In our study, we used a 0.5-McFarland-standard inoculum with the RMM and NRA for oxacillin.
In the absence of vancomycin pressure, vancomycin resistance is unstable and is expressed at a low level (3). This low-level expression of vancomycin resistance in S. aureus may be the reason these strains are difficult to detect clinically. In addition, it is known that automated systems such as MicroScan (Dade Behring) and Vitek or Vitek 2 (bioMérieux) fail to detect low-level vancomycin-resistant strains, especially if the vancomycin-resistant S. aureus (VRSA) has an MIC in the vicinity of 32 μg/ml (3, 12). In this study, we have also observed difficulties in the detection of resistant strains using smaller inocula in the colorimetric methods. When the larger inoculum was used (1 McFarland standard), VRS1 and VRS3 were identified as resistant but not VRS2, which was detected as intermediate. The inoculum did not affect the MICs of VISA strains in the two colorimetric methods.
It has been shown that agar dilution, agar screening, and single-point population analysis have low sensitivity and specificity for detection of low-level vancomycin resistance. The Etest (AB Biodisk, Solna, Sweden) with a normal inoculum (0.5 McFarland standard) on Mueller-Hinton agar or with a high inoculum (2 McFarland standard) on brain heart infusion agar has been found to have higher sensitivity and specificity for the detection of VISA (9). In our study, all three VISA strains tested were correctly detected by the two colorimetric methods.
An indicator similar to resazurin is used in the antibiotic susceptibility test panel of the Phoenix system developed by Becton Dickinson (7), but the system is expensive and can be used only in well-equipped laboratories.
Molecularly-based methods, such as the PBP 2a latex agglutination assay, are optimal for detection of oxacillin resistance. However, laboratories in developing countries (where cost is an important issue) may not have such methods available. In such cases, we feel that our two colorimetric methods may be useful. Both methods are, in our experience, sufficiently robust to allow for their use with clinical isolates. Automated methods are currently not recommended for routine detection of VISA or VRSA strains (3), but the possibility that our two methods may be adapted for this purpose cannot be excluded.
In summary, since oxacillin and vancomycin susceptibility results may be obtained in a shorter period of time by the RMM and NRA than by the standard methodology and results were in concordance with the standard method, we suggest that both methods may be acceptable and cheap, allowing for their routine use in clinical laboratories without access to commercial methodology. The validity of these two methods requires confirmation by testing of more oxacillin-resistant strains by more laboratories and (in time, as these strains are isolated clinically, as they surely will be) by testing of more VISA and VRSA strains.
REFERENCES
- 1.Arbique, J., K. Forward, D. Haldane, and R. Davidson. 2001. Comparison of the Velogene Rapid MRSA identification assay, Denka MRSA-Screen assay, and BBL Crystal MRSA ID system for rapid identification of methicillin-resistant Staphylococcus aureus. Diagn. Microbiol. Infect. Dis. 40:5-10. [DOI] [PubMed] [Google Scholar]
- 2.Baker, C. N., and F. C. Tenover. 1996. Evaluation of alamar colorimetric broth microdilution susceptibility testing method for staphylococci and enterococci. J. Clin. Microbiol. 34:2654-2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bozdogan, B., L. Ednie, K. Credito, K. Kosowska, and P. C. Appelbaum. 2004. Derivatives of a vancomycin-resistant Staphylococcus aureus strain isolated at Hershey Medical Center. Antimicrob. Agents Chemother. 48:4762-4765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Centers for Disease Control and Prevention. 2002. Staphylococcus aureus resistant to vancomycin—United States, 2002. Morb. Mortal. Wkly. Rep. 51:565-567. [PubMed] [Google Scholar]
- 5.Centers for Disease Control and Prevention. 2002. Public health dispatch: vancomycin-resistant Staphylococcus aureus—Pennsylvania, 2002. Morb. Mortal. Wkly. Rep. 51:902. [PubMed] [Google Scholar]
- 6.Centers for Disease Control and Prevention. 2004. Brief report: vancomycin-resistant Staphylococcus aureus—New York, 2004. Morb. Mortal. Wkly. Rep. 53:322-323. [PubMed] [Google Scholar]
- 7.Ferraro, M. J., and J. H. Jorgensen. 2003. Susceptibility testing instrumentation and computerized expert systems for data analysis and interpretation, p. 208-217. In P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller, and R. H. Yolken (ed.), Manual of clinical microbiology, 8th ed. American Society for Microbiology, Washington, D.C.
- 8.Food and Drug Administration. 2003. Class II special controls guidance document: antimicrobial susceptibility test (AST) systems; guidance for industry and FDA. [Online.] http://www.fda.gov/cdrh/ode/631.pdf.
- 9.Howden, B. P., P. B. Ward, P. D. R. Johnson, P. G. P. Charles, and M. L. Grayson. 2005. Low-level vancomycin resistance in Staphylococcus aureus—an Australian perspective. Eur. J. Clin. Microbiol. Infect. Dis. 24:100-108. [DOI] [PubMed] [Google Scholar]
- 10.National Committee for Clinical Laboratory Standards. 2000. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically, 5th ed. Approved standard M7-A5. National Committee for Clinical Laboratory Standards, Wayne, Pa.
- 11.Tunney, M. M., G. Ramage, T. T. Field, T. F. Moriarty, and D. G. Storey. 2004. Rapid colorimetric assay for antimicrobial susceptibility testing of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 48:1879-1881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Whitener, C. J., S. Y. Park, F. A. Browne, L. J. Parent, K. Julian, B. Bozdogan, et al. 2004. Vancomycin-resistant Staphylococcus aureus in the absence of vancomycin exposure. Clin. Infect. Dis. 38:1049-1055. [DOI] [PubMed] [Google Scholar]