Abstract
In this study, the microbial community succession in a thermophilic methanogenic bioreactor under deteriorative and stable conditions that were induced by acidification and neutralization, respectively, was investigated using PCR-mediated single-strand conformation polymorphism (SSCP) based on the 16S rRNA gene, quantitative PCR, and fluorescence in situ hybridization (FISH). The SSCP analysis indicated that the archaeal community structure was closely correlated with the volatile fatty acid (VFA) concentration, while the bacterial population was impacted by pH. The archaeal community consisted mainly of two species of hydrogenotrophic methanogen (i.e., a Methanoculleus sp. and a Methanothermobacter sp.) and one species of aceticlastic methanogen (i.e., a Methanosarcina sp.). The quantitative PCR of the 16S rRNA gene from each methanogen revealed that the Methanoculleus sp. predominated among the methanogens during operation under stable conditions in the absence of VFAs. Accumulation of VFAs induced a dynamic transition of hydrogenotrophic methanogens, and in particular, a drastic change (i.e., an approximately 10,000-fold increase) in the amount of the 16S rRNA gene from the Methanothermobacter sp. The predominance of the one species of hydrogenotrophic methanogen was replaced by that of the other in response to the VFA concentration, suggesting that the dissolved hydrogen concentration played a decisive role in the predominance. The hydrogenotrophic methanogens existed close to bacteria in aggregates, and a transition of the associated bacteria was also observed by FISH analyses. The degradation of acetate accumulated during operation under deteriorative conditions was concomitant with the selective proliferation of the Methanosarcina sp., indicating effective acetate degradation by the aceticlastic methanogen. The simple methanogenic population in the thermophilic anaerobic digester significantly responded to the environmental conditions, especially to the concentration of VFAs.
Anaerobic methanogenic bioreactors have been widely used for the treatment of municipal and industrial wastewater (26, 40, 45). A variety of microorganisms coexist in the methanogenic bioreactor, and their appropriate combination is necessary for the conversion of organic compounds to methane (53). The methanogenic process can be divided into three major metabolic steps, i.e., hydrolysis, acidogenesis, and methanogenesis. The last step is considered to be the rate-limiting step and can be performed only by a specialized functional group of microorganisms, the methanogenic archaea. The physiological characteristics of methanogens, i.e., the inherent low growth rate and high susceptibility to external conditions such as pH, temperature, or chemical substrates, have been thought to make the whole process sensitive to environmental change (5, 15).
Although engineering of reactor design based on empirical investigations has been used to avoid process deterioration (49), unexpected process failures sometimes occur. In most cases of process deterioration, volatile fatty acids (VFAs) such as acetate or propionate accumulate and methane gas production is reduced. The anaerobic microbial degradation of VFAs has been intensively investigated using pure-culture assays (19, 20). Among VFAs, acetate is directly degraded by aceticlastic methanogens. In addition, syntrophic association between methanogens and proton-reducing bacteria converts VFAs to methane (38, 42).
On the other hand, the recent application of molecular ecological techniques enables us to get an overview of the microbial community without cultivation. In-depth knowledge of the methanogenic community succession during the deteriorative and recovery phases is thought to be of particular importance to obtain insight into the stability of the process. However, almost all molecular analyses have focused on batch or continuous culture under steady-state conditions (8, 12, 23). There appears to have been little examination of the microbial succession during the deteriorative and recovery phases. Only one investigation targeting one mesophilic process has been reported so far (11).
The thermophilic methanogenic process has attracted a great deal of attention due to the feasible removal efficiency of high-strength wastewater. Despite the high-level performance of the process, only a little is known about the microbial community structure and dynamics. Recently, investigations concerning the linkage between the microbial population and reactor conditions in thermophilic processes perturbed by sulfate addition or operational failure (i.e., heat shock and pump failure) were reported (7, 30). However, the sampling interval for the community analyses was too long to characterize the microbial community corresponding to each operational phase. More precise monitoring of the community dynamics during each operational phase and the transition state is thought to be of particular significance. The objective in this study was to characterize the microbial community structure and dynamics in a thermophilic process under deteriorative and stable conditions that were induced by acidification and neutralization, respectively. Acidification of the methanogenic bioreactor has been widely understood to be one of the most common problems inducing process deterioration (21, 48). The responses of archaeal and bacterial communities to these reactor conditions were investigated using a combination approach of process analyses and molecular ecological techniques, i.e., PCR-mediated single-strand conformation polymorphism (SSCP) based on the 16S rRNA gene, quantitative PCR, and fluorescence in situ hybridization (FISH).
MATERIALS AND METHODS
Operation of the thermophilic methanogenic bioreactor.
The seed culture used in this study was the anaerobic digestion sludge collected from a thermophilic process for treating garbage wastewater (50). The methanogenic microflora was cultivated at 55°C in a 1.4-liter stirred tank reactor (Labo-controller MDL-8L; B. E. Marubishi, Tokyo, Japan) continuously agitated with a magnetic stirrer at 100 rpm. Synthetic wastewater containing glucose (1%, wt/vol) as a sole carbon and energy source was continuously supplied to the reactor at a dilution rate of 0.1 day−1. The synthetic wastewater (pH 7.2) consisted of (in grams liter−1) KH2PO4, 0.908; Na2HPO4 · 12H2O, 2.39; NH4Cl, 0.5; MgCl2 · 6H2O, 0.18; yeast extract (Difco), 2.0; and glucose, 10.0; as well as 10 ml of a vitamin solution containing (in milligrams liter−1) biotin, 2.0; folic acid, 2.0; pyridoxine HCl, 10.0; thiamine HCl, 5.0; riboflavin, 5.0; nicotinic acid, 5.0; dl-calcium pantothenate, 5.0; vitamin B12, 0.1; p-aminobenzoic acid, 5.0; and lipoic acid, 5.0. A mineral solution was injected directly into the reactor once a day at the following final concentrations (milligrams liter−1): FeSO4 · 7H2O, 5.53; CoCl2 · 6H2O, 0.48; ZnCl2, 0.67; CaCl2 · 2H2O, 0.59; CuCl2 · 2H2O, 0.16; MnCl2 · 4H2O, 2.02; H3BO4, 0.063; Na2MoO4 · 2H2O, 0.0045; and NiCl2 · 6H2O, 0.65. The pH was not controlled for the first 45 days. Thereafter, the pH value was maintained at 7.1 during the remainder of the operation by automatic titration with 5 N NaOH. Gas production was measured daily with a gas meter based on the liquid displacement. Culture solution was taken every 3 days for the following chemical analyses during the 90-day operation. The concentration of VFAs was determined using a liquid chromatograph (L6300; Hitachi, Tokyo, Japan) equipped with a TSKgel Oapak-A column (Tohso, Tokyo, Japan) and a UV spectrophotometric detector (SPD-7A; Shimadzu, Tokyo, Japan). Inorganic carbon (IC) was determined with a total organic carbon analyzer (TOC-V CSN; Shimadzu). For the microbial analyses, the reactor was sampled on days 0, 9, 18, 30, 39, 44, 48, 51, 56, 63, 69, 78, and 90.
DNA extraction and PCR amplification of partial 16S rRNA genes.
The genomic DNA was extracted from the culture sample by the benzyl chloride method (54) and was used as the PCR template. PCR amplification was performed using AmpliTaqGold (Applied Biosystems, Tokyo, Japan) according to the manufacturer’s instructions for single-strand conformation polymorphism analysis. The PCR primers, which have been reported previously (6, 17, 47, 52), were modified to widen the availability on the basis of the nucleotide database updated in 2004 using the ARB program (http://www.arb-home.de/) (Table 1). The availability of the primers was confirmed using the Probe_Match tool of the Ribosomal Database Project II (http://rdp.cme.msu.edu/). The primer sets used in this study are listed in Table 1. The PCR primer set B342If/U806Ir was used to amplify the bacterial 16S rRNA gene. The archaeal 16S rRNA gene was amplified by nested PCR. The primer sets U341If/A1399r and A348If/U806Ir were used for the first and second PCRs, respectively. The reverse primer U806Ir was phosphorylated at the 5′ end for the single-strand digestion. The bacterial PCR or archaeal second PCR was started with an initial denaturation for 10 min at 95°C. A total of 25 cycles, each including 60 s at 93°C, 60 s at 50°C, and 70 s at 72°C, was followed by a final extension step of 5 min at 72°C. The thermal profile of the archaeal first PCR was as described above except that 35 cycles were applied for amplification. The products were examined by electrophoresis on 1.5% agarose gels before being subjected to SSCP.
TABLE 1.
Primers used in this study
| Primera | Sequence (5′-3′)b | Positionsc | Target | Referenced |
|---|---|---|---|---|
| U341If | CCTACGGGIIGCIICAG | 341-357 | Universal | 17 |
| B342If | CTACGGGIGGCIGCAGT | 342-358 | Bacteria | 52 |
| A348If | GGIGCAICAGGCGCGAAA | 348-365 | Archaea | 6 |
| U806Ir | GGACTACCIGGGTITCTAA | 788-806 | Universal | 47 |
| A1399r | GTGTGTGCAAGGAGCAG | 1383-1399 | Archaea | 17 |
| Mc412f | CTGGGTGTCTAAAACACACCCAA | 412-435 | Methanoculleus | This study |
| Mc578r | ATTGCCAGTATCTCTTAG | 578-598 | Methanoculleus | This study |
| Ms413f | CAGATGTGTAAAATACATCTGTT | 413-436 | Methanosarcina | This study |
| Ms578r | TCTGGCAGTATCCACCGA | 578-598 | Methanosarcina | This study |
| Mt392f | ACTCTTAACGGGGTGGCTTTT | 392-413 | Methanothermobacter | This study |
| Mt578r | TCATGATAGTATCTCCAGC | 578-598 | Methanothermobacter | This study |
f, forward primer; r, reverse primer.
I, inosine.
The numbering is based on the E. coli 16S rRNA gene.
The original primers in the literature were modified (see the text for details).
SSCP analysis.
SSCP analysis was carried out according to the procedure of Schmalenberger and Tebbe (37). The PCR products were purified with a QIAquick purification kit (Qiagen, Tokyo, Japan) according to the manufacturer's instructions. The purified PCR product was digested with lambda exonuclease (New England BioLabs, Beverly, MA) at 37°C for 1 h. The single-stranded DNA was purified by phenol-chloroform extraction. Agarose gel electrophoresis was used to confirm the equivalence of the amounts of DNA subjected to SSCP electrophoresis. Prior to SSCP electrophoresis, the DNA suspended in 12 μl of 10 mM Tris-HCl, pH 8.0, and 12 μl of denaturing loading buffer (95% [vol/vol] formamide, 10 mM NaOH, 0.025% [wt/vol] bromophenol blue, and 0.025% [wt/vol] xylene cyanol) was boiled for 2 min and cooled immediately on ice. For electrophoresis, we used a gel matrix (MDE; FMC Bioproducts, Rockland, ME) at a final concentration of 0.675-fold and prepared a gel of 16 cm in length, 16 cm in width, and 1 mm in thickness in 0.5× TBE (45 mM Tris-HCl, 45 mM boric acid, 1 mM EDTA, pH 8.0). Electrophoresis was performed at 20°C and 400 V for 20 h. The running buffer was 0.5× TBE. DNA in the gels was visualized by silver staining.
Statistical analysis of SSCP data.
For statistical analysis, principal-component analysis (PCA) of the SSCP data was performed using the software JMP version 5.1 (SAS Institute, Tokyo, Japan). The SSCP fingerprints were converted to binary data with the migration and presence of bands by using the software Quantity One version 3.1 (Toyobo, Tokyo, Japan), and the data were subjected to PCA. A covariance data matrix was extracted with pairwise deletion and varimax factor rotation. Data reduction provided a two-factorial ordering of the variance of SSCP profiles, which was plotted as a map.
Sequencing and phylogenetic analysis.
The selected bands were excised with a clean razor blade, and the single-stranded DNA was eluted from the gel by the “crush-and-soak” procedure (35). The DNA was recovered by using a QIAEX gel extraction kit (QIAGEN) and then was resuspended in 10 μl of 10 mM Tris-HCl, pH 8.0. PCR reamplification and preparation of single-stranded DNA were performed as described above. The single-stranded DNA was examined by SSCP electrophoresis to confirm that the bands were present at the same positions on the first SSCP profile.
The PCR products amplified from the DNA in the SSCP bands were ligated into the plasmid vector pGEM-T Easy (Promega, Tokyo, Japan), and the ligation mixture was used to transform Escherichia coli JM109 supercompetent cells by a protocol provided by the manufacturer. Cloned 16S rRNA gene segments were sequenced using the ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Applied Biosystems) and ABI PRISM model 377 genetic analyzer (Applied Biosystems). It was confirmed by sequencing more than three times that the clones recovered from one band had the same nucleotide sequence. The primers for sequencing were T7 (5′-TAATACGACTCACTATAG-3′) and SP6 (5′-ATTTGGTGACACTATAGAAT-3′). The 16S rRNA gene sequences obtained were compared with those from the DDBJ nucleotide sequence database by using the BLAST program.
Quantitative PCR of different groups of methanogens.
On the basis of the alignment of the sequences recovered from the three bands detected in the archaeal SSCP profile and of the 16S rRNA gene sequences from methanogens in the database, three sets of specific primers were designed to distinguish the sequences of the respective methanogens (Table 1). The specificity was checked with the Probe_Match tool of the Ribosomal Database Project II and was confirmed by the lack of PCR amplification with the nontargeted cloned bands. The purified DNA fragments amplified from these clones by using the primer set A348If and U806Ir were used as a template to obtain the standard curve. The number of control PCR fragments in the reaction mixture was varied between 0 and 108 copies. The concentration of genomic DNA extracted from the bioreactor was determined spectrophotometrically, and 10 ng of the DNA was used as the template for quantitative PCR.
A LightCycler (Roche Diagnostics, Tokyo, Japan) was used with LightCycler-FastStart DNA Master SYBR Green I (Roche Diagnostics). Each reaction mixture consisted of the following: LightCycler-FastStart DNA Master SYBR Green I, 2 μl; 25 mM MgCl2, 2.4 μl; forward and reverse specific primers (10 pmol μl−1), 1 μl each; PCR grade distilled water, 18 μl; and DNA solution, 2 μl. Real-time PCR was started with an initial denaturation at 95°C for 10 min. Subsequently, the target DNA was amplified in 35 cycles. Each cycle consisted of denaturation for 5 s at 95°C, annealing for 6 s at 65°C (for the band 2 clone [Methanosarcina-related species]) or at 62°C (for the band 1 clone [Methanoculleus-related species] and the band 3 clone [Methanothermobacter-related species]), and extension for 20 s at 72°C. Fluorescence was detected at the end of the extension reaction. The temperature transition rate was 20°C s−1 except in the case of the annealing temperature, where it was 2°C s−1. The specificity of the amplified PCR product was assessed by performing a melting curve analysis, which consisted of denaturation at 95°C, annealing for 15 s at 70°C, and gradual denaturation with a temperature transition rate of 0.1°C s−1 until 95°C with continuous detection of fluorescence.
Fluorescence in situ hybridization.
The culture cells were gently washed with phosphate-buffered saline (0.13 M NaCl, 10 mM Na2HPO4, pH 7.2) and were centrifuged at 12,000 × g for 2 min. The cells were fixed in 4% paraformaldehyde at 4°C for 20 h and stored in 99% ethanol-phosphate-buffered saline (1:1). To allow probes to penetrate the cells in the culture samples, five freeze-and-thaw cycles were performed after fixation (41). Just before hybridization, the samples were incubated in a 99% ethanol-formalin solution (9:1) for 10 min at room temperature (18).
An appropriate volume of the fixed sample was centrifuged at 12,000 × g for 2 min to remove the buffer solution, and the cell pellet obtained was suspended in a hybridization buffer (0.9 M NaCl, 0.01% sodium dodecyl sulfate, 20 mM Tris-HCl, pH 7.2, and 15% formamide) containing the fluorescently labeled probes (0.5 pmol/μl). After incubation at 46°C for 2 h, the buffer was replaced with washing solution (200 mM NaCl, 0.01% sodium dodecyl sulfate, 20 mM Tris-HCl, pH 7.2, and 5 mM EDTA). The sample was incubated at 46°C for 20 min and centrifuged at 12,000 × g for 2 min. The cell pellet was suspended in 50% (vol/vol) glycerol solution, and the sample obtained was observed by confocal laser scanning microscopy (Fluoview FV500; Olympus, Tokyo, Japan). Two domain-specific probes were used in this study. EUB338 (5′-GCTGCCTCCCGTAGGAGT-3′) for detection of almost all Bacteria was 5′ end labeled with rhodamine (3). ARC915 (5′-GTGCTCCCCCGCCAATTCCT-3′) for detection of almost all Archaea was 5′ end labeled with Alexa488 (46).
Nucleotide sequence accession numbers.
The nucleotide sequence data obtained in this study have been deposited in the DDBJ nucleotide sequence database under accession numbers AB200367, AB200368, and AB200369.
RESULTS
Performance of the thermophilic methanogenic bioreactor.
An anaerobic digestion sludge collected by a thermophilic process was cultivated at 55°C with synthetic wastewater containing glucose as the sole energy and carbon source. The pH, gas production rate, and VFA concentration were constantly monitored throughout the operation (Fig. 1). The IC concentration, which primarily represented bicarbonate, was also determined. The gas produced during the operation contained not less than 60% CH4, and the remainder was CO2.
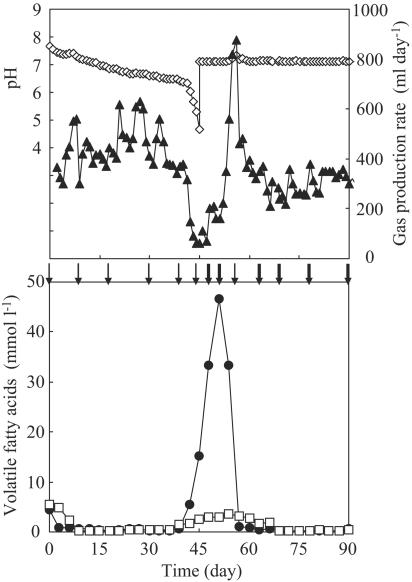
FIG. 1.
Change in reactor performance of the thermophilic methanogenic process: ⋄, pH; ▴, gas production rate; •, acetate concentration; □, propionate concentration. Arrows indicate the sampling points for SSCP analysis and quantitative PCR. Arrows in boldface indicate the sampling points for FISH.
During acclimation to the synthetic wastewater, the pH gradually decreased from 7.7 at day 0 to 6.3 at day 41. The gas production rate varied from 300 to 630 ml day−1 during this period. The propionate and acetate concentrations were kept below 0.5 mmol liter−1 from day 9 to day 36, during which period the reactor performance was considered to be stable. The pH suddenly dropped from 6.3 at day 41 to 4.7 at day 45. The acidification coincided with a decrease in the gas production rate from 320 to 60 ml day−1. The IC concentration also decreased from 1,265 mg liter−1 at day 0 to 54 mg liter−1 at day 45. The propionate concentration increased to 1.4 mmol liter−1 at day 39, and the acetate concentration substantially increased from 0.6 mmol liter−1 at day 39 to 15.1 mmol liter−1 at day 45. Despite the maintenance of the pH at 7.1 by addition of NaOH from day 45, the increase in VFA concentration and low gas production rate continued for the following 6 days. The recovery from this deteriorative condition began at day 51; a rapid increase in the gas production rate was accompanied by a degradation of the accumulated acetate. Following the acetate degradation, the propionate concentration gradually decreased from day 54. Finally a stable performance was achieved again; the gas production rate was stable within the range from 300 to 350 ml day−1 from day 82 to the end of the operation, during which time the acetate and propionate concentrations were less than 0.7 mmol liter−1, and the carbon recovery was well balanced (i.e., the value was over 80%).
Microbial community dynamics determined by PCR-mediated single-strand conformation polymorphism and statistical analysis.
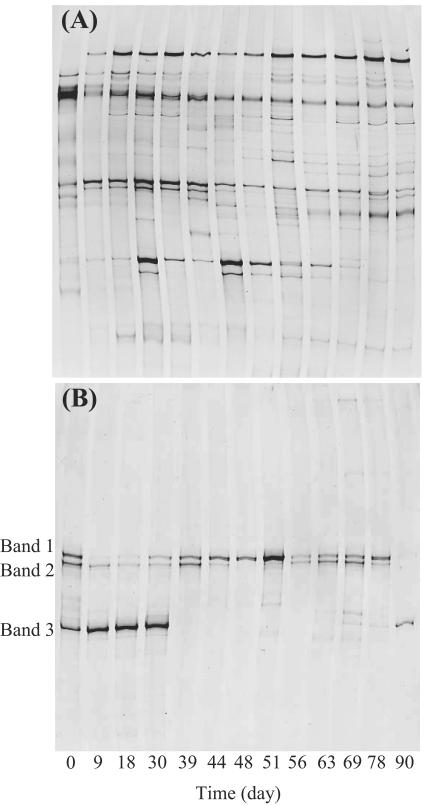
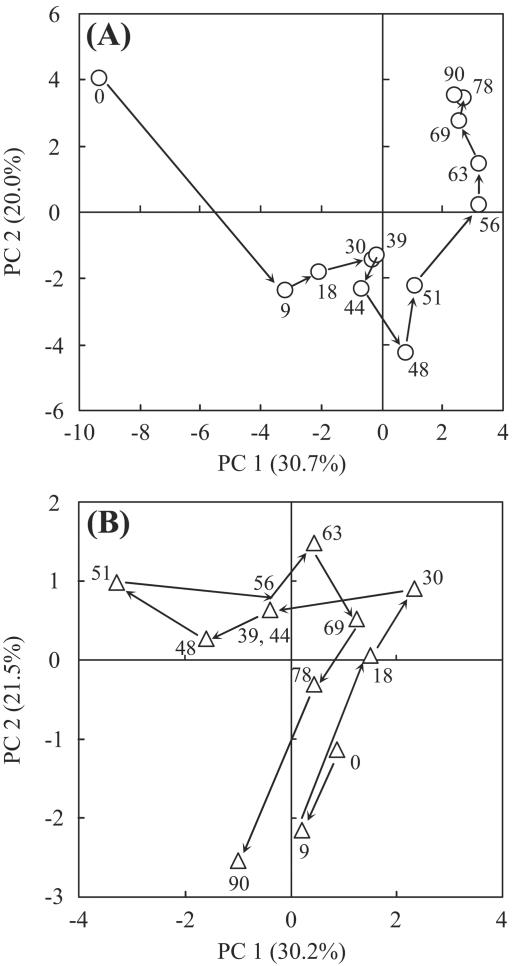
The bacterial and archaeal community structures were analyzed by PCR-SSCP based on the 16S rRNA gene (Fig. 2). On the bacterial SSCP profile (Fig. 2A), a total of 39 bands were detected. The 12 SSCP bands were detected on the profile at day 0, and the subsequent operation produced the disappearance of some of these bands and their reappearance. Some of the bands which newly appeared after day 0 continued to exist and others disappeared during the operation. The succession of the bacterial banding patterns was highly complicated. Statistical analyses of the banding patterns were useful to compare the patterns and their succession. Principal-component analysis, which is applied to DNA fingerprints of a variety of environmental samples (16, 39, 44), was performed to compare among these SSCP banding patterns throughout the operation (Fig. 3A). Day 0 was clearly distinct from any other time points. The time points during the first 45 days of the operation without pH control were clustered in a group on the plot. Thereafter, the banding patterns progressively changed toward a stable condition (i.e., to the upper right on the plot). Finally, the last three time points (i.e., days 69, 78, and 90) were closely located on the plot, indicating that the bacterial community was mostly stable from day 69 in the bioreactor. The PCA plot revealed that the bacterial community structure was influenced intensively by pH and had little relationship to the gas production rate or VFA concentration. The banding patterns of the start and end points were quite different on the plot. This indicates that the bacterial communities after the neutralization when the stable reactor performance was observed again were distinct from those at the first stable operation (days 9 to 30). The change in substrate composition at the beginning of the operation would also affect the bacterial community structures.
FIG. 2.
Microbial community structure in the thermophilic methanogenic process as determined using PCR-SSCP analysis. (A) Bacterial fingerprint; (B) archaeal fingerprint. Bands 1, 2, and 3 in panel B were closely related to Methanothermobacter thermautotrophicus (accession number AE000940; 100% sequence similarity), Methanosarcina thermophila (M59140; 98% sequence similarity), and Methanoculleus thermophilicus (AB065297; 100% sequence similarity), respectively.
FIG. 3.
Principal-component analysis of the SSCP data. (A) PCA plot for the bacterial fingerprint; (B) PCA plot for the archaeal fingerprint. The successive time points (days) are connected by arrows and indicated by numbers.
The archaeal community structure (Fig. 2B) was much simpler than the bacterial community structure. Three predominant bands (indicated as bands 1, 2, and 3 in Fig. 2B) were detectable on the profile. From the phylogenetic affiliation of the three predominant bands, bands 1, 2, and 3 were closely related to Methanothermobacter thermautotrophicus (accession no. AE000940; 100% sequence similarity), Methanosarcina thermophila (M59140; 98% sequence similarity), and Methanoculleus thermophilicus (AB065297; 100% sequence similarity), respectively. PCA analysis of the SSCP banding pattern showed that the time points at which the VFAs accumulated were clustered in a group at the upper left on the plot (Fig. 3B). However, the banding patterns could not be characterized by pH or gas production rate. These findings indicated that the archaeal community structure was correlated with VFA concentration rather than with pH or gas production rate. The second reactor was operated as a control under the same conditions as described above except that neutral pH was maintained to avoid acidification. The control reactor was stably operated, and a predominance of the Methanoculleus sp. was detected during the operation (data not shown).
Transition of methanogenic archaea determined by group-specific quantitative PCR.
The transition of the 16S rRNA gene copy number of each predominant methanogen was monitored using quantitative real-time PCR (Fig. 4). The three primer sets Mc412f/Mc578r, Mt392f/Mt578r, and Ms413f/Ms578r were designed (Table 1).
FIG. 4.
Transition of the 16S rRNA gene copy numbers of the methanogens Methanoculleus sp. (○), Methanosarcina sp. (□), and Methanothermobacter sp. (▵). Two trials were conducted to analyze each sample. The variations between the duplications were less than 1% of the mean.
The Methanoculleus sp. (band 3 in Fig. 2B) was shown to exist at a much higher abundance (4.6 × 103 to 5.6 × 104 copies ng−1 DNA) than the other methanogens during the period from day 0 to 30. Thereafter the copy number of the Methanoculleus sp. started to decrease, and it continued to decrease until day 56. The copy number of the Methanothermobacter sp. (band 1 in Fig. 2B) and the Methanosarcina sp. (band 2 in Fig. 2B) increased significantly from day 30; in particular, a drastic change in the copy number of the Methanothermobacter sp., i.e., an approximately 10,000-fold increase, was observed during the period from day 30 to 44. These dynamic changes in the archaeal population were accompanied by an increase in VFA concentration. The copy numbers of all three methanogens decreased from day 44 to 48, when the lowest gas production rate was observed. The period from day 51 to 56 was characterized by a selective increase in the Methanosarcina sp., indicating the contribution of this methanogen to the vigorous degradation of acetate. The highest population of Methanosarcina sp. was continuously detected until day 78. The copy number of the Methanothermobacter sp. decreased after day 44, although a temporal increase was observed from day 48 to 51. Toward the end of the operation, the population of the Methanothermobacter sp. was the lowest among the methanogens. The copy number of the Methanoculleus sp. was lowest during the period when VFAs accumulated, but it increased from day 56 and became the most abundant methanogen again at day 90.
Spatial distribution of the microbial community.
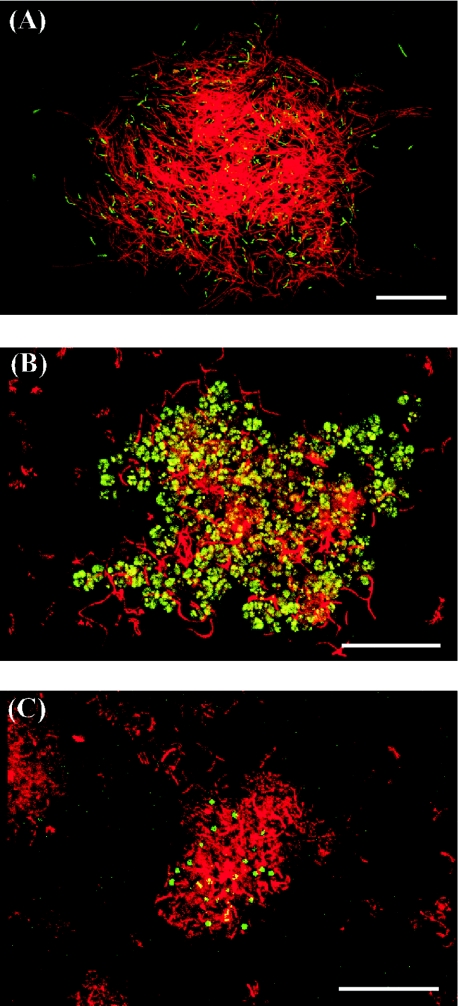
The syntrophic association via hydrogen interspecies transfer requires the juxtaposition of the microbial cells (9). The spatial distributions of Archaea and Bacteria were elucidated by FISH using the fluorescent probes Alexa488-labeled ARC915 for Archaea and rhodamine-labeled EUB338 for Bacteria. Samples taken on days 48, 51, 63, 69, 78, and 90 after starting the pH control (Fig. 1) were selected to investigate the respective operational conditions. Figure 5 shows the microscopic images revealing a representative spatial distribution among these samples. From the morphological characteristics of methanogenic archaea described in the literature (5) and the predominance of each methanogen determined by quantitative PCR, the affiliation of the archaeal cells shown in green in Fig. 5 could be estimated.
FIG. 5.
Fluorescence in situ hybridization of methanogenic microflora viewed by confocal laser scanning microscopy. (A) Day 48; (B) day 63; (C) day 90. The methanogenic microflora was simultaneously hybridized with Alexa488-labeled archaeal probe ARC915 (green) and rhodamine-labeled bacterial probe EUB338 (red). Representative fields of vision are shown. Bars, 20 μm.
The samples from days 48 and 51 contained mainly curved slender rods considered to be a Methanothermobacter sp., which were surrounded by numerous filamentous bacterial cells (Fig. 5A). The slender rods seldom existed as suspended cells in these samples. Spheroid bodies considered to be a Methanosarcina sp. were frequently observed in the culture at day 63, 69, and 78. Some of the spheroid bodies were present in the aggregates with bacterial cells (Fig. 5B). Irregular coccoids considered to be a Methanoculleus sp. surrounded by rod-shaped bacteria were observed in the sample at day 90 (Fig. 5C). Although the irregular coccoids were also observed as suspended cells, their fluorescence was very weak, indicating that the metabolic activity of the suspended microbes was lower than that of the cells within the aggregate (13, 29, 33, 51).
DISCUSSION
In this study, we investigated the archaeal and bacterial community successions in a thermophilic methanogenic bioreactor under deteriorative and stable conditions that were induced by acidification and neutralization, respectively. PCR-mediated single-strand conformation polymorphism based on the 16S rRNA gene rapidly provided an overview of microbial population dynamics, and quantitative PCR revealed a significant transition of the specific methanogens. Furthermore, FISH provided important information about the spatial distribution that could be used to estimate the specific association between Archaea and Bacteria. The combination of the process analyses and these molecular ecological techniques was a powerful approach for investigating the response of the microbial community to the reactor conditions.
The pH in the bioreactor gradually decreased from the start of the operation, although no obvious changes in VFA concentration were observed. The pH is strongly dependent on the buffering capacity of the processes. The most important buffering component within the optimal pH for methanogenic archaea is bicarbonate (4), which was represented mainly by IC during the operation. The IC concentration decreased from 1,265 to 54 mg liter−1 during the first 45 days, when the seed culture was replaced by the synthetic wastewater. This decrease in IC levels was a possible reason for the acidification. The sudden drop of the pH to 4.7 at day 45 was thought to be due to a breakthrough of the buffering capacity of the synthetic wastewater used in this study. By introduction of pH titration to 7.1, the bioreactor successfully recovered from the deteriorative condition induced by the acidification.
The microbial community structure throughout the operation was examined by PCR-SSCP based on the 16S rRNA gene. Here, it is noteworthy that the DNA approach used in this study was sufficient to detect changes of the archaeal community structure; however, in previous reports the population dynamics in the mesophilic processes were not detected by DNA analyses (11, 43). PCA analyses of the SSCP data indicated that populations of both Archaea and Bacteria had no direct relationship to the gas production rate. On the other hand, the archaeal community structure was closely correlated with the VFA concentration, and the pH had an intense impact on the bacterial community structure. The differences in the linkage between the microbial community structure and the reactor parameters could be accounted for by previous investigations revealing the insufficiency of gas phase parameters compared to liquid phase parameters (e.g., pH and VFA concentration) as indicators of the reactor conditions (2, 14, 32, 34).
The dynamic transition of each methanogen in response to the VFA concentration was clarified by quantitative PCR (Fig. 4). The aceticlastic methanogen (i.e., the Methanosarcina sp.) selectively grew during the period from day 51 to 56, during which the acetate was effectively degraded, suggesting that the Methanosarcina sp. contributed significantly to the relief from the high acetate concentration. This is supported by the previous findings that Methanosarcina primarily converts acetate to methane at an acetate concentration higher than 1 mmol liter−1 (22, 55). Another genus of aceticlastic methanogen, Methanosaeta, which grew favorably at acetate concentrations lower than 1 mmol liter−1, could not be detected throughout the operation. A similar observation was confirmed previously in thermophilic bioreactors, where the syntrophic oxidation preferentially played a role of acetate degradation rather than aceticlastic methanogenesis at low acetate concentrations (1, 31). In this study, the Methanoculleus sp. predominated or grew favorably at the low acetate concentration (<1 mmol liter−1); it predominated for the first 30 days and at day 90, and it grew favorably from day 56 to 78. FISH analyses revealed that the Methanoculleus sp. lay adjacent to rod-shaped bacteria. Juxtaposition of the microbial cells has been widely recognized for the syntrophic association (7, 9, 25, 41). Therefore, syntrophic acetate oxidation by the Methanoculleus sp. and proton-reducing bacteria possibly occurred during these periods.
In previous reports on thermophilic processes, one group of hydrogenotrophic methanogen predominated under conditions of low VFA concentration, and the accumulation of VFAs induced the growth of aceticlastic methanogen (1, 31). In this study, two groups of hydrogenotrophic methanogens were detectable. The Methanothermobacter sp. started to grow favorably while the Methanoculleus sp. began to decline during the period from day 30 to 39, when the propionate concentration increased from 0.3 to 1.4 mmol liter−1, although the other liquid phase parameters (i.e., pH or acetate concentration) changed little. The Methanothermobacter sp. became the predominant hydrogenotrophic methanogen during the accumulation of propionate (>1.4 mmol liter−1, from day 39 to 66). It is noteworthy that the predominance of one group of hydrogenotrophic methanogen was replaced by the other in response to propionate concentration. The VFA concentration has been reported to be closely related to the dissolved hydrogen concentration (10, 36). The growth of Methanoculleus spp. is known to be preferable to that of Methanothermobacter spp. at low hydrogen concentrations. These results suggested that the dissolved hydrogen concentration played a direct role in the predominance of the hydrogenotrophic methanogens. The difference between these two hydrogenotrophic methanogens in affinity to hydrogen concentration would induce the specific relationship with the distinct partner bacteria (Fig. 5A and C). Unfortunately, little is known about the ecophysiological role of the association between Methanothermobacter spp. and filamentous bacteria. However, this association may partially contribute to VFA degradation during the deteriorative phase.
Coexistence of aceticlastic and hydrogenotrophic methanogens has also been reported to occur in natural environmental ecosystems, e.g., river floodplains or rice fields (24, 27). It was clarified that the methanogenic community in these ecosystems was largely diverse and that the aceticlastic methanogens responded to the acetate concentration; i.e., the Methanosarcina sp. was adapted to acetate concentrations higher than 1 mmol liter−1, while the Methanosaeta sp. grew preferentially at the lower concentration. However, the community of hydrogenotrophic methanogens did not reveal any transition, and its structure was stable compared to that of aceticlastic methanogens. In upland pasture rhizosphere soil, it was confirmed that a pH change (i.e., acidification) had no effect on the indigenous community of methanogenic archaea (28). No investigation has yet revealed a drastic change in hydrogenotrophic methanogens.
The results of this study revealed that the archaeal population dynamics were closely correlated with the VFA concentration. In particular, the predominance of hydrogenotrophic methanogen associated with specific bacteria dynamically changed in response to the propionate concentration. The aceticlastic methanogen selectively proliferated over the period during which the accumulated acetate effectively was degraded. The simple archaeal population in the thermophilic process significantly responded to the environmental conditions, i.e., the concentration of VFAs. The substrate sensing and metabolic regulation of the simple methanogenic community in this study should be investigated to clarify the mechanism of the dynamic transition of the microbial population.
Acknowledgments
We are grateful to Christoph C. Tebbe (Institut für Agrarökologie, Bundesforschunganstalt für Landwirtschaft) for his significant advice on the PCR-SSCP procedure. We also thank Toru Shigematsu (Department of Applied Chemistry and Biochemistry, Kumamoto University) for his critical reading of the manuscript and Bo H. Svensson (Department of Water and Environmental Studies, Linköping University) and Ed W. J. van Niel (Department of Applied Microbiology, Lund University) for their constructive discussion on the study.
This work was supported by the New Energy and Industrial Technology Development Organization (NEDO), Japan.
REFERENCES
- 1.Ahring, B. K. 1995. Methanogenesis in thermophilic biogas reactors. Antonie Leeuwenhoek 67:91-102. [DOI] [PubMed] [Google Scholar]
- 2.Ahring, B. K., M. Sandberg, and I. Angelidaki. 1995. Volatile fatty acids as indicators of process imbalance in anaerobic digestors. Appl. Microbiol. Biotechnol. 41:559-565. [Google Scholar]
- 3.Amann, R. I., B. J. Binder, R. J. Olson, S. W. Chisholm, R. Devereux, and D. A. Stahl. 1990. Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56:1919-1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bjornsson, L., M. Murto, T. G. Jantsch, and B. Mattiasson. 2001. Evaluation of new methods for the monitoring of alkalinity, dissolved hydrogen and the microbial community in anaerobic digestion. Water Res. 35:2833-2840. [DOI] [PubMed] [Google Scholar]
- 5.Boone, D. R., and R. W. Castenholz. 2001. The Archaea and the deeply branching and phototrophic Bacteria. In G. M. Garrity (ed.), Bergey's manual of systematic bacteriology, 2nd ed., vol. 1. Springer, New York, N.Y.
- 6.Casamayor, E. O., R. Massana, S. Benlloch, L. Ovreas, B. Diez, V. J. Goddard, J. M. Gasol, I. Joint, F. Rodriguez-Valera, and C. Pedros-Alio. 2002. Changes in archaeal, bacterial and eukaryal assemblages along a salinity gradient by comparison of genetic fingerprinting methods in a multipond solar saltern. Environ. Microbiol. 4:338-348. [DOI] [PubMed] [Google Scholar]
- 7.Chen, C. L., H. Macarie, I. Ramirez, A. Olmos, S. L. Ong, O. Monroy, and W. T. Liu. 2004. Microbial community structure in a thermophilic anaerobic hybrid reactor degrading terephthalate. Microbiology 150:3429-3440. [DOI] [PubMed] [Google Scholar]
- 8.Chouari, R., D. Le Paslier, P. Daegelen, P. Ginestet, J. Weissenbach, and A. Sghir. 2005. Novel predominant archaeal and bacterial groups revealed by molecular analysis of an anaerobic sludge digester. Environ. Microbiol. 7:1104-1115. [DOI] [PubMed] [Google Scholar]
- 9.Conrad, R., T. J. Phelps, and J. G. Zeikus. 1985. Gas metabolism evidence in support of the juxtaposition of hydrogen-producing and methanogenic bacteria in sewage sludge and lake sediments. Appl. Environ. Microbiol. 50:595-601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cord-Ruwisch, R., T. I. Mercz, C.-Y. Hoh, and G. E. Strong. 1997. Dissolved hydrogen concentration as an on-line control parameter for the automated operation and optimization of anaerobic digesters. Biotechnol. Bioeng. 56:626-634. [DOI] [PubMed] [Google Scholar]
- 11.Delbès, C., R. Moletta, and J. J. Godon. 2001. Bacterial and archaeal 16S rDNA and 16S rRNA dynamics during an acetate crisis in an anaerobic digestor ecosystem. FEMS Microbiol. Ecol. 35:19-26. [DOI] [PubMed] [Google Scholar]
- 12.Delbès, C., R. Moletta, and J. J. Godon. 2000. Monitoring of activity dynamics of an anaerobic digester bacterial community using 16S rRNA polymerase chain reaction-single-strand conformation polymorphism analysis. Environ. Microbiol. 2:506-515. [DOI] [PubMed] [Google Scholar]
- 13.DeLong, E. F., G. S. Wickham, and N. R. Pace. 1989. Phylogenetic stains: ribosomal RNA-based probes for the identification of single cells. Science 243:1360-1363. [DOI] [PubMed] [Google Scholar]
- 14.Frigon, J. C., and S. R. Guiot. 1995. Impact of liquid-to-gas hydrogen mass transfer on substrate conversion efficiency of an upflow anaerobic sludge bed and filter reactor. Enzyme Microb. Technol. 17:1080-1086. [Google Scholar]
- 15.Garcia, J.-L., B. K. C. Patel, and B. Ollivier. 2000. Taxonomic, phylogenetic, and ecological diversity of methanogenic archaea. Anaerobe 6:205-226. [DOI] [PubMed] [Google Scholar]
- 16.Girvan, M. S., C. D. Campbell, K. Killham, J. I. Prosser, and L. A. Glover. 2005. Bacterial diversity promotes community stability and functional resilience after perturbation. Environ. Microbiol. 7:301-313. [DOI] [PubMed] [Google Scholar]
- 17.Hansen, M. C., T. Tolker-Neilson, M. Givskov, and S. Molin. 1998. Biased 16S rDNA PCR amplification caused by interference from DNA flanking template region. FEMS Microbiol. Ecol. 26:141-149. [Google Scholar]
- 18.Haruta, S., M. Kondo, K. Nakamura, H. Aiba, S. Ueno, M. Ishii, and Y. Igarashi. 2002. Microbial community changes during organic solid waste treatment analyzed by double gradient-denaturing gradient gel electrophoresis and fluorescence in situ hybridization. Appl. Microbiol. Biotechnol. 60:224-231. [DOI] [PubMed] [Google Scholar]
- 19.Hattori, S., Y. Kamagata, S. Hanada, and H. Shoun. 2000. Thermacetogenium phaeum gen. nov., sp. nov., a strictly anaerobic, thermophilic, syntrophic acetate-oxidizing bacterium. Int. J. Syst. Evol. Microbiol. 50:1601-1609. [DOI] [PubMed] [Google Scholar]
- 20.Imachi, H., Y. Sekiguchi, Y. Kamagata, A. Ohashi, and H. Harada. 2000. Cultivation and in situ detection of a thermophilic bacterium capable of oxidizing propionate in syntrophic association with hydrogenotrophic methanogens in a thermophilic methanogenic granular sludge. Appl. Environ. Microbiol. 66:3608-3615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jantsch, T. G., and B. Mattiasson. 2004. An automated spectrophotometric system for monitoring buffer capacity in anaerobic digestion processes. Water Res. 38:3645-3650. [DOI] [PubMed] [Google Scholar]
- 22.Jetten, M. S. M., A. J. M. Stams, and A. J. B. Zehnder. 1992. Methanogenesis from acetate: a comparison of acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol. Rev. 88:181-198. [Google Scholar]
- 23.Karakashev, D., D. J. Batstone, and I. Angelidaki. 2005. Influence of environmental conditions on methanogenic compositions in anaerobic biogas reactors. Appl. Environ. Microbiol. 71:331-338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kemnitz, D., K. J. Chin, P. Bodelier, and R. Conrad. 2004. Community analysis of methanogenic archaea within a riparian flooding gradient. Environ. Microbiol. 6:449-461. [DOI] [PubMed] [Google Scholar]
- 25.Lanthier, M., P. Juteau, F. Lepine, R. Beaudet, and R. Villemur. 2005. Desulfitobacterium hafniense is present in a high proportion within the biofilms of a high-performance pentachlorophenol-degrading, methanogenic fixed-film reactor. Appl. Environ. Microbiol. 71:1058-1065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lettinga, G. 1995. Anaerobic digestion and wastewater treatment systems. Antonie Leeuwenhoek 67:3-28. [DOI] [PubMed] [Google Scholar]
- 27.Lueders, T., and M. W. Friedrich. 2002. Effects of amendment with ferrihydrite and gypsum on the structure and activity of methanogenic populations in rice field soil. Appl. Environ. Microbiol. 68:2484-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nicol, G. W., G. Webster, L. A. Glover, and J. I. Prosser. 2004. Differential response of archaeal and bacterial communities to nitrogen inputs and pH changes in upland pasture rhizosphere soil. Environ. Microbiol. 6:861-867. [DOI] [PubMed] [Google Scholar]
- 29.Odaa, Y., S. Slagmana, W. G. Meijerb, L. J. Forneya, and J. C. Gottschala. 2000. Influence of growth rate and starvation on fluorescent in situ hybridization of Rhodopseudomonas palustris. FEMS Microbiol. Ecol. 32:205-213. [DOI] [PubMed] [Google Scholar]
- 30.Pender, S., M. Toomey, M. Carton, D. Eardly, J. W. Patching, E. Colleran, and V. O'Flaherty. 2004. Long-term effects of operating temperature and sulphate addition on the methanogenic community structure of anaerobic hybrid reactors. Water Res. 38:619-630. [DOI] [PubMed] [Google Scholar]
- 31.Petersen, S. P., and B. K. Ahring. 1991. Acetate oxidation in a thermophilic anaerobic sewage-sludge digestor: the importance of non-aceticlastic methanogenesis from acetate. FEMS Microbiol. Ecol. 86:149-158. [Google Scholar]
- 32.Pind, P. F., I. Angelidaki, B. K. Ahring, K. Stamatelatou, and G. Lyberatos. 2003. Monitoring and control of anaerobic reactors. Adv. Biochem. Eng. Biotechnol. 82:135-182. [DOI] [PubMed] [Google Scholar]
- 33.Poulsen, L. K., G. Ballard, and D. A. Stahl. 1993. Use of rRNA fluorescence in situ hybridization for measuring the activity of single cells in young and established biofilms. Appl. Environ. Microbiol. 59:1354-1360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Puss, A., G. Andre, M. Perrier, and S. R. Guiot. 1990. Liquid-to-gas mass transfer in anaerobic processes: inevitable transfer limitations of methane and hydrogen in the biomethanation process. Appl. Environ. Microbiol. 56:1636-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 36.Schink, B. 1997. Energetics of syntrophic cooperation in methanogenic degradation. Microbiol. Mol. Biol. Rev. 61:262-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schmalenberger, A., and C. C. Tebbe. 2003. Bacterial diversity in maize rhizospheres: conclusions on the use of genetic profiles based on PCR-amplified partial small subunit rRNA genes in ecological studies. Mol. Ecol. 12:251-262. [DOI] [PubMed] [Google Scholar]
- 38.Schnurer, A., G. Zellner, and B. H. Svensson. 1999. Mesophilic syntrophic acetate oxidation during methane formation in biogas reactors. FEMS Microbiol. Ecol. 29:249-261. [Google Scholar]
- 39.Sekiguchi, H., M. Watanabe, T. Nakahara, B. Xu, and H. Uchiyama. 2002. Succession of bacterial community structure along the Changjiang River determined by denaturing gradient gel electrophoresis and clone library analysis. Appl. Environ. Microbiol. 68:5142-5150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sekiguchi, Y., Y. Kamagata, and H. Harada. 2001. Recent advances in methane fermentation technology. Curr. Opin. Biotechnol. 12:277-282. [DOI] [PubMed] [Google Scholar]
- 41.Sekiguchi, Y., Y. Kamagata, K. Nakamura, A. Ohashi, and H. Harada. 1999. Fluorescence in situ hybridization using 16S rRNA-targeted oligonucleotides reveals localization of methanogens and selected uncultured bacteria in mesophilic and thermophilic sludge granules. Appl. Environ. Microbiol. 65:1280-1288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shigematsu, T., Y. Tang, T. Kobayashi, H. Kawaguchi, S. Morimura, and K. Kida. 2004. Effect of dilution rate on metabolic pathway shift between aceticlastic and nonaceticlastic methanogenesis in chemostat cultivation. Appl. Environ. Microbiol. 70:4048-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shigematsu, T., Y. Tang, T. Kobayashi, H. Kawaguchi, K. Ninomiya, J. Kijima, T. Kobayashi, S. Morimura, and K. Kida. 2003. Effect of dilution rate on structure of a mesophilic acetate-degrading methanogenic community during continuous cultivation. J. Biosci. Bioeng. 96:547-558. [DOI] [PubMed] [Google Scholar]
- 44.Sliwinski, M. K., and R. M. Goodman. 2004. Spatial heterogeneity of crenarchaeal assemblages within mesophilic soil ecosystems as revealed by PCR-single-stranded conformation polymorphism profiling. Appl. Environ. Microbiol. 70:1811-1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Speece, R. E. 1996. Anaerobic biotechnology for industrial wastewaters. Archae Press, Nashville, Tenn.
- 46.Stahl, D. A., and R. Amann. 1991. Development and application of nucleic acid probes, p. 205-248. In E. Stackbrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley & Sons, Inc., New York, N.Y.
- 47.Takai, K., and K. Horikoshi. 2000. Rapid detection and quantification of members of the archaeal community by quantitative PCR using fluorogenic probes. Appl. Environ. Microbiol. 66:5066-5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tatara, M., A. Yamazawa, Y. Ueno, H. Fukui, M. Goto, and K. Sode. 2005. High-rate thermophilic methane fermentation on short-chain fatty acids in a down-flow anaerobic packed-bed reactor. Bioprocess Biosyst. Eng. 27:105-113. [DOI] [PubMed] [Google Scholar]
- 49.Tchobanoglous, G., and F. L. Burton. 1991. Wastewater engineering: treatment, disposal, and reuse, 3rd ed. Irwin McGraw Hill, Inc., New York, N.Y.
- 50.Ueno, Y., S. Haruta, M. Ishii, and Y. Igarashi. 2001. Changes in product formation and bacterial community by dilution rate on carbohydrate fermentation by methanogenic microflora in continuous flow stirred tank reactor. Appl. Microbiol. Biotechnol. 57:65-73. [DOI] [PubMed] [Google Scholar]
- 51.Wagner, M., G. Rath, R. Amann, H.-P. Koops, and K.-H. Schleifer. 1995. In situ identification of ammonia-oxidizing bacteria. Syst. Appl. Microbiol. 18:251-264. [Google Scholar]
- 52.Watanabe, K., Y. Kodama, and S. Harayama. 2001. Design and evaluation of PCR primers to amplify bacterial 16S ribosomal DNA fragments used for community fingerprinting. J. Microbiol. Methods 44:253-262. [DOI] [PubMed] [Google Scholar]
- 53.Zehnder, A. J. B. 1978. Ecology of methane formation, p. 349-376. In R. Mitchell (ed.), Water pollution microbiology, vol. 2. John Wiley & Sons, Ltd., London, United Kingdom. [Google Scholar]
- 54.Zhu, H., F. Qu, and L. H. Zh. 1993. Isolation of genomic DNAs from plants, fungi and bacteria using benzyl chloride. Nucleic Acids Res. 21:5279-5280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zinder, S. H. 1990. Conversion of acetic acid to methane by thermophiles. FEMS Microbiol. Rev. 75:125-138. [Google Scholar]