Abstract
Typing of F-specific RNA (FRNA) coliphages has been proposed as a useful method for distinguishing human from animal fecal contamination in environmental samples. Group II and III FRNA coliphages are generally associated with human wastes, but several exceptions have been noted. In the present study, we have genotyped and partially sequenced group III FRNA coliphage field isolates from swine lagoons in North Carolina (NC) and South Carolina (SC), along with isolates from surface waters and municipal wastewaters. Phylogenetic analysis of a region of the 5′ end of the maturation protein gene revealed two genetically different group III FRNA subclusters with 36.6% sequence variation. The SC swine lagoon isolates were more closely related to group III prototype virus M11, whereas the isolates from a swine lagoon in NC, surface waters, and wastewaters grouped with prototype virus Q-beta. These results suggest that refining phage genotyping systems to discriminate M11-like phages from Q-beta-like phages would not necessarily provide greater discriminatory power in distinguishing human from animal sources of pollution. Within the group III subclusters, nucleotide sequence diversity ranged from 0% to 6.9% for M11-like strains and from 0% to 8.7% for Q-beta-like strains. It is demonstrated here that nucleotide sequencing of closely related FRNA strains can be used to help track sources of contamination in surface waters. A similar use of phage genomic sequence information to track fecal pollution promises more reliable results than phage typing by nucleic acid hybridization and may hold more potential for field applications.
In the United States, approximately 13% of assessed river and stream miles and 15% of assessed estuarine waters are out of compliance with fecal pollution standards, as measured by indicator bacteria (14). These bacterial indicators are incapable of distinguishing among sources of pollution by standard detection methods. Moreover, numerous studies have shown that bacterial indicators are unreliable for detecting the presence of pathogenic viruses (8, 25, 28). F-specific RNA (FRNA) coliphages are candidate alternative indicators of viral pathogens (22) that may also allow sources of fecal contamination to be identified (27, 34, 40, 41).
FRNA coliphages, members of the family Leviviridae, are nonenveloped viruses of 26 nm in diameter possessing single-stranded RNA genomes of 3.8 to 4.2 kb enclosed by a 216-nm capsid with icosahedral symmetry (6). FRNA coliphages comprise two genera (Levivirus and Allolevivirus) and three unclassified groups (a, b, and c) (33). The genus Levivirus contains the MS-2-like (group I) and GA-like (group II) viruses, whereas the genus Allolevivirus contains Q-beta-like (group III) and SP-like (group IV) viruses. These subgroups were initially classified through serological typing (15) and have been confirmed by physiochemical data and nucleotide sequencing.
The genomic organizations of the Levivirus and Allolevivirus genera differ significantly. In addition to the genes coding for the maturation protein, coat protein, and replicase protein, viruses of the Allolevivirus genus (group III and IV phages) also contain a region encoding a carboxyl-terminally extended coat protein, named the readthrough protein (4).
Phage ecology studies have revealed that group II and III FRNA phages are generally associated with human waste, whereas groups I and IV are associated with animal waste (15). These observations led to the use of FRNA coliphage typing to help identify sources of fecal pollution in polluted waters and shellfish (1, 5, 9, 20). Group II and III coliphages, those typically associated with humans, have been detected in surface water sites downstream of wastewater treatment facilities (J. R. Stewart, J. W. Daugomah, D. E. Chestnut, D. A. Graves, M. D. Sobsey, and G. I. Scott, submitted for publication), at human-impacted sites (9), and in proximity to a populated day camp (5). However, the association between coliphage groups and sources is not absolute, as exceptions have been noted. Group I FRNA coliphages have been repeatedly isolated from municipal wastewaters (17, 20), despite the typical association of this subgroup with animals. Group II and III FRNA coliphages have been periodically isolated from animal wastes (9, 38). Sequence information for these strains could provide information as to whether they belong to unique genetic clusters different from GA- and Q-beta-like strains. If this is the case, the current genotyping system could be further refined to better discriminate between FRNA phages from different sources.
For this study, we examined the genetic diversity among group III FRNA phage strains isolated from swine lagoons, municipal wastewater, and surface waters.
MATERIALS AND METHODS
FRNA reference viruses.
FRNA prototype strain Qβ (serogroup III) was kindly provided by K. Furuse (Tokai University, Japan), and FRNA strains M11 (serogroup III) and MX1 (serogroup III) were gifts from J. van Duin (Leiden University, The Netherlands) and originated from the Watanabe collection. In total, 32 group III FRNA coliphage field strains were analyzed in this study, including 23 strains from swine lagoons. Ten of the swine lagoon samples were collected in November 2000 in South Carolina (SC), and 13 were collected in May 2000 in North Carolina (NC). In addition, nine strains (four from wastewater and five from surface water) collected between April and November 2000 from the Catawba (CW) and Saluda (S) watersheds in SC were included.
FRNA genotyping.
All strains were genotyped by direct hybridization with group-specific digoxigenin-labeled probes, as described previously (27). Strains that did not hybridize were further tested with the probes described by Beekwilder et al. (2).
Coliphage RNA isolation.
Coliphage RNAs were isolated from high-titer coliphage stocks using a QIAamp viral RNA mini kit (QIAGEN, Valencia, CA). Overnight enrichments were centrifuged at 3,220 × g for 10 min to pellet the host cells and debris. Viral RNA was then isolated from 140 μl of the supernatant following the manufacturer's instructions.
RT-PCR.
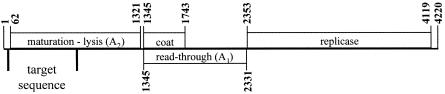
Reverse transcription-PCR (RT-PCR) was conducted using a QIAGEN OneStep RT-PCR kit (QIAGEN, Valencia, CA). Oligonucleotide primers were designed based on the genomic sequences of three prototype group III FRNA coliphages, Q-beta (AY099114), MX1 (AF059242), and M11 (AF052431). The strategy employed was to sequence a region including part of the 5′ untranslated region (UTR) and the 5′ end of the maturation protein (MP) gene (Fig. 1), a region encompassing the targets of the probes described by Hsu et al. (27) and Beekwilder et al. (2). For RT, primer IIIR, which targets a conserved region in the MP genes of all three group III prototype strains, was designed (Table 1). For PCR, two different forward primers, IIF1 (based on strain Q-beta; accession numbers AY099114 and M25014) and IIIF2 (based on strains MX1 and M11; accession numbers AF09242 and AF052431, respectively), were used because the swine lagoon group III isolates did not amplify with primer IIIF1 (Table 1).
FIG. 1.
Genomic organization of group III prototype strain Q-beta (31, 42) showing the region within the maturation protein gene analyzed in this study.
TABLE 1.
Gene-specific primers for RT-PCR and sequencing reactions
| Primer | DNA sequence (5′-3′) | Use(s) | Genomic locationa |
|---|---|---|---|
| IIIF1 | TTT AGG GGG TCA CCT CAC A | RT-PCR, sequencing | 11-29 (Q-beta) |
| IIIF2 | CCC GTA GGG GGG TAC TCT AT | RT-PCR, sequencing | 9 (MX1) |
| IIIR | AGC CAG AGA TTA CCA GCA GTA GC | RT-PCR | 660 (Q-beta and MX1) |
| IIIRQb | TGC GTG TCR GAA GAT TCG | Sequencing | 150 (Q-beta) |
| IIIRMX1 | TTG TCC CAC TCA GCC CTC AT | Sequencing | 347 (MX1) |
Genomic locations are based on sequences presented by Beekwilder et al. (3).
Reverse transcription was conducted at 50°C for 30 min, followed by incubation at 95°C for 15 min. PCR consisted of 30 cycles of 94°C for 45 s, 57°C for 45 s, and 72°C for 60 s. After a final extension of 10 min at 72°C, amplified products (∼650 bp) were analyzed in ethidium bromide-stained 1% agarose gels. Positive Q-beta and MX1 controls and negative water controls were included in each experiment.
DNA sequencing and phylogenetic analysis.
RT-PCR products were gel purified (Qiaquick PCR purification kit; QIAGEN Inc.) and sequenced at the UNC Lineberger Comprehensive Cancer Center sequencing facility (Chapel Hill, NC). DNA sequencing was performed at the UNC-CH Genome Analysis Facility using a 3100 genetic analyzer and an ABI PRISM Big Dye Terminator cycle sequencing ready reaction kit (Applied Biosystems, Foster City, CA). Primers IIIF1 and IIIRQb (for strains that were amplified with IIIF1) or IIIF2 and IIIRMX1 (for strains that were amplified with IIIF2) were employed for sequencing (Table 1).
Nucleotide sequences were aligned using MultAlign software, version 5.4.1 (11, 12). The multiple sequence alignment was then imported into the MEGA 2.1 software package (30) to compute pairwise differences and to create phylogenetic trees. Pairwise differences were computed for 609 nucleotides (nt 42 to 650) for the cluster including M11 and for 588 nucleotides (nt 42 to 629) for strains within the Q-beta cluster. Distance calculations were computed based on a 567-nt alignment of the maturation protein gene. Phylogenetic trees were constructed using neighbor-joining analysis, with distance estimations based on P distances. Confidence values for the internal branches were further analyzed by bootstrap analysis using 500 replications.
Nucleotide sequence accession numbers.
Sequences of 14 FRNA coliphage strains have been deposited into GenBank and assigned accession numbers DQ006814 to DQ006827.
RESULTS
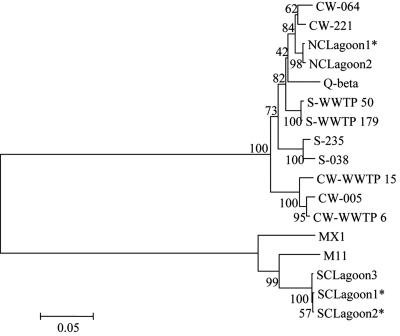
The goal of this study was to determine and interpret the genetic variability among group III FRNA coliphage strains isolated from different sources. Therefore, we sequenced part of the maturation protein gene (Fig. 1) of 32 group III FRNA coliphage strains isolated from swine lagoons, wastewaters, and surface waters. Phylogenetic analysis revealed that the strains could be grouped into two major clusters represented by prototype strains Q-beta and M11. The SC swine lagoon strains could be grouped with M11, whereas the NC swine lagoon isolates and the wastewater and surface water isolates grouped with Q-beta (Fig. 2). Within these two clusters, the nucleotide sequence diversity among the strains was relatively small (0 to 8%). The sequences of the SC swine lagoon isolates displayed 93.1 to 93.3% similarity with the published M11 sequence and 99.7 to 100% similarity with each other. Compared to the published sequence for Q-beta, environmental isolates were 92.2 to 95.9% similar.
FIG. 2.
Phylogenetic relationship among group III FRNA coliphages based on a 567-nt region of the maturation protein gene. The tree includes sequences from Q-beta (AY099114), MX1 (AF059242), and M11 (AF052431). Bootstrap values for the internal nodes are indicated. *, strains that represent multiple isolates with 100% homology (12 for NCLagoon1, 2 for SCLagoon1, and 7 for SCLagoon2). CW, Catawba; S, Saluda; WWTP, wastewater treatment plant.
All of the observed differences in the studied nucleic acid sequences were nucleotide substitutions. No base insertions or deletions were observed. The observed nucleotide substitutions included more transitions (C↔T or A↔G) than transversions. The number of transitional substitutions between the SC swine lagoon isolates and M11 ranged from 33 to 34. For the Q-beta group, the number of transitional substitutions ranged from 22 to 40 compared to the published Q-beta sequence.
Among 10 SC swine lagoon isolates, three unique sequences were observed. One pattern (SC Lagoon 1) was shared by two isolates, another (SC Lagoon 2) was shared by seven isolates, and the third (SC Lagoon 3) was unique to one isolate. Among 13 tested NC swine lagoon isolates, 12 isolates were found to share the same sequence (NC Lagoon 1) and 1 isolate displayed a unique sequence (NC Lagoon 2). Each of the four wastewater and five surface water isolates was found to have a unique sequence. None of the tested isolates had sequences that exactly matched the analogous sequence for Q-beta or M11 (Table 2).
TABLE 2.
Percent nucleotide sequence similarities within a 567-nucleotide stretch of maturation protein genes of group III FRNA coliphage strains analyzed in this studya
| Strain no. | Strain nameb | % Nucleotide sequence similarity with indicated strain
|
||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 16 | 17 | ||
| 1 | Q-beta | 100 | ||||||||||||||||
| 2 | CW-005 | 91.9 | 100 | |||||||||||||||
| 3 | CW-WWTP #6 | 92.6 | 98.9 | 100 | ||||||||||||||
| 4 | CW-WWTP #15 | 91.9 | 97.4 | 97.7 | 100 | |||||||||||||
| 5 | S-235 | 92.9 | 91.7 | 92.1 | 91.7 | 100 | ||||||||||||
| 6 | S-038 | 92.2 | 91.2 | 91.5 | 91.5 | 98.2 | 100 | |||||||||||
| 7 | S-WWTP #50 | 94.9 | 92.9 | 93.3 | 92.6 | 94.5 | 94.2 | 100 | ||||||||||
| 8 | S-WWTP #179 | 95.2 | 92.6 | 92.9 | 92.6 | 94.9 | 94.5 | 99.6 | 100 | |||||||||
| 9 | CW-064 | 94.2 | 92.4 | 93.1 | 92.4 | 93.8 | 93.5 | 96.1 | 96.1 | 100 | ||||||||
| 10 | CW-221 | 95.1 | 92.6 | 93.3 | 92.9 | 94.0 | 93.7 | 96.8 | 96.8 | 97.9 | 100 | |||||||
| 11 | NC Lagoon 1* | 95.6 | 91.7 | 92.4 | 92.4 | 94.5 | 94.2 | 96.5 | 96.8 | 97.5 | 98.2 | 100 | ||||||
| 12 | NC Lagoon 2 | 95.9 | 91.7 | 92.4 | 92.4 | 94.5 | 93.8 | 96.1 | 96.5 | 97.2 | 98.2 | 99.6 | 100 | |||||
| 13 | M11 | 60.0 | 63.1 | 62.6 | 62.1 | 62.8 | 62.8 | 62.4 | 62.1 | 61.9 | 62.3 | 61.2 | 61.4 | 100 | ||||
| 14 | MX1 | 61.4 | 63.7 | 64.2 | 63.3 | 63.7 | 63.7 | 63.7 | 63.7 | 62.4 | 62.4 | 61.9 | 61.7 | 88.5 | 100 | |||
| 15 | SC Lagoon 1* | 61.4 | 64.4 | 64.2 | 64.0 | 63.8 | 63.8 | 63.8 | 63.5 | 63.7 | 63.7 | 62.6 | 62.8 | 92.8 | 90.1 | 100 | ||
| 16 | SC Lagoon 2* | 61.4 | 64.4 | 64.2 | 64.0 | 63.8 | 63.8 | 63.8 | 63.5 | 63.7 | 63.7 | 62.6 | 62.8 | 92.9 | 90.3 | 99.8 | 100 | |
| 17 | SC Lagoon 3 | 61.2 | 64.2 | 64.0 | 63.8 | 64.0 | 64.0 | 64.0 | 63.7 | 63.8 | 63.8 | 62.8 | 63.0 | 92.9 | 90.5 | 99.6 | 99.8 | 100 |
Within-group differences cited in this paper are based on longer sequences and may differ slightly from the numbers shown in this table.
*, strains that represent multiple isolates with 100% homology (12 for NC Lagoon 1, 2 for SC Lagoon 1, and 7 for SC Lagoon 2).
The coliphage isolate from strain CW-005 was included in this study because it was isolated directly downstream of a wastewater treatment plant in the Catawba watershed (CW-WWTP). The sequence for this isolate had the smallest number of pairwise differences with the two analyzed CW-WWTP isolates, differing by 6 bases from one isolate and 15 bases from the other. The same isolate varied from the other Catawba surface water isolates (CW-064 and CW-221) by 44 bases each. Compared to the published Q-beta sequence, this isolate shared 38 nucleotide substitutions with both of the CW-WWTP wastewater isolates. Twelve of these substitutions (at positions 204, 208, 210, 217, 220, 300, 402, 409, 486, 498, 505, and 506) were unique to these three isolates (CW-005, CW-WWTP 6, and CW-WWTP 15), meaning that the base substitutions were not observed among any of the other analyzed isolates or any published sequences. These data genetically link the isolate downstream from the wastewater treatment facility to the wastewater in the facility.
Between the M11 and Q-beta groups, nucleotide sequence divergence ranged from 35.6 to 40.0% (Table 2). Analysis of the deduced amino acid sequences showed that Q-beta and M11 differed in 78 (41%) of the first 190 amino acids. Within the M11 and Q-beta clusters, similarities ranged from 90.5 to 100%. Between the clusters, amino acid sequence similarities ranged from 58.9 to 64.7%.
DISCUSSION
The detection of group II and III coliphages in environmental waters and shellfish is generally considered to be associated with human fecal pollution. This association is based on decades of phage ecology studies, beginning in Asia, where researchers serologically typed thousands of RNA phage isolates from various sources. During the course of these studies, the researchers noted a preferential distribution of RNA phage groups in human and animal hosts (16-18, 36, 37). A similar study in The Netherlands identified only group II and III FRNA coliphages in hospital wastewater and group I and IV coliphages in animal feces (24), and a more recent study conducted on samples from South Africa and Spain demonstrated that the association is significant but not absolute (38). Group III FRNA phages have occasionally been isolated from animal wastes, including waste from swine (9, 38) and gulls (34). For this study, we have partially sequenced group III strains isolated from different sources, including swine lagoons and human-impacted sites, to identify whether specific group III subclusters are associated with specific animal sources. In addition, we identified a close genetic relationship among group III coliphage strains isolated from wastewater effluent and a nearby surface water station, which strongly suggests that DNA sequencing of RT-PCR amplicons from FRNA coliphage genomic targets could help trace the sources of viruses in environmental samples. Genomic targets that could be used for source tracking in this manner include conserved regions of the phage genome (43). Several of the group III SC swine lagoon isolates were unable to hybridize with any of the 5′ UTR probes but did give a signal using a probe that targets a region of the Q-beta maturation protein that is conserved among group III viruses (2). Phylogenetic analysis confirmed that these viruses belong to a different subcluster within group III FRNA viruses, with M11 as the prototype strain. The nucelotide divergence between the two group III clusters was greater than one might expect, even between species, when examining the same gene and likely demonstrates the high mutation rate for these RNA viruses caused by the lack of proofreading ability of virus-encoded RNA polymerase (13).
It is not surprising that all observed variations in the partial maturation protein gene sequences were nucleotide substitutions. This region codes for an expressed protein, and insertions or deletions would lead to frame shifts likely to be lethal to the mutant. Insertion and deletion events were also not observed in the partial sequences of the 5′ UTR. These results contribute indirect evidence to support the hypothesis that the untranslated regions play an important structural role. It is currently understood that the 5′ untranslated region of the RNA coliphages forms a stable 5′-terminal hairpin (21, 35). Olsthoorn et al. (35) speculated that this hairpin is important during replication to ensure that the emerging nascent plus strand folds back on itself instead of annealing with the template strand. Schuppli et al. (39) demonstrated through deletion analysis that the 5′ UTR contains a binding site for Q-beta replicase and so could be the initiation site for plus-strand synthesis.
The direct hybridization system for typing FRNA coliphages described by Hsu et al. (27) has worked well in most field studies, with successful typing of all or most tested isolates (5, 20, 27). These results suggest that M11-like viruses are somewhat rare in the environment or that the swine lagoon group III strains found in our study might be prevalent in swine and overlooked in most studies so far. If future studies confirm that M11-like viruses are more likely to be associated with swine than with humans, the Hsu group III probe may be preferable for microbial source tracking studies aimed at identifying human contamination. For studies in which the identification of all known group III isolates is desired, however, use of the group III DNA probe described by Beekwilder et al. (2) may be more inclusive. Alternatively, a recently described reverse line blot hybridization assay may be used for simultaneous detection and typing of FRNA coliphages. This method employs a reusable membrane to which an array of FRNA typing probes are covalently bound, making the method conducive to standardized high-throughput screening of coliphage isolates (43). A multiplexed PCR for simultaneous detection and typing of FRNA coliphages is also under development, which will allow coliphage typing without isolate cultivation (29).
The identification of group III coliphage isolates associated with swine waste is not consistent with the typical association of group III coliphages with human origins. The fact that the SC swine isolates were more closely related to M11 and the NC swine isolates were more related to Q-beta suggests that distinguishing M11-like from Q-beta-like viruses will not be unconditionally useful for distinguishing group III coliphages with swine origins from those with human origins. The Q-beta-like phages may include viruses of both human and swine origins, and the association of M11-like viruses with swine origins is based on a limited data set. The genetic similarities of human and swine FRNA coliphages, however, are consistent with evidence of considerable similarities in human and swine strains of hepatitis E virus (HEV), a single-stranded RNA virus causing infectious hepatitis in humans and found in both human and swine wastewaters (19).
The similarities observed between isolates within geographic regions could have implications for microbial source tracking. The identification of genetic subclusters within geographic regions may serve to link coliphage isolates in contaminated waters to their origins, such as the clustering of a Catawba surface water isolate with wastewater isolates accomplished with this study. Geographic similarities of human and swine HEV strains have also been observed, indicating a possible zoonotic relationship of swine HEV with human infections (19). It appears that the examination of sequence information is a very useful approach for source tracking, allowing for specific sources to be identified convincingly. A similar use of phage genomic sequence information to track fecal pollution shows promise for field applications, particularly as sequencing large numbers of isolates has become more practical and economical. Commercial and academic sequencing facilities are now generally available to perform high-throughput sequence analysis at a low cost.
Although it is unlikely, a human contribution to swine lagoon water cannot be ruled out as an explanation for the identification of group III coliphages from the water. However, no human waste sources were known to contribute to these lagoons. Schaper et al. (38) identified group III coliphages in pig slurry and wastewater but did not find any group III coliphages among 221 isolates from 37 fecal samples. The inclusion of lagoon samples in microbial source tracking studies is not likely to end soon because of the major use of such lagoons for animal agricultural waste, the magnitude of animal agriculture as a fecal waste source, and the documented impacts of animal waste on water resources (26, 32). Furthermore, the low frequency of FRNA coliphage carriage (7, 10, 23) means that composite samples such as swine lagoons likely contribute higher FRNA coliphage loads to waterways than would the feces of individual pigs. Until the ecology of group III coliphages is better understood, however, it should be assumed that source distinctions based only on genotyping of these viruses are not absolute and that nucleotide sequencing may be needed to better identify specific waste sources.
Acknowledgments
We thank Jan van Duin (University of Leiden, Leiden, The Netherlands) for providing the MX1 and M11 prototype strains, Sharon Long (University of Massachusetts, Amherst) for providing the NC swine lagoon isolates, and Thomas Greig and Lara Adams (NOAA, Charleston, S.C.) for assistance with genetic analysis software.
The National Ocean Service (NOS) does not approve, recommend, or endorse any proprietary product or material mentioned in this publication.
REFERENCES
- 1.Alderisio, K. A., D. A. Wait, and M. D. Sobsey. 1996. Detection and characterization of male-specific RNA coliphages in a New York City reservoir, p. 133-142. In J. J. McDonnell, D. L. Leopold, J. B. Stribling, and L. R. Neville (ed.), Watershed restoration management, New York City water supply studies. American Water Resources Association, Herndon, Va.
- 2.Beekwilder, J., R. Nieuwenhuizen, A. H. Havelaar, and J. van Duin. 1996. An oligonucleotide hybridization assay for the identification and enumeration of F-specific RNA phages in surface water. J. Appl. Bacteriol. 80:179-186. [DOI] [PubMed] [Google Scholar]
- 3.Beekwilder, J., R. Nieuwenhuizen, R. Poot, and J. van Duin. 1996. Secondary structure model for the first three domains of Q beta RNA. Control of A-protein synthesis. J. Mol. Biol. 256:8-19. [DOI] [PubMed] [Google Scholar]
- 4.Bollback, J. P., and J. P. Huelsenbeck. 2001. Phylogeny, genome evolution, and host specificity of single-stranded RNA bacteriophage (family Leviviridae). J. Mol. Evol. 52:117-128. [DOI] [PubMed] [Google Scholar]
- 5.Brion, G. M., J. S. Meschke, and M. D. Sobsey. 2002. F-specific RNA coliphages: occurrence, types, and survival in natural waters. Water Res. 36:2419-2425. [DOI] [PubMed] [Google Scholar]
- 6.Büchen-Osmond, C. (ed.). 2003. Leviviridae. In ICTVdB—The Universal Virus Database, version 3. ICTVdB Management, Columbia University, New York, N. Y.
- 7.Calci, K. R., W. Burkhardt III, W. D. Watkins, and S. R. Rippey. 1998. Occurrence of male-specific bacteriophage in feral and domestic animal wastes, human feces, and human-associated wastewaters. Appl. Environ. Microbiol. 64:5027-5029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chung, H., and M. D. Sobsey. 1993. Comparative survival of indicator viruses and enteric viruses in seawater and sediment. Water Sci. Technol. 27:425-428. [Google Scholar]
- 9.Cole, D., S. C. Long, and M. D. Sobsey. 2003. Evaluation of F+ RNA and DNA coliphages as source-specific indicators of fecal contamination in surface waters. Appl. Environ. Microbiol. 69:6507-6514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cornax, R., M. A. Morinigo, F. Gonzalez-Jaen, M. C. Alonso, and J. J. Borrego. 1994. Bacteriophages presence in human faeces of healthy subjects and patients with gastrointestinal disturbances. Zentbl. Bakteriol. 281:214-224. [DOI] [PubMed] [Google Scholar]
- 11.Corpet, F. 28. March 2000, posting date. Multiple sequence alignment by Florence Corpet. [Online.] http://prodes.toulouse.inra.fr/multalin/multalin.html.
- 12.Corpet, F. 1988. Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res. 16:10881-10890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Drake, J. W., B. Charlesworth, D. Charlesworth, and J. F. Crow. 1998. Rates of spontaneous mutation. Genetics 148:1667-1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.EPA. 2000. 2000 national water quality inventory. Report no. EPA-841-R-02-001. Office of Water, EPA, Washington, D.C.
- 15.Furuse, K. 1987. Distribution of coliphages in the environment: general considerations, p. 87-124. In S. M. Goyal, C. P. Gerba, and G. Bitton (ed.), Phage ecology. John Wiley & Sons, New York, N.Y.
- 16.Furuse, K., A. Ando, S. Osawa, and I. Watanabe. 1981. Distribution of ribonucleic acid coliphages in raw sewage from treatment plants in Japan. Appl. Environ. Microbiol. 41:1139-1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Furuse, K., A. Ando, and I. Watanabe. 1975. Isolation and grouping of RNA phages. V. A survey in the islands in the adjacent seas of Japan. J. Keio. Med. Soc. 52:259-263. [Google Scholar]
- 18.Furuse, K., T. Sakurai, A. Hirashima, M. Katsuki, A. Ando, and I. Watanabe. 1978. Distribution of ribonucleic acid coliphages in south and east Asia. Appl. Environ. Microbiol. 35:995-1002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goens, S. D., and M. L. Perdue. 2004. Hepatitis E viruses in animals and humans. Anim. Health Res. 5:145-156. [DOI] [PubMed] [Google Scholar]
- 20.Griffin, D. W., R. Stokes, J. B. Rose, and J. H. Paul III. 2000. Bacterial indicator occurrence and the use of an F(+) specific RNA coliphage assay to identify fecal sources in Homosassa Springs, Florida. Microb. Ecol. 39:56-64. [DOI] [PubMed] [Google Scholar]
- 21.Groeneveld, H., K. Thimon, and J. van Duin. 1995. Translational control of maturation-protein synthesis in phage MS2: a role for the kinetics of RNA folding? RNA 1:79-88. [PMC free article] [PubMed] [Google Scholar]
- 22.Havelaar, A. H., M. van Olphen, and Y. C. Drost. 1993. F-specific RNA bacteriophages are adequate model organisms for enteric viruses in fresh water. Appl. Environ. Microbiol. 59:2956-2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Havelaar, A. H., K. Furuse, and W. M. Hogeboom. 1986. Bacteriophages and indicator bacteria in human and animal faeces. J. Appl. Bacteriol. 60:255-262. [DOI] [PubMed] [Google Scholar]
- 24.Havelaar, A. H., W. M. Pot-Hogeboom, K. Furuse, R. Pot, and M. P. Hormann. 1990. F-specific RNA bacteriophages and sensitive host strains in faeces and wastewater of human and animal origin. J. Appl. Bacteriol. 69:30-37. [DOI] [PubMed] [Google Scholar]
- 25.Hernroth, B. E., A. C. Conden-Hansson, A. S. Rehnstam-Holm, R. Girones, and A. K. Allard. 2002. Environmental factors influencing human viral pathogens and their potential indicator organisms in the blue mussel, Mytilus edulis: the first Scandinavian report. Appl. Environ. Microbiol. 68:4523-4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill, V. R., and M. D. Sobsey. 2001. Removal of Salmonella and microbial indicators in constructed wetlands treating swine wastewater. Water Sci. Technol. 44:215-222. [PubMed] [Google Scholar]
- 27.Hsu, F. C., Y. S. Shieh, J. van Duin, M. J. Beekwilder, and M. D. Sobsey. 1995. Genotyping male-specific RNA coliphages by hybridization with oligonucleotide probes. Appl. Environ. Microbiol. 61:3960-3966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jiang, S., R. Noble, and W. Chu. 2001. Human adenoviruses and coliphages in urban runoff-impacted coastal waters of Southern California. Appl. Environ. Microbiol. 67:179-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kirs, M., and D. C. Smith. 2005. Presented at the 105th General Meeting of the American Society for Microbiology, Atlanta, Ga., 5 to 9 June 2005.
- 30.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 31.Mekler, P. 1981. Determination of nucleotide sequences of the bacteriophage QB genome: organization and evolution of an RNA virus. Ph.D. dissertation. University of Zurich, Zurich, Switzerland.
- 32.Miner, J. R. 1999. Alternatives to minimize the environmental impact of large swine production units. J. Anim. Sci. 77:440-444. [DOI] [PubMed] [Google Scholar]
- 33.Murphy, F. A., C. M. Fauquet, D. H. L. Bishop, S. A. Ghabrial, A. W. Jarvis, G. P. Martelli, M. A. Mayo, and M. D. Summers. 1995. Virus taxonomy: the classification and nomenclature of viruses. Springer-Verlag, Vienna, Austria.
- 34.Noble, R. T., S. A. Allen, A. D. Blackwood, W. Chu, S. C. Jiang, G. L. Lovelace, M. D. Sobsey, J. R. Stewart, and D. A. Wait. 2003. Use of viral pathogens and indicators to differentiate between human and non-human fecal contamination in a microbial source tracking comparison study. J. Water Health 1:195-204. [PubMed] [Google Scholar]
- 35.Olsthoorn, R. C., G. Garde, T. Dayhuff, J. F. Atkins, and J. Van Duin. 1995. Nucleotide sequence of a single-stranded RNA phage from Pseudomonas aeruginosa: kinship to coliphages and conservation of regulatory RNA structures. Virology 206:611-625. [DOI] [PubMed] [Google Scholar]
- 36.Osawa, S., K. Furuse, and I. Watanabe. 1981. Distribution of ribonucleic acid coliphages in animals. Appl. Environ. Microbiol. 41:164-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Osawa, S., K. Furuse, M. S. Choi, A. Ando, T. Sakurai, and I. Watanabe. 1981. Distribution of ribonucleic acid coliphages in Korea. Appl. Environ. Microbiol. 41:909-911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schaper, M., J. Jofre, M. Uys, and W. O. Grabow. 2002. Distribution of genotypes of F-specific RNA bacteriophages in human and non-human sources of faecal pollution in South Africa and Spain. J. Appl. Microbiol. 92:657-667. [DOI] [PubMed] [Google Scholar]
- 39.Schuppli, D., I. Barrera, and H. Weber. 1994. Identification of recognition elements on bacteriophage Q beta minus strand RNA that are essential for template activity with Q beta replicase. J. Mol. Biol. 243:811-815. [DOI] [PubMed] [Google Scholar]
- 40.Scott, T. M., J. B. Rose, T. M. Jenkins, S. R. Farrah, and J. Lukasik. 2002. Microbial source tracking: current methodology and future directions. Appl. Environ. Microbiol. 68:5796-5803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Simpson, J. M., J. W. Santo Domingo, and D. J. Reasoner. 2002. Microbial source tracking: state of the science. Environ. Sci. Technol. 36:5279-5288. [DOI] [PubMed] [Google Scholar]
- 42.van Duin, J. 1998. Single-stranded RNA bacteriophages, p. 117-167. In R. Calendar (ed.), The bacteriophages. Plenum Press, New York, N.Y.
- 43.Vinjé, J., S. J. Oudejans, J. R. Stewart, M. D. Sobsey, and S. C. Long. 2004. Molecular detection and genotyping of male-specific coliphages by reverse transcription-PCR and reverse line blot hybridization. Appl. Environ. Microbiol. 70:5996-6004. [DOI] [PMC free article] [PubMed] [Google Scholar]