Abstract
The ubiquity of fecal indicator bacteria such as Escherichia coli and Enterococcus spp. in urban environments makes tracking of fecal contamination extremely challenging. A multitiered approach was used to assess sources of fecal pollution in Ballona Creek, an urban watershed that drains to the Santa Monica Bay (SMB) near Los Angeles, Calif. A mass-based design at six main-stem sites and four major tributaries over a 6-h period was used (i) to assess the flux of Enterococcus spp. and E. coli by using culture-based methods (tier 1); (ii) to assess levels of Enterococcus spp. by using quantitative PCR and to detect and/or quantify additional markers of human fecal contamination, including a human-specific Bacteroides sp. marker and enterovirus, using quantitative reverse transcriptase PCR (tier 2); and (iii) to assess the specific types of enterovirus genomes found via sequence analysis (tier 3). Sources of fecal indicator bacteria were ubiquitous, and concentrations were high, throughout Ballona Creek, with no single tributary dominating fecal inputs. The flux of Enterococcus spp. and E. coli averaged 109 to 1010 cells h−1 and was as high at the head of the watershed as at the mouth prior to discharge into the SMB. In addition, a signal for the human-specific Bacteroides marker was consistently detected: 86% of the samples taken over the extent during the study period tested positive. Enteroviruses were quantifiable in 14 of 36 samples (39%), with the highest concentrations at the site furthest upstream (Cochran). These results indicated the power of using multiple approaches to assess and quantify fecal contamination in freshwater conduits to high-use, high-priority recreational swimming areas.
The Santa Monica Bay (SMB), California, is home to some of the most popular beaches in the world. It is located adjacent to metropolitan Los Angeles, and more than 50 million beachgoers visit SMB shorelines every year—more than those visiting all other beaches in California combined (38). However, there are serious concerns about beach water quality because of continued exceedances of water quality thresholds based on fecal indicator bacteria such as total coliforms, fecal coliforms, or Escherichia coli and Enterococcus spp., particularly in areas impacted by urban runoff. Thirteen percent of the shoreline mile-days in the SMB exceeded water quality thresholds between 1995 and 2000, with over 50% of these exceedances located near storm drains (37). The public health risk associated with urban runoff has been directly demonstrated through epidemiology studies. Haile et al. (19) demonstrated that swimmers near storm drain discharges in the SMB had a higher likelihood of respiratory and/or gastrointestinal symptoms than swimmers more than 400 m from a storm drain.
Despite the impairment of water quality and risks to human health, identification and elimination of the sources of bacteria responsible for the beach warnings remain elusive. The difficulty in identifying and eliminating the sources of bacteria results from three important factors. First, the traditional indicators of fecal pollution on the basis of which the water quality thresholds were developed are not specific to humans. These fecal indicator bacteria can be shed from any warm-blooded organism, including wild and domesticated animals (12). Therefore, source tracking turns into a challenging scenario when these diffuse and frequently intermittent or episodic fecal releases occur. The second difficulty in identifying and eliminating sources of fecal indicator bacteria is their ubiquity in urban environments. Finally, unlike many human pathogens of concern, fecal indicator bacteria may survive and even grow in the environment (see, e.g., references 24, 39, and 44).
Viruses are one tool that could prove useful in source-tracking studies, because they include many pathogens of concern, and they are generally species specific. Viruses are known to cause a significant portion of waterborne disease, mostly from ingestion of sewage-contaminated water and seafood (10). Until recently, however, virus detection and quantification have relied on cell culture-based approaches that are much too slow to be effective source-tracking tools. Recently developed molecular techniques, such as quantitative reverse transcriptase PCR (QRT-PCR), can detect and quantify viral genetic material directly from water samples. Tests conducted previously in Southern California (11, 22, 32, 41, 42), Florida (17, 36), and Europe (35) using conventional RT-PCR or PCR have detected genetic material from human-specific viruses, including enterovirus, hepatitis A virus, rotavirus, and adenovirus, in urban runoff discharges or seawater samples.
A different approach would be to use alternative bacterial indicators for source tracking that might be much more abundant in urban discharges. For example, Bacteroides spp. make up approximately one-third of the human fecal microflora, considerably outnumbering fecal coliforms, E. coli, and Enterococcus spp. Bacteroides spp. are obligate anaerobes, so there is little concern over persistence or regrowth in the environment. More importantly, human-specific Bacteroides markers have been developed, increasing the value of this potential indicator (2, 3, 7).
Both viruses and alternative bacterial indicators such as Bacteroides spp. have been shown to be potentially useful source-tracking tools. Griffith et al. (18) and Noble et al. (33) concluded that genetics-based methods, such as PCR, consistently provided the best information in efforts to conduct source tracking on mixed-source samples. To date, however, no single method has all of the traits necessary to be the consummate source-tracking tool. Therefore, a multitiered, multiindicator approach has been recommended by some investigators (4, 40). By using multiple tools, investigators can utilize the strengths of each to ascertain inputs and track fates that will ultimately lead to successful management solutions.
The objective of this study was to identify the contributions and quantify the loading of fecal contamination affecting the SMB by using a multitiered approach. The first tier included traditional measurements of fecal indicator bacteria. The second tier included molecular assays developed and conducted for Enterococcus spp., a human-specific Bacteroides marker, and enterovirus. These methods rely on conventional PCR, quantitative PCR (QPCR), or QRT-PCR, which have not previously been applied in conjunction with one another for source-tracking studies in urban watersheds. The third tier involved sequencing of the enterovirus from the field samples with the greatest concentrations so as to determine the likely type of enterovirus amplified in the assay. The multitiered approach was applied using a mass-based design to quantify inputs and flux through an urban watershed to the beach.
MATERIALS AND METHODS
Study site.
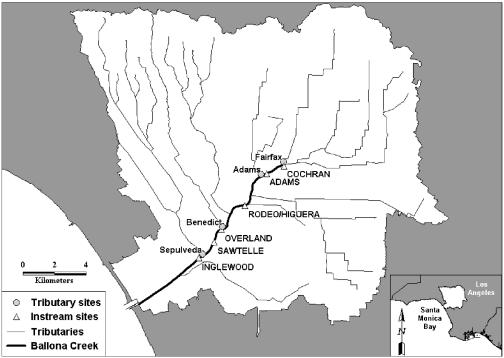
This study quantified inputs of flow, bacterial concentrations, and virus genomic equivalents and then tracked them through an urban watershed over time. This mass-based design was applied in the watershed of Ballona Creek, the largest tributary to the SMB. Ballona Creek is over 85% developed and currently has the largest inputs of fecal indicator bacteria to the SMB (Fig. 1).
FIG. 1.
Map of the Ballona Creek watershed in Los Angeles, Calif. Tributary and main-stem sampling sites for the water quality study are indicated. (Inset) Santa Monica Bay, in Southern California.
Sample collection and filtration.
Samples were collected at six main-stem sites and four major tributaries to Ballona Creek. The six main-stem sites extended from Cochran Ave. (where the system daylights from the underground storm drainage system) to Inglewood Ave. (located at the head of tide just prior to discharge into the SMB) (Table 1). The four tributaries represented the four largest hydrodynamic inputs to the system and were located in reaches between each of the main-stem sampling sites. Flow was calculated as the product of the flow rate and the wetted cross-sectional area (43). Doppler area-velocity sensors (Teledyne ISCO, Los Angeles, CA) were used to measure flow rate. Pressure transducers that measure stage, along with verified as-built cross sections, were used to estimate the wetted cross-sectional area. One-minute instantaneous flow was logged electronically during the entire 6-h sampling period. Both the area-velocity sensors and the pressure transducers were calibrated prior to sampling.
TABLE 1.
Sampling sites along the main stem and major tributaries of Ballona Creek
| Site | Description | GPS coordinates (NAD 83 datum)a |
|---|---|---|
| Cochran Ave. | Main stem/tributary | 34°02.662"N, 118°21.237"W |
| Fairfax drain | Tributary | 34°02.298"N, 118°22.136"W |
| Adams Ave. | Main stem | 34°02.009"N, 118°22.494"W |
| Adams drain | Tributary | 34°02.009"N, 118°22.494"W |
| Rodeo/Higuera | Main stem | 34°01.305"N, 118°22.693"W |
| Benedict Box Channel | Tributary | 34°00.925"N, 118°23.432"W |
| Overland Ave. | Main stem | 33°00.429"N, 118°23.771"W |
| Sawtelle Ave. | Main stem | 33°59.816"N, 118°24.164"W |
| Sepulveda channel | Tributary | 33°59.512"N, 118°24.693"W |
| Inglewood Ave. | Main stem | 33°59.394"N, 118°24.696"W |
GPS, global positioning system; NAD 83 datum, North American datum of 1983.
One-hour composite water samples were collected immediately downstream of flow measurement devices at each site (Table 1, GPS coordinates) between 8:00 and 14:00 on 26 August 2004. The 6-h sampling period corresponds to the approximate hydrodynamic travel time from Cochran Ave. to Inglewood Ave. (1). The hourly 4-liter composite samples at each site were created after combining 10 individual 400-ml grab samples collected every 6 min into a single container. In total, 60 composite samples were collected at Ballona Creek as a result of sampling 6 h at 10 different sites.
After collection, samples were placed on ice and transported immediately to the University of Southern California for processing. For each composite sample, 100 ml of water was devoted to indicator bacteria analysis, and 200 to 600 ml of the sample volume was vacuum filtered through replicate 47-mm-diameter, 0.4-μm-pore-size polycarbonate filters (Poretics, Inc., Livermore, CA) using a filter funnel and receiver (Millipore, Inc., Bedford, MA) for Enterococcus sp. analyses by QPCR or Bacteroides sp. analysis by conventional PCR, as suggested by Haugland et al. (20). In addition, replicate filtrations were also conducted using 47-mm-diameter, 0.45-μm-pore-size type HA (Millipore, Inc., Bedford, MA) mixed cellulose ester filters for enterovirus analysis as suggested by Fuhrman et al. (11). For each filter, the volume filtered was the maximum filterable within ca. 10 min of the start of filtration. The total volume filtered was dependent on the location and turbidity of each individual sample, and the filter volumes were carefully recorded to the nearest 1 ml. All filters were placed in microcentrifuge tubes and stored at −80°C for further analysis.
Analysis of indicator bacteria.
Concentrations of E. coli and Enterococcus spp. were measured by defined substrate technology using kits supplied by IDEXX Laboratories, Inc. (Westbrook, ME) according to the manufacturer's instructions. Briefly, 10-fold and 100-fold dilutions of the water samples were made with deionized water containing the appropriate media and sodium thiosulfate, mixed to dissolve, dispensed into trays (Quanti-Tray/2000), and heat sealed. E. coli was measured using the Colilert-18 reagents, while Enterococcus spp. were measured using Enterolert reagents. Samples were incubated overnight according to the manufacturer's instructions and inspected for positive wells. Conversion of positive wells from these tests to a most probable number was done following Hurley and Roscoe (21).
Extraction of DNA.
The polycarbonate filters were processed for DNA extraction using the UltraClean fecal DNA isolation kit (MoBio Laboratories, Inc., Carlsbad, CA) according to the manufacturer's alternative protocol for maximum yield. Eluted DNA extracts were stored at −20°C until use.
The concentration of the extracted DNA was measured using the Quant-iT Picogreen double-stranded DNA reagent (Invitrogen, Carlsbad, CA) according to the manufacturer's instructions. Standard curves were generated in duplicate using Lambda DNA standards between 2.5 ng/ml and 600 ng/ml and a negative control (0 ng/ml). Fluorometric measurements were made using a Bio-Rad VersaFluor fluorometer.
Bacterial analyses using QPCR.
The total Enterococcus sp. primers and probe are described by Ludwig and Schleifer (28) and were constructed using the 23S rRNA gene regions around the target site of a well-established Enterococcus group-specific primer (ENC854R). Primer ECST748F targets Enterococcus spp., lactococci, and several clostridia. The target site of probe GPL813TQ is present in the 23S rRNA genes from a variety of representatives of gram-positive bacteria with low G+C DNA contents (28).
The master mix contained 1× Taq buffer, 4 mM MgCl2, 3 mM deoxynucleoside triphosphates (dNTPs), 2.5 U Ex Taq R polymerase (R PCR kit for quantitative PCR; TaKaRa Mirus Bio, Madison, WI), 1 μM ENC854R, 1 μM ECST748F, 0.1 μM GPL813 TQ Cy3 Probe (synthesized by MWG Biotech, High Point, NC), and nuclease-free water, yielding a final volume of 20 μl, to which 5 μl of sample (either DNA extract from an environmental sample, ranging from 1 to 76 ng genomic DNA, or 5 μl of lysed cell suspension or genomic equivalents) was added for a final volume of 25 μl. The samples were run under the following optimized assay conditions for PCR: 1 cycle consisting of an initial hold at 95°C for 2 min and 45 cycles of denaturation at 94°C for 15 s and annealing/extension at 60°C for 30 s (the optics were turned on during the annealing step). The Cepheid Smart Cycler II was set with the following specific parameters for this assay. The Dye Set was set for FCTC25. The cycle threshold analysis mode was set for growth curve (linear) analyses, with a manual threshold typically set at 5 to 15 fluorescence units. The background subtract level was set at a minimum of 12 and a maximum of 40. The BoxCar averaging feature was set at zero. The assay was previously optimized for Taq, Mg2+, and dNTP concentrations as well as all cycling parameters (data not shown). For quality control, Enterococcus faecalis ATCC 29212 and Enterococcus faecium ATCC 35667 combined were used as our calibration strains for the total-Enterococcus primer and probe set. Control bacterial preparations were prepared by boiling bacteria for 5 min, centrifuging for 2 min at 12,000 × g at 4°C, and storing immediately on ice. E. faecalis and E. faecium cells were enumerated using SYBR Green I epifluorescence microscopy (30). Serial dilutions of the standards were made in diethyl pyrocarbonate-treated sterile water, and four-point standard curves were run in duplicate in concert with the unknown samples on the Smart Cycler II instrument. Total-Enterococcus primers were tested with all 19 validly described species of the genus Enterococcus and demonstrated amplification of rRNA genes of all strains, with varying efficiencies (28).
Analysis of Bacteroides spp. using conventional PCR.
Amplification of the human-specific Bacteroides/Prevotella marker generally followed the procedure of Bernhard and Field (2), with PCR primers that amplify partial 16S rRNA from the human feces-specific group (BAC708R and HS183F). A range of extracted DNA quantities (representing 1 to 70 ng per assay, with most samples in the range of 5 to 20 ng) was tested to avoid problems with inhibition of the PCR. DNA was amplified with the Bacteroides-Provotella-specific primers described by Bernhard and Field (2). The PCR mixture was 1× Taq polymerase buffer, 1μm each primer, 200 μm dNTPs, 1.25 U Taq polymerase, 640 ng μl−1 BSA, and 1.5 mM MgCl2. The PCR conditions were specifically optimized for this study and differed from those in the original publication: 2 min at 95°C, then 25 cycles of 95°C for 30 s, 60°C for 30 s, and 72°C for 30 s, followed by a 5-min extension at 72°C. Then 1 μl of each PCR product was reamplified using the conditions given above for another 25 cycles. PCR was performed on a 3000MX thermal cycler (Stratagene). PCR products were visualized in a 2% agarose gel stained with 1× SYBR Gold (Molecular Probes, Eugene, OR) and compared to a 100-bp DNA ladder (Promega). Positive results yielded 525-bp amplicons. The positive control was a human fecal sample extracted with a QIAamp DNA stool kit (QIAGEN, Valencia, CA). Negative controls contained water instead of sample. All samples were initially run with 5 μl of extracted material. After the initial analyses, all negative samples (14 of 60) were spiked with 0.1 ng of positive-control DNA and reanalyzed to determine possible inhibition. Three of the 14 negative samples (two from Benedict Box Channel and one from Adams) were determined to be inhibited. Inhibited samples were reanalyzed using 2 μl of sample, but all three remained negative.
Enterovirus analyses.
Samples, along with negative controls, were extracted using a modified RNeasy plant minikit (QIAGEN Plant and Fungi RNA isolation protocol). The RLT homogenization buffer supplied was supplemented with polyvinyl pyrrolidone 40 (PVP-40) at a final concentration of 2%. Prior to the extraction, fresh β-mercaptoethanol (Sigma Chemical Co.) was added to the extraction buffer in the exact concentration recommended by the manufacturer. Filters (type HA) were manually homogenized with a pipette tip, and 700 μl of the RLT/filter slurry was applied to a QiaShredder column (QIAGEN) and spun at maximum speed, ≥8,000 × g, for 2 min to aid in viral lysis as well as to separate filter particles from the filtrate (11). The filtrate was then carefully removed without disturbing the pelleted material and was placed into a new 1.5-ml tube. The volume of solution in each tube was estimated by pipetting, and 0.4 volume of 5 M potassium acetate (pH 6.5) was added. Tubes were mixed by inversion and incubated on ice for 15 min. The mixture was then spun (12,000 × g) at 4°C for 15 to 30 min and the supernatant transferred to a new 1.5-ml microcentrifuge tube. Subsequently, the Plant and Fungi RNA isolation protocol was followed starting at step 5. RNA was eluted with 50 μl of the supplied RNase-free water. A one-step TaqMan QRT-PCR was performed on the extracted RNA, with final reaction volumes of 25 μl, using a QIAGEN One-Step RT-PCR kit. Five microliters of extracted RNA was added to 20 μl of master mix containing 1× RT buffer, 6 mM MgCl2, 500 nM dNTPs, 700 nM EV1 reverse primer (5′-TGTCACCATA AGCAGCCA-3′), 700 nM EV1 forward primer (5′-CCCTGAATGCGGCTAAT-3′), 30 μg bovine serum albumin, 20 U of recombinant RNasin (Promega Corp.), 1.5% PVP-25 (29) (Sigma Chemical Co.), 100 genomic equivalent units of a competitive internal positive control (CIPC) developed in-house (16) by following the general approach of Kleiboeker (25), plus 300 nM CIPC probe 5′-Cy5-TGTGCTGCAAGGCGATTAAGTTGGGT-BHQ-2-3′, and 300 nM EV-BHQ probe 5′-6-carboxyfluorescein-ACGGACACCCAAAGTAGTCGGTTC-BHQ-1-3′, and 1 μl of enzyme mix (containing both reverse transcriptase and DNA polymerase). The probe and primers were synthesized by MWG Biotech, Inc. The Cepheid Smart Cycler II was programmed as follows: a 1-h reverse transcription step at 50°C followed by a 15-min hold at 95°C for DNA polymerase activation, then 45 cycles of 94°C for 15 s (denaturation) and 60°C for 1 min (annealing and extension, with optics on).
QRT-PCR results were available 3 h after the start of analysis, making the total RNA extraction, QRT-PCR preparation, and analysis time less than 5 h. After the first analyses, those samples that appeared to have inhibition of the QRT-PCR (as indicated by the CIPC) had RNA sample volumes reduced in half and were rerun. No inhibition was observed at this lower RNA concentration. Standard curves were generated using a synthetic enterovirus transcript that was quantified using fluorometric analysis, and sample genome concentrations were interpolated from the standard curve using the manufacturer's curve-fitting software. Quantitative results are reported per liter sample volume.
Sequencing of enterovirus QRT-PCR positives.
QRT-PCR-positive samples from the enterovirus analyses were sequenced to assess the specificity and fidelity of our enterovirus QRT-PCR and to elucidate the identities of the enterovirus genomes being amplified by the assay. Following the initial enteroviral analysis of the Ballona Creek samples, RNA samples identified as positive for enterovirus genomes were again amplified using the QRT-PCR protocol. However, after numerous repetitions of freezing and thawing of the extracted RNAs for various analyses, only Cochran Ave. samples from 9:00, 10:00, and 11:00 contained amplifiable enterovirus genetic material (11). The 144-bp enterovirus QRT-PCR product was distinguished from the 126-bp internal positive control product using a 10% polyacrylamide-1× Tris-borate-EDTA gel. The gel was stained with ethidium bromide and visualized with a 254-nm UV light. The larger 144-bp enterovirus QRT-PCR products were excised, homogenized in 50 μl of 1× QIAGEN One-Step RT-PCR buffer using a microcentrifuge pestle, and incubated overnight at 37°C with shaking. The enterovirus products were purified (Wizard SV gel and PCR Clean Up System; Promega), cloned into the 3.9-kilobase pCR 2.1 TOPOVector (TOPO TA cloning kit; Invitrogen, Carlsbad, CA), transformed into TOP10 chemically competent E. coli, and plated on Luria-Bertani agar plates containing 100 μg/ml of ampicillin. Bacterial clones were screened using QPCR, and positive colonies from each of the three sites were selected and grown individually in 3 ml of 2× medium overnight at 37°C. Plasmid DNA was isolated (PerfectPrep plasmid minikit; Eppendorf, Westbury, CT), and the DNA concentration was calculated using yeast extract-tryptone Ribogreen (Molecular Probes). Plasmid DNA was sequenced bidirectionally by MWG Biotech, and the chromatograms were inspected using Sequencher, version 4.2 (Gene Codes Corp., Ann Arbor, MI). The sequences were aligned to sequences in the NCBI GenBank using BLAST and are available by searching nucleotide accession numbers DQ196482 through DQ196487.
Calculations and statistical analyses.
Data analysis comprised four steps. First, the hydrologic budget was evaluated to determine if the majority of the flow was sampled. This evaluation was conducted by comparing the volumetric inputs from each of the tributaries to the volumetric discharges along the main stem of Ballona Creek. The second step was to examine temporal and spatial trends in the flux of fecal indicator bacteria. The flux of indicator bacterial cells per hour was calculated by multiplying the concentration of the indicator bacteria (per deciliter [100 ml]) by the flow rate (in deciliters per hour). The mean hourly flux (temporal analysis) was calculated by averaging the flux of indicator bacteria at all main-stem locations for each hourly interval (n = 6). The mean flux at each site (spatial analysis) was calculated by averaging the flux at each main-stem or tributary site for all hourly samples (n = 6). Statistical analysis of the differences in bacterial flux between hourly time periods or, alternatively, between main-stem sites was conducted using analysis of variance (46). The third data analysis step was to examine spatial and temporal patterns in the frequency of Bacteroides detection. The Bacteroides method used in this study was a presence/absence end point. This examination was conducted by tabulating the locations and time periods of Bacteroides detection to detect patterns moving downstream, adjacent to tributaries, or over time. The fourth data analysis step was to examine the spatial and temporal extent of enterovirus concentrations. Unlike that of Bacteroides, the presence of enterovirus was quantified, so the magnitude of enterovirus concentrations was tabulated among the different locations across the different time periods. As in the Bacteroides data analysis, patterns moving downstream, adjacent to tributaries, or over time were examined.
RESULTS
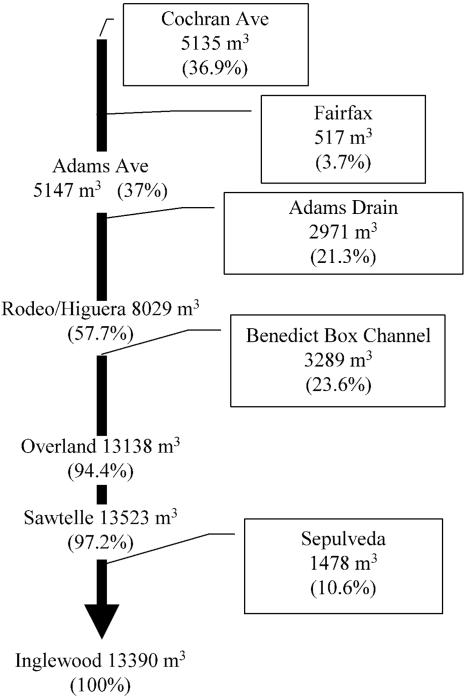
The total volume discharged from Ballona Creek during the 6-h sampling period was 13,390 m3 (Fig. 2). Of this volume, 97% was attributed to monitored inputs from Cochran, Fairfax, Adams, Benedict, and Sepulveda tributaries. The largest volume was contributed at Cochran Ave., where the creek emerges into daylight from beneath downtown Los Angeles. Flow remained relatively stable over the study period at all sites, with little variation or pattern in discharge. For example, the coefficient of variation for flow at the most-downstream site, Inglewood Ave., was less than 8%, approaching the resolution of our flow-monitoring devices.
FIG. 2.
Schematic diagram depicting additive flow in the main channel of Ballona Creek and the percentage contributed by each tributary sampled.
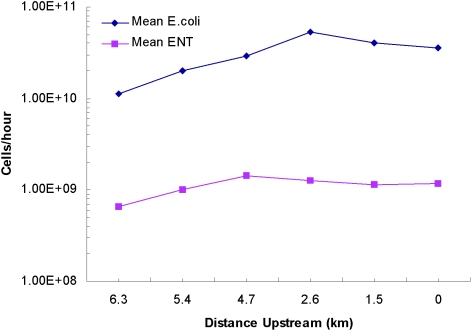
There was no observed spatial trend in the flux of fecal indicator bacteria in Ballona Creek during this study (Fig. 3). The average flux of E. coli ranged from 1.1 × 1010 to 5.3 × 1010 cells h−1 at the six main-stem sites. The average flux of Enterococcus spp. ranged from 6.6 × 108 to 1.4 × 109 cells h−1 at the six main-stem sites. In both cases, there was no discernible increase in bacterial flux as one moved downstream; no two main-stem sites were significantly different from one another in the flux of either E. coli or Enterococcus spp. (P > 0.05 by analysis of variance).
FIG. 3.
Mean flux of E. coli and Enterococcus cells (expressed as cells per hour) at main-channel sampling sites of Ballona Creek (26 August 2004).
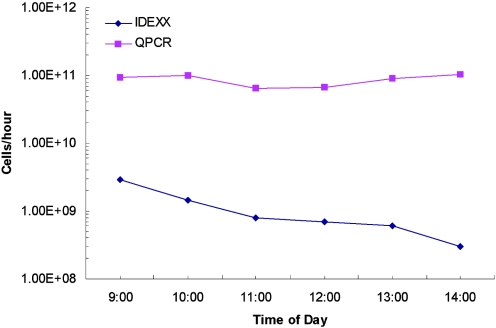
A temporal trend in the flux of fecal indicator bacteria in Ballona Creek was observed during this study (Fig. 4). The average flux of Enterococcus spp. was highest at 9:00 (2.9 × 1010 cells h−1) and decreased monotonically throughout the study period. The lowest flux was measured at 14:00 (3.0 × 109 cells h−1). Similar patterns were observed for E. coli (data not shown). In contrast to the results obtained by the culture-based methods, the QPCR results for Enterococcus spp. did not decrease over time. The flux of Enterococcus spp. ranged from 2.7 × 1010 to 4.7 × 1010 cells h−1, and the 9:00 and 14:00 samples were nearly equivalent (Fig. 4).
FIG. 4.
Mean hourly flux of Enterococcus spp. (expressed as cells per hour) along the main channel of Ballona Creek, measured by using either an IDEXX chromogenic substrate (Enterolert) or QPCR methods on 26 August 2004.
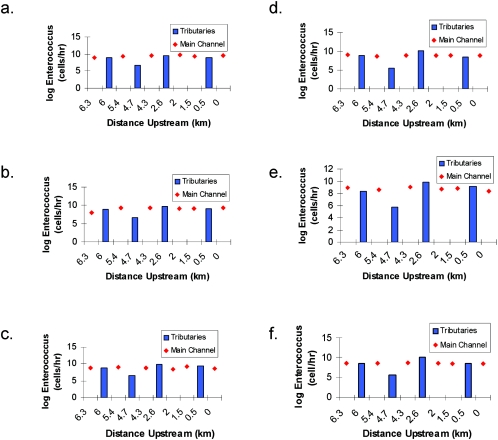
The relative patterns of Enterococcus contributions from the tributaries were similar at all time periods (Fig. 5). Benedict tributary always had the greatest flux of fecal indicator bacteria, followed by Sepulveda, Fairfax, and Adams tributaries. A similar pattern was also observed for E. coli. The flux of Enterococcus spp. from Benedict tributary ranged from 4.1 × 109 to 1.4 × 1010 cells h−1 throughout the sampling period while the flux of Enterococcus spp. from Adams tributary ranged from 3.7 × 105 to 4.4 × 106 cells h−1. On average, Benedict tributary contributed 81% of the Enterococcus spp. loading from all four tributaries.
FIG. 5.
Loading of Enterococcus spp. (expressed as cells per hour) in the main channel and tributaries of Ballona Creek at 9:00 (a), 10:00 (b), 11:00 (c), 12:00 (d), 13:00 (e), and 14:00 (f) on 26 August 2004.
The hourly flux of Enterococcus spp. (determined by culture-based methods) from each of the four main tributaries approximated the load being passed down Ballona Creek (Fig. 5). Regardless of the hour, the flux from each of the tributaries was within a factor of 101 compared to its nearest downstream site on the main stem of Ballona Creek. The only exception was the Adams tributary, for which the flux was as much as 4 orders of magnitude less than that for the nearest downstream site. The main stem showed virtually no response to any of these tributary inputs, including that of Adams. The flux of Enterococcus spp. remained virtually unchanged from upstream to downstream of each of the tributary inputs (Fig. 3 and 5).
The human-specific Bacteroides marker was positively detected throughout the main stem of Ballona Creek; its presence in 31 of the 36 main-stem samples tested (86%) was confirmed (Table 2). A positive signal was observed at all sites during the early morning hours, but during periods of heightened UV radiation (midday and late afternoon), a decrease in the number of positive results for the human-specific Bacteroides marker was observed. For example, only three of six samples at 14:00 were positive for the human-specific Bacteroides marker and exceeded the water quality threshold for E. coli.
TABLE 2.
Number of enterovirus genomes per liter along the main stem of Ballona Creek in Los Angeles, CA
| Distance upstream from Santa Monica Bay (km) (name of site) | No. of enterovirus genomes/liter and presence or absence of the human-specific Bacteroides markera at the following time of day:
|
|||||
|---|---|---|---|---|---|---|
| 9:00 | 10:00 | 11:00 | 12:00 | 13:00 | 14:00 | |
| 6.3 (Cochran) | 3,255* | 1,391* | 1,714* | 1,440* | 1,336* | * |
| 5.4 (Adams) | * | 630* | 200b | 290* | * | * |
| 4.7 (Rodeo/Higuera) | * | 96* | * | 1,641* | 579 | |
| 2.6 (Overland) | * | * | * | 926* | * | * |
| 1.5 (Sawtelle) | * | * | * | 61* | * | 384 |
| 0 (Inglewood) | * | * | * | * | * | * |
Asterisks indicate presence.
PCR inhibited for human-specific Bacteroides marker.
Enteroviruses were detected and quantified in 14 of the 36 main-stem samples tested (39%) (Table 2). Moreover, spatial and temporal patterns in enterovirus concentrations were also evident in the Ballona Creek system. Main-channel locations in the upper reaches of the study area were more likely to be positive for enteroviruses than downstream sites. The most consistently positive site was Cochran Ave., where 89% of the samples contained measurable levels of enterovirus. In addition, some of the highest concentrations of enterovirus were measured at Cochran during four of the six time periods (concentrations ranging from 1,336 to 3,255 enterovirus genomes per liter). A general pattern in enterovirus detection was observed during the course of the day. Enterovirus was detected during the early morning and at midday at upstream sites but was detected most frequently late in the day at the downstream sites. The 12:00 sampling interval had the most frequent detection of enterovirus, with the highest concentrations observed, at the middle sites in the watershed. In nearly all of the tributary samples, no enterovirus was detected (data not shown); only Adams Drain tributary had any detectable enterovirus.
The highest sequence homology observed for the three enterovirus-positive Cochran Ave. samples that were sequenced (>95%) was with human coxsackievirus A22 (GenBank accession no. AF499643), human coxsackievirus A19 (accession no. AF499641), and human enterovirus 90 (accession no. AY773285). The 144-bp QRT-PCR product corresponded to nucleotides 453 to 596 of human coxsackievirus A22, nucleotides 457 to 600 of human enterovirus 90, and nucleotides 454 to 597 of human coxsackievirus A19.
DISCUSSION
The Ballona Creek watershed is a system impacted by fecal pollution. The flux of fecal indicator bacteria was as high at the head of the watershed as it was at the mouth of the creek, where it discharges into the SMB. Although we focused on the flux of these fecal indicator bacteria, it should be noted that 92% of all samples collected from Ballona Creek in this study exceeded the water quality thresholds established by the State of California (CA State Assembly Bill 411). The presence of human enterovirus and of human-specific markers of Bacteroides spp. further characterizes the fecal inputs and should increase an environmental manager's awareness of the human health risks associated with these discharges.
Our study is not the first to examine the presence of viruses in urban runoff entering shorelines in the SMB and other Southern California urban watersheds. For example, Gold and colleagues (13, 14) found viruses in repeated samples from multiple storm drains to the SMB by using both cell culture and RT-PCR techniques. Haile et al. (19) detected human-specific viruses in all three storm drains tested in their epidemiological study of the SMB. Noble and Fuhrman (32) found human enteric virus genomes in the near-shore marine waters of the SMB. Jiang et al. (22) found human adenovirus in samples collected at 12 sites between Malibu and the Mexican border, and Fuhrman et al. (11) previously found human enterovirus genomes in Ballona Creek.
The multitiered approach used in this study can assist watershed managers in determining sources and efficiently abating the most significant inputs of fecal indicator bacteria. If managers relied solely on the patterns in fecal indicator bacteria from Ballona Creek, then the only option would be to treat the entire 37-m3 s−1 discharge furthest downstream at Inglewood Ave., because the flux of fecal indicator bacteria was similar at all sources. The use of multiple tools, however, allows managers to prioritize the most important sources. In this case, the presence of human enterovirus was greatest at the Cochran Ave. site, where the system daylights from the underground storm drain system beneath Los Angeles and the discharge volume is one-third of the volume at Inglewood Ave. Previous studies of Southern California storm drains have detected a human-pathogenic virus signal (22, 23, 31, 32). Since Cochran Ave. had the most frequent occurrence and highest concentrations of enterovirus, plus a consistent co-occurrence of the human-specific Bacteroides marker, this source would appear to be the most likely candidate for future management actions. The sequencing results that confirmed the presence of several potential risks to human health (human coxsackievirus and enterovirus) should provide the reassurance most managers would need before planning future management steps.
The lack of correlation between bacterial indicator levels and levels of human-pathogenic viruses has been observed in previous studies (8, 9) and demonstrates the value of the multitiered approach used here for source identification. For example, analysis of wild shellfish from the Atlantic coast of France indicated no significant correlation between fecal coliforms and enteroviruses or hepatitis A virus (26, 27), and viruses have sometimes been found in oysters without coliform contamination (15, 45). Noble and Fuhrman (32) detected enterovirus in 35% of the 50 shoreline samples they examined over a 5-year period, and no significant statistical relationship to any of the standard bacterial indicators was found. Virus and fecal indicator bacteria were measured in dry-weather urban runoff in drains along 300 km of shoreline from Santa Barbara to San Diego, CA (31). Although 40% of the storm drains contained detectable enterovirus, there was no correlation with concentrations of fecal indicator bacteria. It is also possible that differential rates of degradation of viruses and bacteria can explain much of the discordant relationship between viral pathogens and indicator bacteria (see, e.g., reference 34).
The use of QPCR to measure fecal indicator bacteria presents unique opportunities and challenges. An advantage of using QPCR for measuring fecal indicator bacteria is speed; the method potentially provides measurements in less than 3 h (18). However, culture-based methods quantify only viable bacteria, while QPCR measures the DNA from both cultivable and noncultivable microbes. This was most apparent in the temporal trends from Ballona Creek. Levels of Enterococcus spp. determined using culture-based methods generally decreased as the day progressed, most likely as a result of photoinactivation of cells by sunlight (5, 6, 34). Ballona Creek is a 40-m-wide concrete-lined channel, concentrating solar energy into the shallow creek in the channel invert. The QPCR results, however, remained steady, indicating that the bacterial DNA was still intact and detectable, even though the Enterococcus spp. were not viable.
Overall, the use of multiple approaches provided convincing evidence of the extent and types of microbial contamination in this urban watershed. We believe such studies should provide invaluable information for researchers and managers trying to balance regulatory burdens and public safety.
Acknowledgments
Funding for this project has been provided in full through agreement 02-039-254-0 with the State Water Resources Control Board (SWRCB) pursuant to the Costa-Machado Water Act of 2000 (Proposition 13) and any amendments thereto for the implementation of California's Nonpoint Source Pollution Control Program. Additional support was provided by a USC Sea Grant.
The contents of this document do not necessarily reflect the views and policies of the SWRCB, nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
We thank Orange County Public Health Laboratories (Douglas Moore) for their high-titer poliovirus stock and Ian Hewson, Joshua Steele, Mike Schwalbach, and Sheila O'Brien for assistance with sample collection and processing. We also thank the volunteers of Santa Monica Baykeeper for assistance with sample collection and environmental parameter data collection.
REFERENCES
- 1.Ackerman, D., K. Schiff, and S. B. Weisberg. 2003. Evaluating HSPF in an arid urban watershed, p. 78-85. In S. B. Weisberg (ed.), Southern California Coastal Water Research Project Annual Report 2000-2002. Southern California Coastal Water Research Project, Westminster, Calif.
- 2.Bernhard, A. E., and K. G. Field. 2000. A PCR assay to discriminate human and ruminant feces on the basis of host differences in Bacteroides-Prevotella genes encoding 16S rRNA. Appl. Environ. Microbiol. 66:4571-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernhard, A. E., and K. G. Field. 2000. Identification of nonpoint sources of fecal pollution in coastal waters by using host-specific 16S ribosomal DNA genetic markers from fecal anaerobes. Appl. Environ. Microbiol. 66:1587-1594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boehm, A. B., J. A. Fuhrman, R. D. Morse, and S. B. Grant. 2003. Tiered approach for identification of a human fecal pollution source at a recreational beach: case study at Avalon Bay, Catalina Island, California. Environ. Sci. Technol. 37:673-680. [DOI] [PubMed] [Google Scholar]
- 5.Davies, C. M., and L. M. Evison. 1991. Sunlight and the survival of enteric bacteria in natural waters. J. Appl. Microbiol. 70:265-274. [DOI] [PubMed] [Google Scholar]
- 6.Davies-Colley, R., R. Bell, and A. Donnison. 1994. Sunlight inactivation of enterococci and fecal coliforms in sewage effluent diluted in seawater. Appl. Environ. Microbiol. 60:2049-2058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dick, L., and K. G. Field. 2004. Rapid estimation of numbers of fecal Bacteroidetes by use of a quantitative PCR assay for 16S rRNA genes. Appl. Environ. Microbiol. 70:5695-5697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dufour, A. P. 1984. Bacterial indicators of recreational water quality. Can. J. Public Health 75:49-56. [PubMed] [Google Scholar]
- 9.Elliott, E. L., and R. R. Colwell. 1985. Indicator organisms for estuarine and marine waters. FEMS Microbiol. Rev. 32:61-79. [Google Scholar]
- 10.Fogarty, J., L. Thornton, C. Hayes, M. Laffoy, D. O'Flanagan, J. Devlin, and R. Corcoran. 1995. Illness in a community associated with an episode of water contamination with sewage. Epidemiol. Infect. 114:289-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fuhrman, J. A., X. Liang, and R. T. Noble. 2005. Rapid detection of enteroviruses from small volumes of natural waters by real-time RT-PCR. Appl. Environ. Microbiol. 71:4523-4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Geldreich, E. E. 1978. Bacterial populations and indicator concepts in feces, sewage, stormwater and solid wastes, p. 51-97. In G. Berg (ed.), Indicators of viruses in water and food. Ann Arbor Science Publishers, Ann Arbor, Mich.
- 13.Gold, M., M. Bartlett, J. H. Dorsey, and C. D. McGee. 1990. An assessment of inputs of fecal indicator organisms and human enteric viruses from two Santa Monica storm drains. Santa Monica Bay Restoration Project Commission, Monterey Park, Calif.
- 14.Gold, M., M. Bartlett, C. D. McGee, and G. Deets. 1992. Pathogens and indicators in storm drains within the Santa Monica Bay Watershed. Santa Monica Bay Restoration Project Commission, Monterey Park, Calif.
- 15.Goyal, S. M., W. N. Adams, M. L. O'Malley, and D. W. Lear. 1984. Human pathogenic viruses at sewage sludge disposal sites in the Middle Atlantic region. Appl. Environ. Microbiol. 48:757-763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gregory, J. B., R. W. Litaker, and R. T. Noble. A rapid one step quantitative reverse transcriptase PCR assay with competitive internal positive control for environmental enteroviral detection. Submitted for publication. [DOI] [PMC free article] [PubMed]
- 17.Griffin, D. W., C. J. Gibson III, E. K. Lipp, K. Riley, J. H. Paul III, and J. B. Rose. 1999. Detection of viral pathogens by reverse transcriptase PCR and of microbial indicators by standard methods in the canals of the Florida Keys. Appl. Environ. Microbiol. 65:4118-4125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Griffith, J. F., S. B. Weisberg, and C. D. McGee. 2003. Evaluation of microbial source tracking methods using mixed fecal sources in aqueous test samples. J. Water Health 1:141-151. [PubMed] [Google Scholar]
- 19.Haile, R. W., J. S. Witte, M. Gold, R. Cressey, C. McGee, R. C. Millikan, A. Glasser, N. Harawa, C. Ervin, P. Harmon, J. Harper, J. Dermand, J. Alamillo, K. Barrett, M. Nides, and G.-Y. Wang. 1999. The health effects of swimming in ocean water contaminated by storm drain runoff. Epidemiology 10:355-363. [PubMed] [Google Scholar]
- 20.Haugland, R. A., S. C. Siefring, L. J. Wymer, K. P. Brenner, and A. P. Dufour. Comparison of Enterococcus measurements in freshwater at two recreational beaches by quantitative polymerase chain reaction and membrane filter culture analysis. Water Res. 39:559-568. [DOI] [PubMed]
- 21.Hurley, M. A., and M. E. Roscoe. 1983. Automated statistical analysis of microbial enumeration by dilution series. J. Appl. Bacteriol. 55:159-164. [Google Scholar]
- 22.Jiang, S., R. T. Noble, and W. Chu. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 67:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang, S. C., and W. Chu. 2004. PCR detection of pathogenic viruses in southern California urban rivers. J. Appl. Microbiol. 97:17-28. [DOI] [PubMed] [Google Scholar]
- 24.Kinzelman, J., K. Pond, K. Longmaid, and R. Bagley. 2004. The effect of two mechanical beach grooming strategies on Escherichia coli density in beach sand at a southwestern Lake Michigan beach. Aquat. Ecosyst, Health Management 7:425-432. [Google Scholar]
- 25.Kleiboeker, S. B. 2003. Applications of competitor RNA in diagnostic reverse transcription-PCR. J. Clin. Microbiol. 41:2055-2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LeGuyader, F., V. Apaire-Marchais, J. Brillet, and S. Billaudel. 1993. Use of genomic probes to detect hepatitis A virus and enterovirus RNAs in wild shellfish and relationship of viral contamination to bacterial contamination. Appl. Environ. Microbiol. 59:3963-3968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Leguyader, F., E. Dubois, D. Menard, and M. Pommepuy. 1994. Detection of hepatitis A virus, rotavirus, and enterovirus in naturally contaminated shellfish and sediment by reverse transcription-seminested PCR. Appl. Environ. Microbiol. 60:3665-3671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ludwig, W., and K. H. Schleifer. 2000. How quantitative is quantitative PCR with respect to cell counts? Syst. Appl. Microbiol. 23:556-562. [DOI] [PubMed] [Google Scholar]
- 29.Monpoeho, S., A. Maul, B. Mignotte-Cadiergues, L. Schwartzbrod, S. Billaudel, and V. Ferré. 2001. Best viral elution method available for quantification of enteroviruses in sludge by both cell culture and reverse transcription-PCR. Appl. Environ. Microbiol. 67:2484-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Noble, R. T., and J. A. Fuhrman. 1998. Use of SYBR Green I for rapid epifluorescence counts of marine viruses and bacteria. Aquat. Microb. Ecol. 14:113-118. [Google Scholar]
- 31.Noble, R. T., and J. A. Fuhrman. 2001. Enterovirus detection by reverse transcriptase polymerase chain reaction from the coastal waters of southern California, p. 226-233. In S. B. Weisberg (ed.), Southern California Coastal Water Research Project Annual Report 1999-2000. Southern California Coastal Water Research Project, Westminster, Calif.
- 32.Noble, R. T., and J. A. Fuhrman. 2001. Enteroviruses detected by reverse transcriptase polymerase chain reaction from the coastal waters of Santa Monica Bay, California: low correlation to bacterial indicator levels. Hydrobiologia 460:175-184. [Google Scholar]
- 33.Noble, R. T., S. M. Allen, A. D. Blackwood, W.-P. Chu, S. C. Jiang, G. L. Lovelace, M. D. Sobsey, J. R. Stewart, and D. A. Wait. 2003. Use of viral pathogens and indicators to differentiate between human and non-human fecal contamination in a microbial source tracking comparison study. J. Water Health 1:195-207. [PubMed] [Google Scholar]
- 34.Noble, R. T., I. M. Lee, and K. Schiff. 2004. Inactivation of indicator bacteria from various sources of fecal contamination in seawater and freshwater. J. Appl. Microbiol. 96:464-472. [DOI] [PubMed] [Google Scholar]
- 35.Pina, S., M. Puig, F. Lucena, J. Jofre, and R. Girones. 1998. Viral pollution in the environment and in shellfish: human adenovirus detection by PCR as an index of human viruses. Appl. Environ. Microbiol. 64:3376-3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rose, J. B., X. T. Zhou, D. W. Griffin, and J. H. Paul. 1997. Comparison of PCR and plaque assay for detection and enumeration of coliphage in polluted marine waters. Appl. Environ. Microbiol. 63:4564-4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schiff, K., J. Morton, and S. Weisberg. 2003. Retrospective evaluation of shoreline water quality along Santa Monica Bay beaches. Marine Environ. Res. 56:245-253. [DOI] [PubMed] [Google Scholar]
- 38.SMBRC. 2005. State of Santa Monica Bay: 2004 progress and challenges. Santa Monica Bay Restoration Commission, Los Angeles, Calif.
- 39.Solo-Gabriele, H. M., M. A. Wolfert, T. R. Desmarais, and C. J. Palmer. 2000. Sources of Escherichia coli in a coastal subtropical environment. Appl. Environ. Microbiol. 66:230-237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Stewart, J. R., R. D. Ellender, J. A. Gooch, S. Jiang, S. P. Myoda, and S. B. Weisberg. 2003. Recommendations for microbial source tracking: lessons from a methods evaluation study. J. Water Health 1:225-231. [PubMed] [Google Scholar]
- 41.Tsai, Y., M. D. Sobsey, L. R. Sangermano, and C. J. Palmer. 1993. Simple method of concentrating enteroviruses and hepatitis A virus from sewage and ocean water for rapid detection by reverse transcriptase-polymerase chain reaction. Appl. Environ. Microbiol. 59:3488-3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tsai, Y. L., B. Tran, L. R. Sangermano, and C. J. Palmer. 1994. Detection of poliovirus, hepatitis A virus, and rotavirus from sewage and ocean water by triplex reverse transcriptase PCR. Appl. Environ. Microbiol. 60:2400-2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Viessman, W., G. Lewis, and J. Knapp. 1989. Introduction to hydrology, 3rd ed. Harper & Row, Publishers, Inc., New York, N.Y.
- 44.Weiskel, P. K., B. L. Howes, and G. R. Heufelder. 1996. Coliform contamination of a coastal embayment: sources and transport pathways. Environ. Sci. Technol. 30:1872-1881. [Google Scholar]
- 45.Yamashita, T., K. Sakae, Y. Ishihara, and S. Isomura. 1992. A 2-year survey of the prevalence of enteric viral infections in children compared with contamination in locally harvested oysters. Epidemiol. Infect. 108:155-163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zar, J. 1984. Biostatistics. Prentice-Hall, Pub., Englewood Cliffs, N.J.