Abstract
Rock varnish from Arizona's Whipple Mountains harbors a microbial community containing about 108 microorganisms g−1 of varnish. Analyses of varnish phospholipid fatty acids and rRNA gene libraries reveal a community comprised of mostly Proteobacteria but also including Actinobacteria, eukaryota, and a few members of the Archaea. Rock varnish represents a significant niche for microbial colonization.
Rock varnish (also known as desert varnish) is a dark, thin (usually 5 to 500 μm thick), layered veneer composed of clay minerals cemented together by oxides and hydroxides of manganese and iron (11, 20, 56, 63, 64). Nineteenth century references to rock varnish include those of Humboldt (42) and Darwin (14). Modern observations of varnish were initiated with the studies of Laudermilk (49) and Engel and Sharp (25); however, despite decades of study, the nucleation and growth mechanisms of rock varnish remain a mystery (11, 18, 37, 44, 57, 58).
Mn(II) is the soluble form of manganese that is available to organisms. It is stable between pH 6 and 9. Mn(III) and Mn(IV) primarily form insoluble oxides and oxyhydroxides. Microbial Mn(II) oxidation could thus result in the formation of manganese oxides as mineral phases in varnishes, as occurs in other environments (23, 39). Like manganese oxidation, iron oxidation (95) occurs at the exterior of the cell surface. Iron hydroxides are often deposited on the remains of biogenic structures (24). The extracellular deposition of ferric hydroxides is a way for iron-oxidizing organisms to prevent encrustation in iron oxide precipitates (88). Such precipitates might be incorporated in a varnish matrix through the activities of iron-oxidizing bacteria.
Rock varnish may hold a record of the microclimate in which it is found (7, 10, 11, 30), a hypothesis that has been questioned previously (67, 68). Some investigators suggest that rock varnish may harbor a historical record of important environmental processes such as long-term climate change (51). Bao et al. (7) studied preservation of atmospheric signatures in rock varnish and concluded that rock varnishes or other surface deposits may provide a record of paleoclimatic information and sulfur biogeochemical cycles. As a deposit of submicrometer layering, rock varnish may record the activity of dust storms, moisture and temperature fluctuations, biological activity, and the occurrence of fires over thousands of years. Rock varnish forms very slowly at rates thought to be between <1 to about 40 μm per 1,000 years (50), thus archeologists have been interested in dating the age of varnishes to place petroglyphs etched into varnish by ancient cultures into their full historical context (22, 90). Unfortunately, radiocarbon dating of varnish has proven difficult, and results must be used with caution (7, 9, 17, 22, 62).
It has been suggested that varnish or varnish-like materials may exist on Mars (2, 36, 44, 65). If so, varnish may be a niche for colonization by extraterrestrial life forms such as bacteria. Microorganisms are ubiquitous within varnishes on Earth. Thus, the study of Earthly varnishes may lead to the proper design of experiments in coming decades for detection of life on other planets. For example, iron and manganese oxidation by microbes cultivated from varnish has been extensively investigated (1, 19, 26, 33, 43, 46, 47, 54, 56, 77, 78, 80, 83). Perry et al. (59) observed a variety of amino acids in rock varnish and suggested that this is evidence for an intimate association of bacteria with the varnish material. A large variety of bacterial genera have been cultivated from rock varnish. These include Bacillus (43, 56), Geodermatophilus, Arthrobacter, Micrococcus, Curtobacterium, Cellulomonas (43, 48), Pedomicrobium, and a Metallogenium-like strain (19, 20). Eppard et al. (26) isolated several actinomycete species including Geodermatophilus. Staley et al. (78, 79) observed microcolonial fungi on rock varnish. Taylor-George et al. (83), Gorbushina et al. (32), and Perry (55) have provided evidence that these fungi may be involved in the formation of varnish.
To our knowledge, only two studies to date have investigated the microbial phylogenetics of varnish (26, 57). Eppard et al. (26) used 16S rRNA gene sequencing techniques to examine phylotypic characteristics of bacteria that had previously been cultured from various rock varnishes. Kuhlman et al. (48) employed 16S rRNA gene sequencing to identify several UV light-resistant bacteria isolated from rock varnish obtained in the Whipple Mountains of the U.S. Mojave Desert. These strains included representatives of the genera Geodermatophilus, Arthrobacter, Curtobacterium, and Cellulomonas. There are several reports of “microcolonial” fungi living within rock varnish or on the surfaces of rocks in hot, dry deserts (45, 55, 56, 78, 79, 83). The first scanning electron microscope images of such microcolonial fungi were reported from samples collected in the Sonoran Desert in 1978 (55). The fungi observed were thought to be representatives of the ascomycetes. It has been suggested that these fungi are the predominant biological forms observed on rock varnish coatings (32, 55, 60, 79, 80) and may be involved in the formation of varnish (83).
Most organisms in nature are refractory to cultivation (4, 8, 85, 89); therefore, many organisms that occupy the varnish habitat remain to be discovered. We report here the characterization of microbial rock varnish communities from the Whipple Mountains of the American Sonoran Desert via preparation of 16S and 18S rRNA gene clone libraries. These analyses are leading us to a greater understanding of the microbial diversity within these communities and their relevance to the possible occurrence of microbial life on other planets, such as Mars, where life forms face exposure to high fluxes of damaging UV light (13) and extremes of temperature and desiccation, such as are seen in locations on Earth where rock varnishes form. Microorganisms such as those that live or survive in varnish may also be potential forward contaminants on spacecraft structures (73).
Varnish samples were collected from alluvial fan deposits surrounding the Whipple Mountains, California. The Whipple Mountains lie west of Parker, Arizona, along the eastern boundary of the Mojave Desert. Previous studies conducted at the site demonstrated relatively thick varnishes (5 to 100 μm thick) on various rock types (5). Varnished rocks and surrounding soil samples were collected in February 2003 when the ground was still wet from winter rains. Only clasts with thick coatings (greater than 50 μm) on relatively large, flat surfaces were collected. It was critical that the samples be collected as aseptically as possible. Sample purity can never be completely verified, since it is very difficult to characterize contamination associated with local wildlife. However, contaminant potential was monitored during phylogenetic investigations by preparing control DNA libraries from soil adjacent to the point of collection of varnished rock. Varnished rocks were approached from the downwind direction, photographed in situ, picked up at arm's length using sterile gloves, and placed within sterile Whirl-pak bags large enough to hold entire rocks, and bags were sealed. Loose dirt on the undersides of the varnished rocks was brushed off in the field. The bags were then wrapped in protective material to prevent damage and subsequent contamination. The varnish was harvested from the host rock in a laminar flow bench. A Dremel grinding tool with flame-sterilized coarse bit was used to grind the varnish from the host rock into a sterile container. This approach minimized possibilities for atmospheric contamination of the varnish and ensured that the majority of bacteria removed came from within the varnish matrix.
Microbial biomass for enumeration and phospholipid fatty acid (PLFA) analysis was obtained from powdered rock varnish (0.1 g) obtained by aseptically grinding the varnish from the rock surface and then adding the varnish to 1 ml of sterile, double-distilled water (18 MΩ MilliQ water) in a 1.5-ml sterile microcentrifuge tube. Serial 1:10 dilutions were made, giving a range of dilutions from 10−1 to 10−3. The dilution samples were fixed with 2% ice-cold, high-performance liquid chromatography-grade methanol, vortexed until the sample appeared homogeneous, and incubated at room temperature for 30 min. The samples were stained with 60 μl ml−1 of a stock 4′,6′-diamidino-2-phenylindole (DAPI) or acridine orange (50 μg ml−1) solution. DAPI-stained samples were incubated at room temperature for 30 min in the dark before filtration. The most appropriate volume for analysis was determined to be 500 μl of the 10−3 dilution. This dilution contained >25 but <250 cells per field and had a low enough mineral content to both count cells in about one microscopic plane and not have cells obscured by the varnish minerals. Samples were filtered onto 25-mm Millipore Isopore 0.22-μm pore-size black polycarbonate filters (Millipore, Billerica, Mass.) with Whatman 25-mm GF/F filters used for support. Fluorescing cells were counted on a Zeiss Research epifluorescence microscope equipped with an Osram xenon short arc photo optic lamp XBO 75W and Chroma no. 31000 filter set for DAPI/Hoechst/AMCA (Zeiss, Inc., Thornwood, N.Y.). The mean number of fields counted per sample (n = 15) was 57.16. The standard deviation per sample was 7.48 fields. The average DAPI direct count of the rock varnish was 9.0 × 107 cells g−1 (standard deviation = 1.2 × 107). There was no difference between DAPI and acridine orange direct counts.
PLFA analyses were carried out by Microbial Insights, Inc. (Rockford, Tenn.). Total lipids were extracted (91) and the polar lipids separated by column chromatography (35). The polar lipid fatty acids were derivatized to fatty acid methyl esters, which were quantified using gas chromatography (69). Fatty acid chemical structures were verified by chromatography/mass spectrometry and equivalent chain length analysis. PLFA are essential components of the membranes of all cells except those of the Archaea, so their profiles allow for examination of most of the important members of many microbial communities. Since phospholipids break down rapidly upon cell death in environments examined thus far (91, 92), PLFA analysis should be also an accurate method for determining the amount of viable microbial biomass in an environment (92). The sum of the PLFA expressed in pmol is proportional to the number of cells, sensu lato. The proportion used here was 20,000 cells pmol−1 total PLFA (Microbial Insights, Inc., Rockford, TN). The number of cells within the microbial community from a sample of rock varnish was thus determined from its PLFA content (31.253 pmol g−1 [dry weight]). The microbial content of the varnish was ∼108 cells g−1 (dry weight), in excellent agreement with the direct microscopic counts of 9.0 × 107 ± 1.2 × 107 cells g−1. The PLFA profile of biomass within the varnish indicated a relatively simple community structure primarily composed of monoenoic PLFA, indicative of Proteobacteria with smaller but significant populations of Actinobacteria and Eukarya (Table 1).
TABLE 1.
PLFA analysis of Whipple Mountain rock varnisha
| Parameter (unit) | Value |
|---|---|
| Biomass (pmol PLFA g−1 dry weight) | 31,253 |
| Cells (g−1 dry weight) | 6.25 × 108 |
| % Gram negatives and firmicutes (terminally branched, saturated PLFA) | 2.4 |
| % Proteobacteria (monoenoic PLFA) | 81.8 |
| % Actinomycetes and sulfate-reducing bacteria (mid-chain branched, saturated PLFA) | 2.3 |
| % General (normal saturated PLFA) | 13.0 |
| % Eukaryotes (polyenoic PLFA) | 0.5 |
PLFA signatures of anaerobic metal reducers were not observed (branched monosaturated PLFA); physiological status is nonstressed (cyclopropyl acid/cis acid ratios and trans/cis ratios were both 0.00).
Three rRNA gene libraries from the Whipple Mountains rock varnish community DNA and two control libraries from soil lacking visible varnish adjacent to the varnished rock were generated using DNA isolated directly from 0.5 g ground varnish and 0.5 g adjacent, unvarnished soils with an Ultraclean soil DNA kit (Mo Bio, Solana Beach, Calif.). We were careful to avoid collecting rock fragments with our soil samples. Varnish 16S rRNA gene libraries were prepared for Bacteria and Archaea, and an 18S rRNA gene library for was prepared for Eukarya. Genes for 16S and 18S rRNA genes were PCR amplified from purified DNA using primers specific for Bacteria (338f/907r) (84), Archaea (A21f/A958r) (29), and Eukarya (NS3/NS8) (94). Clone libraries were prepared using a TA cloning kit (Invitrogen, Carlsbad, Calif.); 100 to 200 clones were prepared for each library. To better assess the diversity of the varnish community with the available resources, analysis of more partial-length sequences was favored over analysis of fewer, full-length sequences. Partial sequences are a reasonable compromise for the assessment of diversity which does not require precise taxonomic placement (12, 76). Each clone library was further classified by placing clones into restriction fragment length polymorphism (RFLP) restriction group patterns by digesting the DNA of individual clones with HhaI followed by electrophoresis on a 2% SeaPlaque agarose gel. Banding patterns were compared (34) by visual inspection of restriction digests of each clone, and a representative clone from each distinct RFLP group was sequenced. Sequencing was performed using an ABI Prism 3100 automated DNA sequencer (Applied Biosystems, Foster City, Calif.). Individual sequencing reactions were performed using the T7 and T3 primers, and a contiguous sequence was generated using ContigExpress software (Vector NTI-Informax; Invitrogen Corp., Carlsbad, Calif.). Clone sequences were evaluated using the Chimera Check program implemented in the Ribosomal Database Project (52). Two chimeras were found in both the Bacteria and Eukarya sequence libraries, while one chimera was found in the surrounding soil bacterial library as well as the varnish-associated Archaea library. These chimeras were excluded from further analysis. No chimeras were found in the surrounding soil Archaea library. Nonchimeric sequences were matched by a standard BLAST search (3) within the NCBI GenBank database to determine closest matches. Sequences were submitted to GenBank for rock varnish Bacteria, Archaea, and Eukarya and nonvarnished soil Bacteria and Archaea (see “Nucleotide sequence accession numbers” below).
Nine distinct RFLP groups obtained from 107 bacterial clones, 13 groups from 57 Eukarya clones, and two groups from 76 Archaea clones were analyzed. Rarefaction analysis (see Fig. S1 in the supplemental material) of the Archaea and the surrounding soil Archaea libraries reached saturation, indicating these two communities were well sampled and of low diversity. In contrast, rarefaction curves for the Bacteria, Eukarya, and surrounding soil Bacteria did not yet reach a plateau, indicating these populations were even more diverse than our initial phylogenetic analyses suggest.
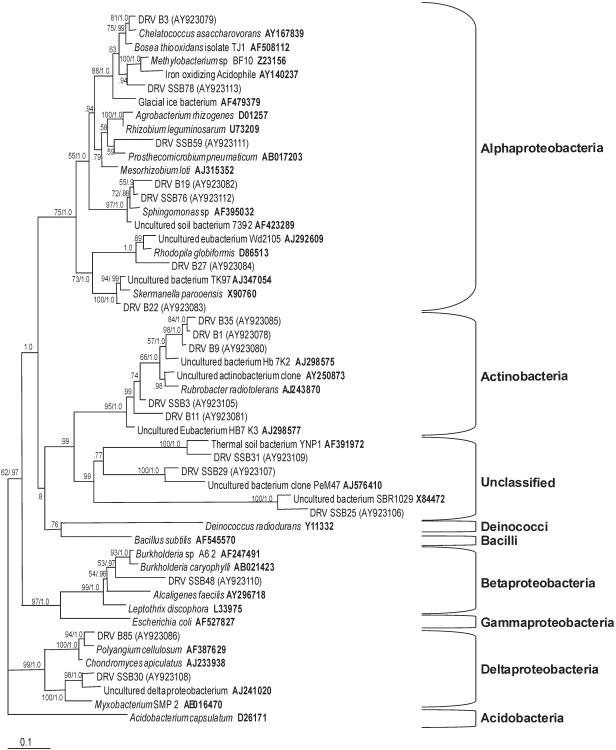
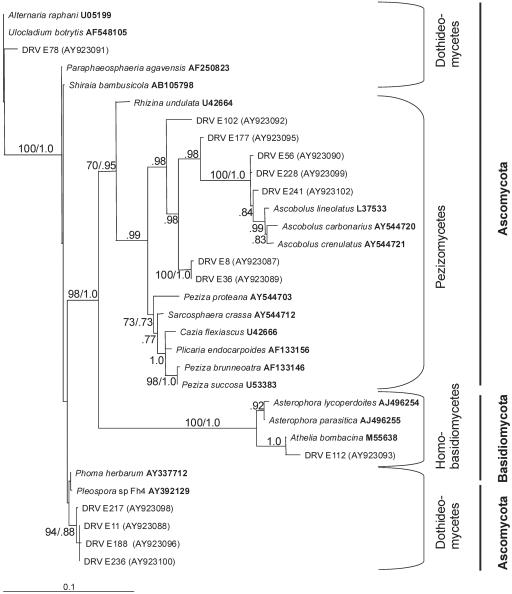
The phylogenetic relationships of bacterial and eukaryotic rRNA gene sequences are shown in Fig. 1 and 2, respectively. The closest database relatives of all sequences were chosen as reference sequences for phylogenetic analysis based on BLAST (3) comparisons. The 16S and 18S rRNA gene sequences were aligned using MegAlign (DNASTAR, Inc., Madison, Wis.). The model for the 16S data set (GTR+I+Γ) was selected based on relative goodness of fit. The relative goodness of fit of 16 alternative models of sequence evolution was compared using the likelihood ratio test (81). The model for the 18S data set (TrNef+I+Γ) was selected using DT-ModSel (53). Maximum likelihood searches were conducted using PAUP* (version 4.0b10) (82) with stepwise addition (10 random sequence additions) and tree bisection-reconnection branch swapping. Nodal support was estimated using bootstrap analysis (100 replicates) under the appropriate model. Posterior probabilities (107 generations, sampling every 100 generations) were determined under the GTR+I+Γ model of sequence evolution using MrBayes (MRBAYES 3.0b4) (40). Multiple independent runs were performed, and the parameter values were visually inspected to ensure convergence. The diversity of the ribotypes in each clone library were compared by rarefaction analysis calculated in DOTUR (72).
FIG. 1.
Phylogenetic tree showing the relationships of bacterial 16S rRNA gene sequences to one another and to known microbial groups using 1,573 aligned characters. Nodal support values with bootstrap support (100 replicates) and posterior probabilities (107 generations), respectively, are listed next to the branches of the unrooted maximum likelihood tree (ln L = −4,404.18186 [TrNef+I+Γ model]; pinv = 0.512289; α = 0.599910). Line length, 0.1 substitutions/site; DRV, desert rock varnish isolate. Accession numbers are shown in boldface type or in parentheses.
FIG. 2.
Phylogenetic tree showing the relationships of eukaryotic 18S rRNA gene sequences to one another and to known microbial groups using 1,201 aligned characters. Nodal support values with bootstrap support (100 replicates) and posterior probabilities (107 generations), respectively, are listed next to the branches of the unrooted maximum likelihood tree (ln L = −4,404.18186 [TrNef+I+Γ model]; pinv = 0.512289; α = 0.599910). Line length, 0.1 substitutions/site; DRV, desert rock varnish isolate. Accession numbers are shown in boldface type or in parentheses.
PLFA analysis (Table 1) provides indirect evidence that these bacteria are physiologically active (see below). It is possible that products produced by varnish bacteria (e.g., pigments) may play important roles in varnish morphogenesis. We did not observe organisms known to be involved in metal oxidation. Since rarefaction analysis (see Fig. S1 in the supplemental material) for the bacterial varnish clone libraries failed to reach saturation, it is possible that ribotypes related to metal oxidizers were simply not sampled from the larger, diverse varnish community. It is possible that ribotypes related to known metal oxidizers may have been found if the total diversity within the varnish had been assessed. Within the 16S rRNA gene libraries of the uncultivated microbial rock varnish community, we observed a number of sequences related to known environmental strains but not to known metal oxidizers. rRNA gene analyses are not sufficient in themselves to determine which organisms are an integral part of the rock varnish community over geologic time scales but do give an indication of the phylogenetic groups that can occupy this niche at a particular moment in time. We previously confirmed by pure culture techniques that viable radiation-tolerant bacteria are present in Whipple Mountains rock varnish. We isolated five UV-resistant bacterial cultures on tryptic soy agar plates after exposure to UV irradiation treatment. All of these strains were found to be related to known strains of Actinobacteria lineages (48). However, since most members of the varnish microbial community observed by molecular techniques were not cultivated (Fig. 1 and 2; see Tables S2 and S3 in the supplemental material), it is possible that these organisms are indeed able to oxidize iron and/or manganese. We cannot know for certain unless the observed strains are eventually cultivated for physiological studies or direct in situ techniques are developed to observe microbe-associated metal oxidation within the intact varnish matrix.
Within the 16S rRNA gene libraries of the uncultivated microbial community of Whipple Mountains rock varnish, we observed a number of sequences related to known environmental strains. Among the most common sequences observed were those closely related to the genus Rubrobacter (clones DRV B1, DRV B9, and DRV B35) (see Table S2 in the supplemental material). Van de Kamp et al. (87) reported in an abstract that actinobacterial 16S rRNA gene sequences were amplified by PCR from rock varnish obtained near Socorro, New Mexico. Rubrobacter species are of actinobacterial lineage and are known to inhabit masonry and lime wall paintings, where they cause a rosy discoloration (71). Diverse, yet-to-be-cultured members of the Rubrobacter subdivision are also widespread in Australian arid soils (38). The sequences of uncultivated bacteria we observed are related to Rubrobacter radiotolerans and Rubrobacter taiwanensis, bacteria known for their exceptional resistance to gamma radiation (28). One rock varnish-inhabiting genus observed in clone DRV B85 was related to Chondromyces or Polyangium (both 96% similarities). These genera are in the lineage of Myxococcales (75), which contains many members known to inhabit extreme environments (15) and to make bioactive substances (66).
A likely representative of the genus Sphingomonas (clone DRV B19) was observed in the uncultivated rock varnish community. This genus inhabits environments such as the deep subsurface (6) and has recently been observed within an endolithic community in Antarctica where it was found within translucent gypsum crusts on the surface of ice-free sandstone boulders (41). This, like rock varnish, represents an environment exposed to high levels of UV irradiation. The genus Sphingomonas contains many representatives that are able to degrade compounds such as polynuclear aromatic or halogenated molecules that might be present at low concentrations in the atmosphere (93).
Clone DRV B27 is a potential member of the genus Rhodopila (93% similarity), an obligatory aerobic, bacteriochlorophyll a-containing bacterium genus. While 93% similarity is not sufficient for unequivocal identification at the genus level, where 95% is the commonly accepted, albeit contentious, standard (4, 27, 70, 72), this GenBank match hints at a possible mechanism of bacterial survival in rock varnish. The environment of rock varnish would obviously be conducive to the development of photosynthetic life forms such as Rhodopila.
As is normal for this type of investigation, several sequences (Fig. 1) (clones DRV B9, DRV B11, DRV B19, DRV B27, and DRV B35) showed similarities to uncultured bacteria. This is confirmation that, as in other environments, many or perhaps most of the bacteria in rock varnish have never been cultured.
To our knowledge, PLFA analyses of rock varnish microbial communities have not been performed previously. Lipid analyses are a very useful supplement to DNA analyses when characterizing a complex microbial community such as the one studied here (21, 61). PLFA analysis of the rock varnish microbial community (Table 1) supports the conclusions made from our rRNA gene analyses. The PLFA profiles of biomass within the varnish indicate a relatively simple community structure primarily composed of monoenoic PLFA, indicative of Proteobacteria. The PLFA profiles also show the presence of a smaller but significant population of Actinobacteria (mid-chain branched saturated PLFA). Proteobacteria and Actinobacteria are of particular interest in that they represent organisms that have the ability to utilize a wide range of carbon sources and adapt quickly to environmental change, such as would be the situation in the rock varnish environment. PLFA also indicated the presence of eukaryota (e.g., fungi), and this was confirmed by 18S rRNA gene fingerprinting (Fig. 2; see Table S2 in the supplemental material).
The membranes of microorganisms adapt to the changing conditions of an environment, particularly under stressful conditions. These changes are reflected in the profiles of PLFA, with a change of cis fatty acids toward more trans-configured acids (35). Also, the Proteobacteria respond to starvation by making cyclopropyl (35) or mid-chain branched fatty acids (86). The “physiological status” of a microbial community can thus be assessed by dividing the amount of the stress-induced fatty acids by the amount of their precursors (Microbial Insights, Rockford, Tenn.). Biomarker ratios calculated for the rock varnish community harvested during the winter (February 2003) season (wet at the time of sampling) were indicative of a community not experiencing starvation or unusual stress. Cyclopropyl acid/cis acid ratios and trans/cis ratios were both 0.00.
Other microbial populations inhabit the surface environment of rocks in what is termed an “endolithic” lifestyle. Endoliths are found within the rock matrix just below the surface and occur in both hot and cold deserts (31). Endolithic communities are invariably colonized by cyanobacterial representatives of the Bacteria that, through photosynthesis, provide the fundamental source of carbon and energy that supports the endolithic community (16, 74). It is important while sampling rock varnish to be certain that endolithic communities are not also collected inadvertently. We observed no 16S rRNA gene sequences indicative of cyanobacteria in our samples of varnish, even though we employed conventional primers targeting conserved regions of 16S rRNA genes that should have detected cyanobacterial sequences. This is support that we sampled true varnish communities and not endolithic communities and also implies that other photosynthetic forms (e.g., bacteriochlorophyll a-containing bacteria such as Rhodopila, as represented by clone DRV 27) might be found in varnish-based ecosystems. This hypothesis merits testing at a variety of varnish sites within both hot and cold deserts.
Sequence analyses of representative clones of RFLP groups within an Archaea rRNA gene library (see Table S1 in the supplemental material) revealed only two RFLP types with 76 clones analyzed. Both types were related to previously observed uncultivated Archaea. Rarefaction curves obtained from the archaeal clone library reached saturation (see Fig. S1 in the supplemental material). Thus, Archaea appear to be present in low diversity in Whipple Mountains rock varnish.
Eukaryota observed by preparation of an 18S rRNA gene library from rock varnish (Fig. 2; see Table S2 in the supplemental material) were dominated by RFLP groups of fungal representatives of the ascomycota and included representatives of the genera Phoma, Plicaria, Ascobolus, Alternaria, Sarcosphaera, and Rhizina. One RFLP group representative most closely aligned with the basidiomycota (the genus Athelia in the family Corticiaceae; clone DRV E112). The dominance of ascomycota in our library is in accord with the microscopic observations of Perry (55), who characterized by scanning electron microscopy a microcolonial fungus from varnish also collected in the Sonoran Desert. Staley and colleagues identified this organism as an ascomycete within the family Dematiaceae (78, 79). The 18S rRNA gene sequences we observed were not of this family but fell into families such as Pleosporaceae (clone DRV E78), Rhizinaceae (clone DRV E102), Pezizaceae (clones DRV E8 and DRV E36), and Ascobolaceae (clone DRV E56).
Sequence analyses of representative clones of bacterial and archaeal rRNA gene libraries of nonvarnished soil (see Table S3 in the supplemental material) revealed mostly lineages different from those seen in rock varnish libraries. Only two nonvarnish clones overlapped. These were a Rubrobacter radiotolerans-like sequence and a Sphingomonas-like sequence. Both of these groups could reasonably be expected to inhabit both soil and varnish habitats.
The analyses reported here are not quantitative, so little can be said of the overall importance of individual RFLP groups or phylogenetic taxa observed to the overall community composition. In this regard, it is of interest that sequences similar to those of UV-resistant pure cultures isolated from the same Whipple Mountains rock varnish used in work reported here (e.g., Geodermatophilus, Arthrobacter, Curtobacterium, and Cellulomonas) (48) were not observed in varnish 16S rRNA gene libraries. This indicates that these strains, though present, probably do not dominate the community, a result that is supported by rarefaction analysis that revealed that the varnish contains a higher diversity of both Bacteria and Eukarya than was sampled in this study. It should be possible in future work to design PCR primers targeting specific RFLP groups (e.g., the Rubrobacter or ascomycota groups) and employ these using quantitative PCR techniques, such as real-time PCR, to examine the quantitative compositions of rock varnish microbial communities. In addition, quantitative real-time PCR using primers specific for enzymes known to be involved in metal oxidation may shed light on the roles that the microbial communities found within rock varnish play with respect to the formation and maintenance of the varnish.
Nucleotide sequence accession numbers.
Sequences were submitted to GenBank under the following accession numbers: rock varnish Bacteria, AY923078 to AY923086; rock varnish Archaea, AY923076 and AY923077; rock varnish Eukarya, AY923087 to AY923102; nonvarnished soil Bacteria and Archaea, AY923105 and AY92310.
ADDENDUM IN PROOF
Since we submitted our paper another article that is relevant to work reported here was published (R. T. Schelble, G. D. McDonald, J. A. Hall, and K. H. Nealson, Geomicrobiol. J. 22:353-360, 2005). In that paper, the authors report on their analyses of fatty acid methyl esters of samples collected from the Mojave Desert and compare the microbial community structure of desert varnish with that of adjacent desert soil. Their analyses indicated that prokaryotic and fungal communities are present in both desert varnish and soil samples. Fatty acid methyl esters specific to gram-positive bacteria were found more often and in greater abundance in varnish samples than in adjacent soils.
Supplementary Material
Acknowledgments
The research described in this publication was carried out at the Jet Propulsion Laboratory, California Institute of Technology, under a contract with the National Aeronautics and Space Administration and at the Environmental Biotechnology Institute at the University of Idaho. This work was supported by an award from the Director's Research and Development Fund at JPL (JPL CREI contract number NAS 7-1260).
We thank Cornelia Sawatzky for editorial assistance.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Adams, J. B., F. Palmer, and J. T. Staley. 1992. Rock weathering in deserts: mobilization and concentration of ferric iron by microorganisms. Geomicrobiol. J. 10:99-115. [Google Scholar]
- 2.Allen, C. C., F. Westall, and R. T. Schelble. 2001. Importance of a martian hematite site for astrobiology. Astrobiology 1:111-123. [DOI] [PubMed] [Google Scholar]
- 3.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 4.Amann, R., W. Ludwig, and K. Schleifer. 1995. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol. Rev. 59:143-169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Anderson, R. C. 1995. Spectral mapping of quaternary geomorphic surfaces in the Whipple and Piute Mountains of southeastern California using visible and near infrared datasets. Ph.D. dissertation. University of Pittsburgh, Pittsburgh, Pa.
- 6.Balkwill, D. L., R. H. Reeves, G. R. Drake, J. Y. Reeves, F. H. Crocker, M. B. King, and D. R. Boone. 1997. Phylogenetic characterization of bacteria in the subsurface microbial culture collection. FEMS Microbiol. Rev. 20:201-216. [DOI] [PubMed] [Google Scholar]
- 7.Bao, H. M., G. M. Michalski, and M. H. Thiemens. 2001. Sulfate oxygen-17 anomalies in desert varnishes. Geochim. Cosmochim. Acta 65:2029-2036. [Google Scholar]
- 8.Barns, S. M., R. E. Fundyga, M. W. Jeffries, and N. R. Pace. 1994. Remarkable archaeal diversity detected in a Yellowstone-National-Park hot-spring environment. Proc. Natl. Acad. Sci. USA 91:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beck, W., D. J. Donahue, A. J. T. Jull, S. Burr, W. S. Broecker, G. Bonani, I. Hajdas, and E. Malotki. 1998. Ambiguities in direct dating of rock surfaces using radiocarbon measurements. Science 280:2132-2135. [Google Scholar]
- 10.Bengang, Z., T. Z. Liu, and Y. Zhang. 2000. Rock varnish microlaminations from northern Tianshan, Xinjiang and their palioclimatic implications. Chin. Sci. Bull. 45:372-379. [Google Scholar]
- 11.Broecker, W. S., and T. Z. Liu. 2001. Rock varnish: recorder of desert wetness? Geol. Today 11:4-10. [Google Scholar]
- 12.Chelius, M. K., and J. C. Moore. 2004. Molecular phylogenetic analysis of Archaea and Bacteria in Wind Cave, South Dakota. Geomicrobiol. J. 21:123-134. [Google Scholar]
- 13.Cockell, C. A. 2001. The martian and extraterrestrial UV radiation environment part II: further considerations on materials and design for artificial ecosystems. Acta Astronaut. 49:563-644. [DOI] [PubMed] [Google Scholar]
- 14.Darwin, C. 1871. Natural history and geology. Appleton and Company, New York, N.Y.
- 15.Dawid, W. 2000. Biology and global distribution of myxobacteria in soils. FEMS Microbiol. Rev. 24:403-427. [DOI] [PubMed] [Google Scholar]
- 16.de los Rios, A., J. Wierzchos, L. G. Sancho, and C. Ascaso. 2003. Acid microenvironments in microbial biofilms of antarctic endolithic microecosystems. Environ. Microbiol. 5:231-237. [DOI] [PubMed] [Google Scholar]
- 17.Dorn, R. I. 1998. Ambiguities in direct dating of rock surfaces using radiocarbon measurements—response. Science 280:2135-2139. [Google Scholar]
- 18.Dorn, R. I. 1998. Rock coatings. Dev. Earth Surf. Process 6:444. [Google Scholar]
- 19.Dorn, R. I., and T. M. Oberlander. 1981. Microbial origin of desert varnish. Science 213:1245-1247. [DOI] [PubMed] [Google Scholar]
- 20.Dorn, R. I., and T. M. Oberlander. 1982. Rock varnish. Prog. Geogr. 6:317-367. [Google Scholar]
- 21.Dowling, N. J. E., F. Widdel, and D. C. White. 1986. Phospholipid ester-linked fatty add biomarkers of acetate-oxidizing sulfate reducers and other sulfide forming bacteria. J. Gen. Microbiol. 132:1815-1825. [Google Scholar]
- 22.Dragovich, D. 2000. Rock engraving chronologies and accelerator mass spectrometry radiocarbon age of desert varnish. J. Archaeol. Sci. 27:871-876. [Google Scholar]
- 23.Ehrlich, H. L. 1996. How microbes influence mineral growth and dissolution. Chem. Geol. 132:5-9. [Google Scholar]
- 24.Emerson, D. 2000. Microbial oxidation of Fe(II) at circumneutral pH, p. 31-52. In D. R. Lovley (ed.), Environmental microbe-metal interactions. ASM Press, Washington, D.C.
- 25.Engel, C. G., and R. S. Sharp. 1958. Chemical data on desert varnish. Geol. Bull. 69:487-518. [Google Scholar]
- 26.Eppard, M., W. E. Krumbein, C. Koch, E. Rhiel, J. T. Staley, and E. Stackebrandt. 1996. Morphological, physiological, and molecular characterization of actinomycetes isolated from dry soil, rocks, and monument surfaces. Arch. Microbiol. 166:12-22. [DOI] [PubMed] [Google Scholar]
- 27.Everett, K., R. Bush, and A. Andersen. 1999. Emended description of the order Chlamydiales, proposal of Parachlamydiaceae fam. nov. and Simkaniaceae fam. nov., each containing one monotypic genus, revised taxonomy of the family Chlamydiaceae, including a new genus and five new species, and standards for the identification of organisms. Int. J. Syst. Bacteriol. 49:415-440. [DOI] [PubMed] [Google Scholar]
- 28.Ferreira, A. C., M. F. Nobre, E. Moore, F. A. Rainey, J. R. Battista, and M. S. da Costa. 1999. Characterization and radiation resistance of new isolates of Rubrobacter radiotolerans and Rubrobacter xylanophilus. Extremophiles 3:235-238. [DOI] [PubMed] [Google Scholar]
- 29.Ficker, M., K. Krastel, S. Orlicky, and E. Edwards. 1999. Molecular characterization of a toluene-degrading methanogenic consortium. Appl. Environ. Microbiol. 65:5576-5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fleisher, M., T. Z. Liu, W. S. Broecker, and W. Moore. 1999. A clue regarding the origin of rock varnish. Geophys. Res. Lett. 26:103-106. [Google Scholar]
- 31.Friedmann, E. I. 1980. Endolithic microbial life in hot and cold deserts. Orig. Life 10:223-235. [DOI] [PubMed] [Google Scholar]
- 32.Gorbushina, A., W. E. Krumbein, C. H. Hamman, L. Panina, S. Soukharjevsky, and U. Wollenzien. 1993. On the role of black fungi in colour change and biodeterioration of antique marbles. Geomicrobiol. J. 11:205-221. [Google Scholar]
- 33.Grote, G., and W. E. Krumbein. 1992. Microbial precipitation of manganese by bacteria and fungi from desert rock and rock varnish. Geomicrobiol. J. 10:49-57. [Google Scholar]
- 34.Gu, A. Z., B. P. Hedlund, J. T. Staley, S. E. Strand, and H. D. Stensel. 2004. Analysis and comparison of the microbial community structures of two enrichment cultures capable of reductively dechlorinating TCE and cis-DCE. Environ. Microbiol. 6:45-54. [DOI] [PubMed] [Google Scholar]
- 35.Guckert, J. B., C. P. Antworth, P. D. Nichols, and D. C. White. 1985. Phospholipid ester-linked fatty acid profiles as reproducible assays for changes in prokaryotic community structure of estuarine sediments. FEMS Microbiol. Ecol. 31:147-158. [Google Scholar]
- 36.Guinness, E. A., R. E. Arvidson, I. H. D. Clark, and M. K. Shepard. 1997. Optical scattering properties of terrestrial varnished basalts compared with rocks and soils at the Viking lander sites. J. Geophys. Res. Planets 102:28687-28703. [Google Scholar]
- 37.Hodge, V. F., D. E. Farmer, T. Diaz, and R. L. Orndorff. 2005. Prompt detection of alpha particles from 210Po: another clue to the origin of rock varnish? J. Environ. Radioact. 78:331-342. [DOI] [PubMed] [Google Scholar]
- 38.Holmes, A. J., J. Bowyer, M. P. Holley, M. O'Donoghue, M. Montgomery, and M. R. Gillings. 2000. Diverse, yet-to-be-cultured members of the Rubrobacter subdivision of the Actinobacteria are widespread in Australian arid soils. FEMS Microbiol. Ecol. 33:111-120. [DOI] [PubMed] [Google Scholar]
- 39.Huang, P. M. 1991. Kinetics of redox reactions on manganese oxides and its impact on environmental quality, p. 191-230. In D. L. Sparks (ed.), Rates of soil chemical processes. Soil Science Society of America, Madison, Wis.
- 40.Huelsenbeck, J. P., and F. Ronquist. 2001. MrBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 17:754-755. [DOI] [PubMed] [Google Scholar]
- 41.Hughes, K. A., and B. Lawley. 2003. A novel Antarctic microbial endolithic community within gypsum crusts. Environ. Microbiol. 5:555-565. [DOI] [PubMed] [Google Scholar]
- 42.Humboldt, A. 1852. Personal narrative of travels to the equinoctial regions of America during the years 1799-1804. Henry Bohn, London, England.
- 43.Hungate, B., A. Danin, N. B. Pellerin, J. Stemmler, P. Kjellander, J. B. Adams, and J. T. Staley. 1987. Characterization of manganese-oxidizing (Mn(II)-Mn(IV)) bacteria from Negev Desert rock varnish-implications in desert varnish formation. Can. J. Microbiol. 33:939-943. [Google Scholar]
- 44.Israel, E. J., R. E. Arvidson, A. Wang, J. D. Pasteris, and B. L. Jolliff. 1997. Laser Raman spectroscopy of varnished basalt and implications for in situ measurements of martian rocks. J. Geophys. Res. Planets 102:28705-28716. [Google Scholar]
- 45.Jones, C. E. 1991. Characteristics and origin of rock varnish from the hyperarid coastal deserts of Northern Peru. Quat. Res. 35:116-129. [Google Scholar]
- 46.Krumbein, W. E., and H. J. Altman. 1973. A new method for the detection and enumeration of manganese oxidizing and reducing microorganisms. Helgol. Wiss. Meeresunters 25:347-356. [Google Scholar]
- 47.Krumbein, W. E., and K. Jens. 1981. Biogenic rock varnishes of the Negev Desert (Israel): an ecological study of iron and manganese transformation by cyanobacteria and fungi. Oecologia 50:25-38. [DOI] [PubMed] [Google Scholar]
- 48.Kuhlman, K. R., L. B. Allenbach, C. L. Ball, W. G. Fusco, M. T. La Duc, G. M. Kuhlman, R. C. Anderson, I. K. Erickson, T. Stuecker, J. Benardini, and R. L. Crawford. 2005. Ultraviolet (UV-C) resistant bacteria isolated from rock varnish in the Whipple Mountains, California. Icarus 174:585-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Laudermilk, J. D. 1931. On the origin of desert varnish. Amer. J. Sci. 21:51-66. [Google Scholar]
- 50.Liu, T. H., and W. S. Broecker. 2000. How fast does rock varnish grow? Geol. Bull. 28:183-186. [Google Scholar]
- 51.Liu, T. Z. 2003. Blind testing of rock varnish microstratigraphy as a chronometric indicator: results on late quaternary lava flows in the Mojave Desert, California. Geomorphology 53:209-234. [Google Scholar]
- 52.Maidak, B. L., J. R. Cole, C. T. Parker, Jr., G. M. Garrity, N. Larsen, B. Li, T. G. Lilburn, M. J. McCaughey, G. J. Olsen, R. Overbeek, S. Pramanik, T. M. Schmidt, J. M. Tiedje, and C. R. Woese. 1999. A new version of the RDP (Ribosomal Database Project). Nucleic Acids Res. 27:171-173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Minin, V., Z. Abdo, P. Joyce, and J. Sullivan. 2003. Performance-based selection of likelihood models for phylogeny estimation. Syst. Biol. 52:674-683. [DOI] [PubMed] [Google Scholar]
- 54.Palmer, F. E., J. T. Staley, R. G. E. Murray, T. Counsell, and J. B. Adams. 1986. Identification of manganese-oxidizing bacteria from desert varnish. Geomicrobiol. J. 4:343-360. [Google Scholar]
- 55.Perry, R. S. 1979. Chemistry and structure of desert varnish. M.S. Thesis. University of Washington, Seattle, Wash.
- 56.Perry, R. S., and J. B. Adams. 1978. Desert varnish—evidence for cyclic deposition of manganese. Nature 276:489-491. [Google Scholar]
- 57.Perry, R. S., J. Dodsworth, J. T. Staley, and M. H. Engel. 2004. Bacterial diversity of desert varnish, p. 259-260. In R. A. Harris and L. Duwehand (ed.), Third European workshop on exo/astrobiology. Mars: the search for life. SP-545. ESA Publications, Noordwijk, The Netherlands.
- 58.Perry, R. S., J. Dodsworth, J. T. Staley, and A. Gillespie. 2003. Molecular analyses of microbial communities in rock coatings and soils from Death Valley, California, abstr. 12624, p. 313. Proceedings of the NASA Astrobiology Institute General Meeting: living links through time and space: meeting the challenges of interdisciplinary science. NASA Astrobiology Institute, Tempe, Ariz.
- 59.Perry, R. S., M. H. Engel, O. Botta, and J. T. Staley. 2003. Amino acid analyses of desert varnish from the Sonoran and Mojave Deserts. Geomicrobiol. J. 20:427-438. [Google Scholar]
- 60.Perry, R. S., and V. M. Kolb. 2003. Biological and organic constituents of desert varnish: review and new hypotheses, p. 202-217. In R. B. Hoover and A. Y. Rozanov (ed.), Instruments, methods, and missions for astrobiology VII, vol. 5163. SPIE, Bellingham, Wash. [Google Scholar]
- 61.Pinkart, H. C., D. B. Ringelberg, Y. M. Piceno, S. J. Macnaughton, and D. C. White. 2002. Biochemical approaches to biomass measurements and community structure analysis, p. 101-113. In C. J. Hurst, R. L. Crawford, M. J. McInerney, G. R. Knudsen, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 62.Pope, G. A. 2000. Weathering of petroglyphs: direct assessment and implications for dating methods. Antiquity 74:833-843. [Google Scholar]
- 63.Potter, R. M., and G. R. Rossman. 1977. Desert varnish—importance of clay-minerals. Science 196:1446-1448. [DOI] [PubMed] [Google Scholar]
- 64.Potter, R. M., and G. R. Rossman. 1979. The manganese- and iron-oxide mineralogy of desert varnish. Chem. Geol. 25:79-94. [Google Scholar]
- 65.Probst, L. W., C. C. Allen, K. L. Thomas-Keprta, S. J. Clemett, T. G. Longazo, M. A. Nelman-Gonzalez, and C. Sams. 2002. Desert varnish: preservation of biofabrics and implications for Mars, abstr. 1764, p. 78. Proceedings of the 33rd Lunar and Planetary Science Conference. Lunar and Planetary Institute, Houston, Texas.
- 66.Reichenbach, H. 2001. Myxobacteria, producers of novel bioactive substances. J. Ind. Microbiol. Biotechnol. 27:149-156. [DOI] [PubMed] [Google Scholar]
- 67.Reneau, S. L. 1993. Manganese accumulation in rock-varnish on a desert piedmont, Mojave Desert, California, and application to evaluating varnish development. Quat. Res. 40:309-317. [Google Scholar]
- 68.Reneau, S. L., R. Raymond, and C. D. Harrington. 1992. Elemental relationships in rock varnish stratigraphic layers, Cima Volcanic Field, California—implications for varnish development and the interpretation of varnish chemistry. Amer. J. Sci. 292:684-723. [Google Scholar]
- 69.Ringelberg, D. B., G. T. Townsend, K. A. DeWeerd, J. M. Sulita, and D. C. White. 1994. Detection of the anaerobic dechlorinating microorganism Desulfomonile tiedjei in environmental matrices by its signature lipopolysaccharide branch-long-chain hydroxy fatty acids. FEMS Microbiol. Ecol. 14:9-18. [Google Scholar]
- 70.Sait, M., P. Hugenholtz, and P. H. Janssen. 2002. Cultivation of globally distributed soil bacteria from phylogenetic lineages previously only detected in cultivation-independent surveys. Environ. Microbiol. 4:654-666. [DOI] [PubMed] [Google Scholar]
- 71.Schabereiter-Gurtner, C., G. Pinar, D. Vybiral, W. Lubitz, and S. Rolleke. 2001. Rubrobacter-related bacteria associated with rosy discolouration of masonry and lime wall paintings. Arch. Microbiol. 176:347-354. [DOI] [PubMed] [Google Scholar]
- 72.Schloss, P. D., and J. Handelsman. 2005. Introducing DOTUR, a computer program for defining operational taxonomic units and estimating species richness. Appl. Environ. Microbiol. 71:1501-1506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schuerger, A. C., R. L. Mancinelli, R. G. Kern, L. J. Rothschild, and C. P. McKay. 2003. Survival of endospores of Bacillus subtilis on spacecraft surfaces under simulated Martian environments: implications for the forward contamination of Mars. Icarus 165:253-276. [DOI] [PubMed] [Google Scholar]
- 74.Sigler, W. V., R. Bachofen, and J. Zeyer. 2003. Molecular characterization of endolithic cyanobacteria inhabiting exposed dolomite in central Switzerland. Environ. Microbiol. 5:618-627. [DOI] [PubMed] [Google Scholar]
- 75.Sproer, C., H. Reichenbach, and E. Stackebrandt. 1999. The correlation between morphological and phylogenetic classification of myxobacteria. Int. J. Syst. Bacteriol. 49:1255-1262. [DOI] [PubMed] [Google Scholar]
- 76.Stackebrandt, E., and F. A. Rainey. 1996. Partial and complete 16S rDNA sequences, their use in generation of 16S rDNA phylogenetic trees and their implications in molecular ecological studies, p. 1-17. In A. D. L. Akkermans, J. D. van Elsas, and F. J. De Brujin (ed.), Molecular microbiology and ecology manual, vol. 3.1.1. Kluwer Academic Publishers, Boston, Mass. [Google Scholar]
- 77.Staley, J. T., J. B. Adams, and F. E. Palmer. 1992. Desert varnish: a biological perspective, p. 173-195. In G. Stotzky and J. M. Bollag (ed.), Soil biochemistry, vol. 7. Marcel Dekker, New York, N.Y. [Google Scholar]
- 78.Staley, J. T., M. J. Jackson, F. E. Palmer, J. B. Adams, D. J. Borns, D. J. Curtiss, and S. Taylor-George. 1983. Desert varnish coatings and microcolonial fungi on rocks of the Gibson and Great Victoria Deserts, Australia. BMR J. Aust. Geol. Geophys. 8:83-87. [Google Scholar]
- 79.Staley, J. T., F. E. Palmer, and J. B. Adams. 1982. Microcolonial fungi: common inhabitants of desert rock. Science 215:1093-1095. [DOI] [PubMed] [Google Scholar]
- 80.Sterflinger, K., W. E. Krumbein, T. Lellau, and J. Rullkotter. 1999. Microbially mediated orange patination of rock surfaces. Anc. Biomol. 3:51-65. [Google Scholar]
- 81.Sullivan, J., J. A. Markert, and C. W. Kilpatrick. 1997. Phylogeography and molecular systematics of the Peromyscus aztecus group (Rodentia: Muridae) inferred using parsimony and maximum likelihood. Syst. Biol. 46:426-440. [DOI] [PubMed] [Google Scholar]
- 82.Swofford, D. L. 2002. PAUP*. Phylogenetic analysis using parsimony (*and other methods), 4th ed. Sinauer Associates, Sunderland, Mass.
- 83.Taylor-George, S., F. Palmer, J. T. Staley, D. J. Borns, B. Curtiss, and J. B. Adams. 1983. Fungi and bacteria involved in desert varnish formation. Microb. Ecol. 9:227-245. [DOI] [PubMed] [Google Scholar]
- 84.Thomsen, T. R., K. Finster, and N. B. Ramsing. 2001. Biogeochemical and molecular signatures of anaerobic methane oxidation in a marine sediment. Appl. Environ. Microbiol. 67:1646-1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Torsvik, V., J. Goksoyr, and F. L. Daae. 1990. High diversity in DNA of soil bacteria. Appl. Environ. Microbiol. 56:782-787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Tsitko, I. V., G. M. Zeltsev, A. G. Lobneok, and M. S. Salkinoja-Salonen. 1999. Effect of aromatic compounds on cellular fatty composition of Rhodococcus opacus. Appl. Environ. Microbiol. 65:853-855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.van de Kamp, J. L., M. N. Spilde, J. R. Snider, D. L. Pham, P. J. Boston, and D. E. Northup. 2005. Abstr. 105th Gen. Meet. Am. Soc. Microbiol., N-081. American Society for Microbiology, Washington, D.C.
- 88.van Veen, W. L., E. G. Mulder, and M. H. Deinema. 1978. The Sphaerotilus-Leptothrix group of bacteria. Microbiol. Rev. 42:329-356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ward, D. M., R. Weller, and M. M. Bateson. 1990. 16S ribosomal-RNA sequences reveal numerous uncultured microorganisms in a natural community. Nature 345:63-65. [DOI] [PubMed] [Google Scholar]
- 90.Watchman, A. 2000. A review of the history of dating rock varnishes. Earth Sci. Rev. 49:261-277. [Google Scholar]
- 91.White, D. C., W. M. Davis, J. S. Nickels, J. D. King, and R. J. Bobbie. 1979. Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oecologia 40:51-62. [DOI] [PubMed] [Google Scholar]
- 92.White, D. C., H. C. Pinkart, and D. Ringelberg. 1997. Biomass measurements: biochemical approaches, p. 91-101. In C. J. Hurst, G. R. Knudsen, M. J. McInerney, L. D. Stetzenbach, and M. V. Walter (ed.), Manual of environmental microbiology. ASM Press, Washington, D.C.
- 93.White, D. C., S. D. Sutton, and D. Ringelberg. 1996. The genus Sphingomonas: physiology and ecology. Curr. Opin. Biotechnol. 7:301-306. [DOI] [PubMed] [Google Scholar]
- 94.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, Inc., New York, N.Y.
- 95.Winogradsky, S. 1988. Ueber eisenbacterian. Bontan. Ztg. 46:261-270. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.