Abstract
Although microbes associated with shallow-water corals have been reported, deepwater coral microbes are poorly characterized. A cultivation-independent analysis of Alaskan seamount octocoral microflora showed that Proteobacteria (classes Alphaproteobacteria and Gammaproteobacteria), Firmicutes, Bacteroidetes, and Acidobacteria dominate and vary in abundance. More sampling is needed to understand the basis and significance of this variation.
The most abundant corals on Gulf of Alaska seamounts are octocorals (9), which create a habitat structure for mobile fauna (4). Concerns about the benthic impacts of commercial fishing have renewed interest in habitat-forming deep-sea corals (4). Studies of shallow-water scleractinian corals (12) have revealed a diverse microflora and evidence of host-microbe interactions. Although studies of the deep-sea octocoral microflora are under way (10), there have been no published reports describing the microbial community composition.
Three Gulf of Alaska seamounts were visited during research cruise AT7-15/16 aboard the R/V Atlantis. The biological objectives of the cruise included sampling of deep-sea octocorals for studies of their dispersal and reproductive strategies, with a particular focus on the abundant bamboo corals (Isididae). We took advantage of available coral specimens to examine their associated microflora.
Coral, rock, and water column samples (Table 1) were collected from the Warwick, Murray, and Chirikof seamounts using the deep-submergence vehicle Alvin. Corals and rocks were harvested using the submersible's manipulators and stored in a closed box during ascent to minimize physical disturbance by surface waters. The water adjacent to coral colonies was sampled using a Niskin bottle fired at depth. After submersible recovery, freshly extruded coral exopolysaccharide and scrapings of coral and rock surfaces were transferred to sterile cryovials. Water samples were prefiltered through 20-μm-pore-size Nitex, concentrated using a TFF apparatus (Millipore), and vacuum filtered (1.0-μm and 0.2-μm pore size). The 0.2-μm filter retentate was resuspended in sterile saline solution. Samples were frozen immediately at −70°C and shipped frozen for subsequent processing.
TABLE 1.
Summary of Gulf of Alaska samples from which 16S rRNA gene sequences were obtained
| Sample | Origin | Seamount | Alvin dive no. | Dive depth (m) for sample collection | No. of sequences | Taxonomy (no. of members)
|
FastGroup result (no. of sequences)b
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Phyla | Classes | Orders | Families | Sing | Doub | Trip | Cluster | ||||||
| CGOA | Bamboo coral | Murray | 3805 | 1,100-1,950a | 99 | 11 | 13 | 21 | 23 | 30 | 3 | 1 | 7 |
| CGOD | Black coral | Murray | 3805 | 1,100-1,950a | 47 | 1 | 2 | 2 | 2 | 0 | 0 | 0 | 2 |
| NISA | Water column | Murray | 3798 | 760 | 236 | 7 | 5 | 20 | 22 | 24 | 2 | 1 | 5 |
| BRRA | Rock biofilm | Murray | 3798 | 670-1,094a | 58 | 6 | 7 | 10 | 13 | 27 | 5 | 1 | 1 |
| CGOC | Bamboo coral | Chirikof | 3803 | 2,660-3,300a | 57 | 6 | 8 | 10 | 10 | 12 | 5 | 1 | 2 |
| CGOF | Bamboo coral | Warwick | 3808 | 758 | 343 | 13 | 15 | 22 | 28 | 46 | 13 | 3 | 12 |
| CGOG | Bamboo coral | Warwick | 3806 | 634 | 45 | 5 | 8 | 10 | 11 | 27 | 5 | 2 | 0 |
Depths covered during entire dive (specific sample collection depth not available).
Results of sorting sequences using FastGroup. Sing, unique sequences; Doub, doubletons (two identical sequences); Trip, tripletons (three identical sequences); Cluster, cluster of more than three identical sequences.
Genomic DNAs were extracted using Ultra Clean soil DNA kits (MoBio), and 16S rRNA genes were PCR amplified using primers 27F and 1525R (11) and PlatTaq PCR supermix (Invitrogen). Amplifications were performed with an initial denaturation of 2 min at 94°C, followed by 29 cycles of 30 s at 94°C, 30 s at 55°C, and 2 min at 72°C, with a final extension of 5 min at 72°C. PCR products were cloned using a TOPO TA cloning kit (Invitrogen), and primers M13F and M13R were used to sequence positions 9 to 1545 of the 16S rRNA gene.
BLASTN (1) was used to compare our query sequences with reference sequences from the RDP2 (3) database. Representative sequences from the BLASTN output were aligned with our query sequences, using an RDP2-provided profile alignment. Neighbor-joining trees were created using PHYLIP (6) and used to assign putative taxonomy down to the family level. Detailed phylogenetic trees were constructed using the relevant sequences from each clone library, two reference sequences most closely related to the query sequence, and additional reference sequences. Alignments were generated using the RDP2 profile alignment, and bootstrapped neighbor-joining trees were reconstructed using PHYLIP (6).
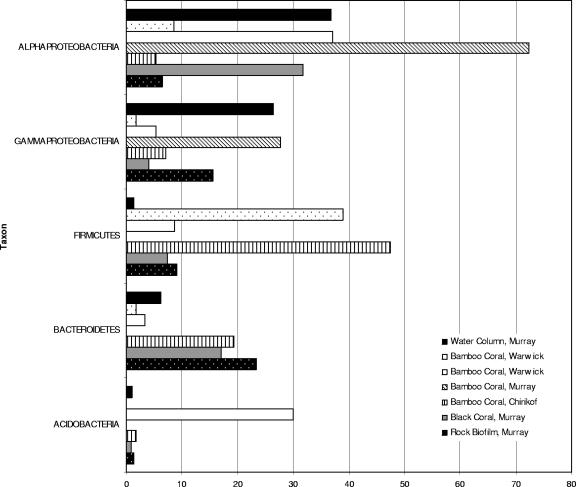
The clones sequenced comprised 19 phyla (see Table S1 in the supplemental material), dominated by Proteobacteria (classes Alphaproteobacteria and Gammaproteobacteria), Firmicutes, Bacteroidetes, and Acidobacteria (Fig. 1). The relative proportions of these groups varied widely across the five coral samples, as did the degree to which a given library was dominated by a single group (Fig. 1; see Table S1 in the supplemental material). At the subphylum level, families occurring in major proportions included Rhizobiaceae, Rhodobacteraceae, and Sphingomonadaceae (Alphaproteobacteria); Pseudomonadaceae, Alteromonadaceae, and Halomonadaceae (Gammaproteobacteria); Bacillaceae, Clostridiaceae, and Mycoplasmataceae (Firmicutes); and Flexibacteraceae and Flavobacteraceae (Bacteroidetes).
FIG. 1.
Histogram showing percentages of composition (by taxon) for 16S rRNA gene libraries generated for this study, showing only taxa comprising at least 20% of sequences in at least one clone library.
Members of the family Rhodobacteraceae and the family Pseudomonadaceae were selected for further analysis, based on their relative abundance and on the previous finding (13) that shallow-water corals contain significantly larger numbers of these bacteria than the surrounding water. Members of the family Rhodobacteraceae comprised 23 to 100% of the alphaproteobacterial sequences in those libraries containing at least 10% alphaproteobacteria. The majority of Rhodobacteraceae sequences obtained in this and previous (12, 13) studies fall within the marine roseobacters (see Fig. S1 in the supplemental material), a major clade of culturable marine heterotrophs (7), many of which play a role in sulfur cycles (e.g., see reference 8). One clade of six CGOF sequences is most closely related to NAC11-6 from a dimethylsulfoniopropionate-producing algal bloom (8), while CGOCA38 groups closely with NAC11-7 (from the same algal bloom study [8]) and an uncultivated marine bacterium, ZD0207, associated with dimethylsulfoniopropionate uptake (15). CGOAB33 is most similar to one (slope strain EI1*) of a group of thiosulfate-oxidizing bacteria from marine sediments and hydrothermal vents (14).
Members of the family Pseudomonadaceae comprised 23 to 69% of the gammaproteobacterial sequences in those samples containing at least 10% gammaproteobacteria. Sequences falling within the pseudomonad tree (see Fig. S2 in the supplemental material) appear most closely related to the oligotrophic marine gammaproteobacteria (OMG) (2). The lack of a close phylogenetic relationship between representatives of the described major OMG clades and our coral sequences suggests that the latter represent new OMG clades.
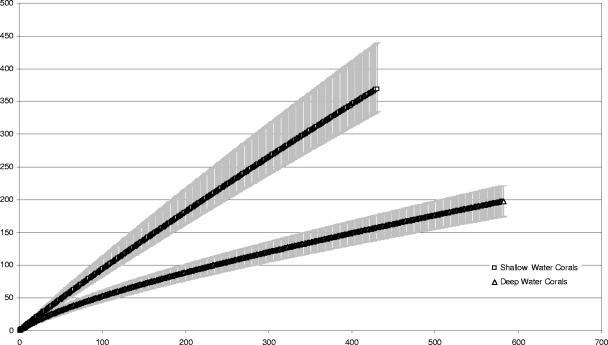
Rarefaction analysis of our data and of sequence data from shallow-water scleractinian coral communities (12) suggested that our accumulated deepwater octocoral samples showed less diversity than their shallow-water counterparts (Fig. 2); with 350 sequences sampled, the shallow-water data set contained approximately twice as many observed operational taxonomic units (97% threshold for operational taxonomic unit definition) as the deepwater set.
FIG. 2.
Rarefaction curves for the accumulated coral-associated 16S rRNA gene sequences generated for this study (CGOA, -C, -D, -F, and -G) and the sequences of Rohwer et al. (12, 13). Bars indicate 95% confidence intervals. Statistical resampling was performed using EstimateS.
This study provides a first glimpse of the deep-sea octocoral microflora. The results suggest that these populations are dominated by several major groups but that the relative proportions of these groups vary (bearing in mind that known methodological biases [5] limit the extent to which clone library compositions reflect community compositions). Phylotypes clustered according to sample origin, and we did not observe much overlap between coral-associated phylotypes and those recovered from the water column and rock surfaces (see Fig. S1 and S2 in the supplemental material), suggesting characteristic coral-associated assemblages with minimal influence of transient water-column microbes. Future sampling of multiple individuals and their immediate environment is clearly needed to perform a more comprehensive survey and to address questions regarding the nutritional relationships, evolution, and biogeography of these populations.
Supplementary Material
Acknowledgments
We thank R/V Atlantis and DSV Alvin personnel, NOAA's Ocean Exploration Program, and Brad Stevens, Randy Keller, Tom Shirley, and Tom Guilderson for help with data acquisition.
Phylogenetic analysis was supported in part by NSF Assembling the Tree of Life grant 0228651 to J.A.E. and N.W.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S., T. Madden, A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cho, J. C., and S. J. Giovannoni. 2004. Cultivation and growth characteristics of a diverse group of oligotrophic marine Gammaproteobacteria. Appl. Environ Microbiol. 70:432-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cole, J., B. Chai, T. Marsh, R. Farris, Q. Wang, S. Kulam, S. Chandra, D. McGarrell, T. Schmidt, G. Garrity, and J. Tiedje. 2003. The Ribosomal Database Project (RDP-II): previewing a new autoaligner that allows regular updates and the new prokaryotic taxonomy. Nucleic Acids Res. 31:442-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Etnoyer, P., and L. Morgan. December. 2003, posting date. Occurrences of habitat-forming deep sea corals in the northeast Pacific Ocean: a report to NOAA's Office of Habitat Conservation. [Online.] http://www.mcbi.org/destructive/DSC_occurrences.pdf.
- 5.Farrelly, V., F. A. Rainey, and E. Stackebrandt. 1995. Effect of genome size and rrn gene copy number on PCR amplification of 16S rRNA genes from a mixture of bacterial species. Appl. Environ. Microbiol. 61:2798-2801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Felsenstein, J. 1989. PHYLIP—phylogeny inference package (version 3.2). Cladistics 5:164-166. [Google Scholar]
- 7.Giovannoni, S., and M. Rappé. 2000. Evolution, diversity, and molecular ecology of marine prokaryotes, p. 47-84. In D. L. Kirchman (ed.), Microbial ecology of the oceans. John Wiley & Sons, New York, N.Y.
- 8.Gonzalez, J. M., R. Simo, R. Massana, J. S. Covert, E. O. Casamayor, C. Pedros-Alio, and M. A. Moran. 2000. Bacterial community structure associated with a dimethylsulfoniopropionate-producing North Atlantic algal bloom. Appl. Environ. Microbiol. 66:4237-4246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Heifetz, J. 2002. Coral in Alaska: distribution, abundance, and species associations. Hydrobiologia 471:19-27. [Google Scholar]
- 10.Kellogg, C., and R. Stone. 2004. A pilot study of deep-water coral microbial ecology. Presented at the ASLO/TOS Ocean Research Conference, Honolulu, Hawaii.
- 11.Rainey, F. A., N. Ward-Rainey, R. M. Kroppenstedt, and E. Stackebrandt. 1996. The genus Nocardiopsis represents a phylogenetically coherent taxon and a distinct actinomycete lineage: proposal of Nocardiopsaceae fam. nov. Int. J. Syst. Bacteriol. 46:1088-1092. [DOI] [PubMed] [Google Scholar]
- 12.Rohwer, F., M. Breitbart, J. Jara, F. Azam, and N. Knowlton. 2001. Diversity of bacteria associated with the Caribbean coral Montastraea franksi. Coral Reefs 20:85-89. [Google Scholar]
- 13.Rohwer, F., V. Seguritan, A. Farooq, and N. Knowlton. 2002. Diversity and distribution of coral-associated bacteria. Mar. Ecol. Prog. Ser. 243:1-10. [Google Scholar]
- 14.Teske, A., T. Brinkhoff, G. Muyzer, D. P. Moser, J. Rethmeier, and H. W. Jannasch. 2000. Diversity of thiosulfate-oxidizing bacteria from marine sediments and hydrothermal vents. Appl. Environ. Microbiol. 66:3125-3133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zubkov, M. V., B. M. Fuchs, S. D. Archer, R. P. Kiene, R. Amann, and P. H. Burkill. 2001. Linking the composition of bacterioplankton to rapid turnover of dissolved dimethylsulphoniopropionate in an algal bloom in the North Sea. Environ. Microbiol. 3:304-311. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.