Abstract
d-Amino acid aminotransferases (d-AATs) from Geobacillus toebii SK1 and Geobacillus sp. strain KLS1 were cloned and characterized from a genetic, catalytic, and structural aspect. Although the enzymes were highly thermostable, their catalytic capability was approximately one-third of that of highly active Bacilli enzymes, with respective turnover rates of 47 and 55 s−1 at 50°C. The Geobacillus enzymes were unique and shared limited sequence identities of below 45% with d-AATs from mesophilic and thermophilic Bacillus spp., except for a hypothetical protein with a 72% identity from the G. kaustophilus genome. Structural alignments showed that most key residues were conserved in the Geobacillus enzymes, although the conservative residues just before the catalytic lysine were distinctively changed: the 140-LRcD-143 sequence in Bacillus d-AATs was 144-EYcY-147 in the Geobacillus d-AATs. When the EYcY sequence from the SK1 enzyme was mutated into LRcD, a 68% increase in catalytic activity was observed, while the binding affinity toward α-ketoglutarate decreased by half. The mutant was very close to the wild-type in thermal stability, indicating that the mutations did not disturb the overall structure of the enzyme. Homology modeling also suggested that the two tyrosine residues in the EYcY sequence from the Geobacillus d-AATs had a π/π interaction that was replaceable with the salt bridge interaction between the arginine and aspartate residues in the LRcD sequence.
d-Amino acid aminotransferase (d-AAT; EC 2.6.1.21) catalyzes the conversion of various α-keto acid substrates into their respective d-amino acids, some of which are indispensable for bacteria as peptidoglycan components of the cell wall (25). As such, the enzyme has been applied as a catalyst to produce optically pure d-amino acids (1, 3, 30) that act as intermediates of semisynthetic antibiotics, bioactive peptides, and other physiologically active compounds (20, 21).
d-AAT activity is found in various gram-positive bacteria, including Bacillus (8, 28, 29), Staphylococcus (22), and Listeria (31), and yet recent biotechnology studies have mainly focused on thermophilic or mesophilic Bacillus enzymes due to their high catalytic activity and broad substrate specificity (4, 9, 10, 23). For example, the d-AAT from thermophilic Bacillus sp. strain YM1 was remarkable in its activity and thermal stability and showed a high identity of 67% with a mesophilic Bacillus sphaericus enzyme, while representing a limited identity of less than 50% with other Bacillus enzymes.
16S rRNA analyses are useful for comparing phenotypically close and yet genetically different microorganisms (6). For instance, in the phylogenetic analysis of bacilli, the thermophilic YM1 has been assigned to genetic group II, together with B. sphaericus, whereas most other thermophilic bacillus species, such as B. stearothermophilus, B. thermodenitrificans, and B. kaustophilus, have been positioned in genetic group V (2), recently renamed as the genus Geobacillus (17, 19).
Consequently, Geobacillus species would seem to be a remarkable resource for new d-AATs with unique sequences and enzymatic properties. Accordingly, the present study presents new thermostable d-AATs from the genus Geobacillus, including a characterization of their genetic and catalytic properties, and mutational studies of the conservative β5-α5 loop (bearing the cofactor-binding lysine) (26). In addition, homology modeling is conducted to analyze the functional relevance of the distinctive loop structure of the Geobacillus d-AATs.
MATERIALS AND METHODS
Strains and plasmids.
The thermophilic bacillus collections, including Bacillus sp. strain YM1 and different soil isolates, was cultivated at 55 or at 65°C in a MY medium, as specified previously (12). The E. coli WM335 strain (leu pro his arg thyA met lac gal rspL hsdM hsdR murI), a d-glutamate auxotroph, was used as the host strain for the genetic complementation method (13). The plasmid pBluescript II SK and E. coli XL1-Blue were purchased from Stratagene; plasmids pUC118 and pUC119 were purchased from Takara Shuzo, Japan; and plasmid pHCE IIB purchased from Bohan Biomedicals (South Korea).
DNA manipulation and mutagenesis.
The genomic DNA of the Geobacillus strains was partially digested with Sau3AI. DNA fragments of 3 to 10 kb were then isolated by centrifugation on a sucrose gradient (5 to 40% [wt/vol]) for 20 h at 25,000 rpm in a Beckman SW40 rotor and ligated into the BamHI site of pUC118 at 16°C for 12 h with a T4 DNA ligase. Thereafter, the E. coli WM335 was transformed with the ligation mixture with electroporation.
The site specific mutagenesis to introduce the desired mutations into the target DNA sequences was performed by using the megaprimer PCR method. The mutagenic internal primer 5-ACAAGAGATGTCCGCTGGCTACGTTGCGATATTAAGAGTTTAAATCTTCTA-3 is designed to bear an LRcD residue (underlined) instead of the wild-type sequence, EYcY, whereas the C-terminal primer 5-GCCGGATCCTTATTTTGCGTTTTTGACAGC-3 is designed to have a BamHI site (underlined). The product of the first PCR with the mutagenic primer and C-terminal primer was purified and then used as a megaprimer for a second PCR, along with the N-terminal primer 5-GCATTAAAGCTGTACGTACTA-3. The final PCR product contained the desired mutation and 3-terminal BamHI site in the DNA sequence. Plasmid pHCE19T(II) was used for the expression of the LRcD mutant: the plasmid vector was digested with NcoI, blunt ended by Klenow treatment, and sequentially digested with BamHI. The resulting vector was ligated with the second PCR product by a blunt-cohesive ligation at 16°C with a T4 DNA ligase.
Expression and purification.
E. coli XL1-Blue cells bearing the plasmid pDSK2 or pHKLS23 were cultivated at 37°C for 16 h in 1 liter of LB medium containing 100 μg of ampicillin/ml. After being harvested by centrifugation, the cells were disrupted by sonification in a standard buffer, including 30 mM Tris-HCl (pH 8.0), 0.01% β-mercaptoethanol, and 0.05 mM pyridoxal-5′-phosphate. The active d-AAT was recovered from the supernatant of the cell lysate and incubated at 60°C for 20 min to remove heat-labile E. coli proteins. The resulting enzyme solution was then loaded onto a Resource Q ion exchange (Pharmacia, Sweden), washed with the standard buffer, and eluted by using a potassium chloride gradient from 0 to 0.5 M. Next, the active fractions were collected, adjusted to include 1.7 M ammonium sulfate, and loaded onto a Phenyl Superose (Pharmacia, Sweden). The elution was carried out by using a reverse gradient of ammonium sulfate from 1.7 to 0 M, and then the active fractions were pooled and dialyzed against the standard buffer and stored in a deep freezer. All of the column procedures were carried out by using an AKTA System (Amersham Bioscience, Sweden) at room temperature.
Sequence analysis.
The nucleotide sequences of the d-AAT and 16S rRNA genes were determined at Solgent Co. (Daejon, South Korea) by primer walking on both DNA strands and then analyzed by using the Vector NTI suite (Informax, Maryland).
The PCR amplification of the 16S rRNA gene was performed with two universal primers, as previously described (33). The 16S rRNA gene sequences were then aligned with reference sequences from related taxa by using CLUSTAL W software (32). Gaps at the 5′ and 3′ ends of the alignment were omitted from further analyses. A phylogenetic tree was constructed by using the neighbor-joining method (as implemented with the NEIGHBOR program of the PHYLIP package) (24).
Homology modeling and structural analysis.
The d-AAT structures of the PDB entries 1DAA, 2DAA, and 3DAA were aligned with the 3-D Align module from DS modeling 1.1 (Accelrys, San Diego, CA). The resulting consensus structure was then used as the template for the 2-D Align with the d-AAT sequences. Three-dimensional homology models of the SK1 d-AAT were generated by using the Modeler module (7), and the model with the best loop conformations determined by using the Profiles-3-D verification method of the Protein Health module for further modeling (14). A coenzyme analog, d-cycloserine pyridoxalphosphate, was extracted from the 2DAA and fitted to the PLP-binding site of each monomer. Energy minimization and molecular dynamics (MD) to optimize the structure were carried out by using the DS CHARMm module in vacuo, using the steepest descent method first, followed by the conjugate gradient method (15). During the minimization process, the protein backbone was restrained by a harmonic constraint. The lowest energy conformer resulting from the MD run was then finally relaxed without restraints.
Enzyme and protein assay.
The d-AAT activity was calculated from the pyruvate formation rate using a coupling assay with lactate dehydrogenase (Roche Diagnostics, Switzerland) or the salicylaldehyde method (5). The assay mixture contained 0.05 mM pyridoxal-5′-phosphate, 0.5 mM NADH, 10 μg of lactate dehydrogenase/ml, and various concentrations of d-alanine and α-ketoglutarate in a 100 mM Tris-HCl buffer (pH 8.5). One unit of enzyme was defined as the activity catalyzing the formation of 1 μmol of pyruvate per min at 50°C. The protein concentration was determined by the Bradford method with bovine serum albumin as the standard.
RESULTS
Thermostable d-AATs from Geobacillus species.
A collection of thermophilic isolates from South Korean soil was examined for d-AAT activity using d-alanine and α-ketoglutarate as the substrates. The screening of d-AAT activity at 55°C identified two strains with the highest activity, subsequently designated LK1 and LK2. However, these highly active strains were unable to grow at 65°C, the temperature at which most Geobacillus species grow, and in microscopic and physiological examinations were found to be very close to the well studied Bacillus sp. strain YM1 (12). Thus, three d-AAT-producers (SK1, KLS1, and SD1) growing at 65°C were selected for further experiments. The average activity of the selected strains, 0.02 U/mg of protein, was approximately one-fifth the level of the highly active Bacillus enzymes growing at 55°C.
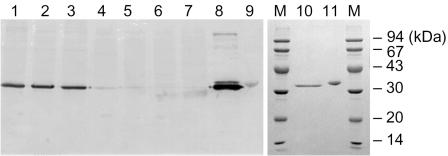
A Western blotting analysis with a polyclonal YM1 d-AAT antibody exhibited strong immuno-signals toward the LK1 and LK2 proteins (Fig. 1, lanes 1 to 3), coinciding with their microbial similarity to the YM1 strain. However, no immuno-signals were detected against SK1, KLS1, and SD1 (lanes 4 to 7 in Fig. 1). Since the depth of the immunoblotting reflects the relevancy between the primary sequences, the three d-AATs isolated at 65°C were determined as sequentially distant from the YM1 d-AAT.
FIG. 1.
Comparison of d-amino acid aminotransferases from thermophilic Bacilli strains using Western blotting analyses. Lanes 1 to 7 are the immunostains for the cell extracts from different Bacillus strains: 1, YM1; 2, LK1; 3, LK2; 4, SK1; 5, KLS1; 6, KL1; and 7, SD1. The polyclonal antibody was prepared from an antiserum boosted by the YM1 d-AAT. Lanes 8 to 11 are the purified d-AATs: 8 and 10, Bacillus sp. strain YM1; 9 and 11, G. toebii SK1. Lanes 10 and 11 are the sodium dodecyl sulfate-polyacrylamide gel electrophoresis analysis of the purified YM1 and SK1 proteins.
Based on the genetic and chemotaxonomic analyses, the SK1 strain was identified as a new species from the genus Geobacillus (27). In addition, the complete 16S rRNA sequences of strain YM1 (gb: DQ100073) and KLS1 (gb: DQ100071) were determined, and imported to the PHYLIP program, as described in Materials and Methods. A phylogenetic analysis of the 16S rRNA sequences also revealed that SK1 and KLS1 were related and belonged to genetic group V, namely, Geobacillus (see Fig. S1 in the supplemental material). Meanwhile, YM1 was positioned in group II, which includes mesophilic B. sphaericus.
Cloning and expression of d-AATs.
For a genetic complementation assay, Escherichia coli WM335 cells containing the genomic DNA of the Geobacillus strains were spread on LB-ampicillin plates without the supplementation of d-glutamate (13). After being incubated overnight, one or two colonies were picked from each plate and transferred to fresh culture media to isolate the plasmids. The resulting pUC118 plasmids were retransformed into E. coli XL1-Blue cells to examine the d-AAT activity. A clone pDSK2 (insert size 3.6 kb) from the SK1 library showed the highest activity of 35 U per mg of protein (1,750-fold increase from the original activity of the SK1 strain), the activity attributed to the 1.7-kb EcoRI-XhoI fragment based on a restriction analysis. Thus, the 1.7-kb fragment was subcloned into pBluescript II SK to construct pDSK231. Meanwhile, a clone pKLS1 was selected from the KLS1 library, and restriction analyses attributed the enzyme activity to the 1.86-kb HindIII-SacI fragment. Thus, the 1.86-kb fragment was subcloned into pBluescript II SK to construct pHKLS23.
Nucleotide sequencing of the 1.7-kb insert in pDSK231 showed an 864-bp open reading frame (gb: DQ100074) that encoded a protein of 288 amino acid residues with a molecular weight of 33,198. These results were in accordance with the results from sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The 1.86-kb insert in pHKLS23 was also sequenced and the open-reading frame (gb: DQ100075) homologous to the SK1 d-AAT was designated KLS1 d-AAT.
Sequence analysis and structural alignment.
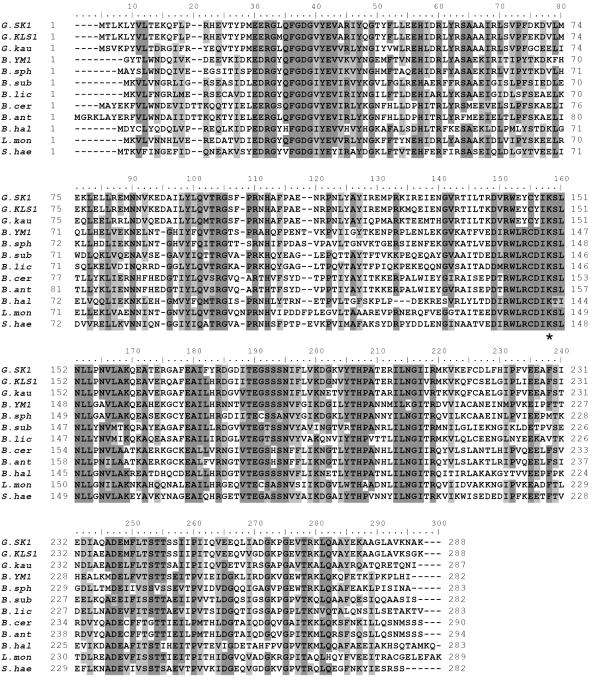
The alignment of the d-AAT sequences showed that almost all of the residues with key roles in the YM1 enzyme (26) were conserved in the Geobacillus enzymes, including the catalytic K145 (K149 in the SK1 d-AAT), carboxylate traps that define the position and orientation of the substrate-binding Y31 (Y35), R98 (R103), and H100 (H105), and cofactor-binding residues R50 (R54), I204 (I208), T241 (T245), E177 (E181), and R138 (R142), all shown in Fig. 2. The loop from Ser240 to Ser243, thought to be important in substrate specificity, was also conserved.
FIG. 2.
Sequence alignment of Bacillus d-AATs. Abbreviations: G.SK1, G. toebii SK1; G.KLS1, Geobacillus sp. strain KLS1; G.kau, G. kaustophilus; B.YM1, Bacillus sp. strain YM1, B.sph, B. sphaericus; B.sub, B. subtilis; B.lic, B. licheniformis; B.cer, B. cereus; B.ant, B. anthracis; B.hal, B. halodurans; L.mon, Listeria monocytogenes; S.hae, Staphylococcus haemolyticus. The conserved residues are shaded dark, while similar residues are shaded light. The asterisk indicates the lysine residue that forms a sciff base with pyridoxal 5′-phosphate.
The most interesting feature in the alignment was the difference in the loop between β5 and α5 covering the catalytic Lys residue (26). Although the sequence of the loop, 138-RWLRCDIKSLNLL-150, has been well conserved in the d-AAT sequences identified to date, it was distinctively changed in the Geobacillus d-AATs, where L140, R141, and D143 in the YM1 d-AAT were substituted with residues 144E, 145Y, and 147Y, respectively (Fig. 2).
The overall sequence of the SK1 d-AAT exhibited a 90.2% identity with the KLS1 enzyme and 71.5% identity with a hypothetical correspondent from the G. kaustophilus genome. However, the identity level with other d-AATs, including Bacillus sp. strain YM1, was lower than 45%.
When the alignments of the Bacilli d-AATs were imported into the PHYLIP program, the Geobacillus enzymes from the present study were located in the same branch of the tree phenogram (Fig. S2 in the supplemental material). The enzymes from YM1 and B. sphaericus in group II were located in the same group, revealing a 67% identity with each other, whereas the enzymes from B. cereus and B. anthracis, with an 89.7%-identity, formed a separate branch to the enzymes from B. subtilis and B. licheniformis that belong to the same genetic group I.
Purification and characterization of kinetic properties.
The E. coli XL1-Blue cells bearing the plasmid pDSK2 exhibited a thick protein band with a molecular mass of 34 kDa. The expressed protein was estimated to be more than 30% of the total E. coli proteins when quantified with an image analyzer (Bio-Rad, California). After heat treatment at 60°C for 20 min and two sequential chromatographies (ion exchange and hydrophobic), the SK1 d-AAT was purified to homogeneity (lane 11 in Fig. 1) at a recovery yield of 59%. The purification protocols were also successfully applied to the site-directed mutant of the SK1 d-AAT, the KLS1 enzyme, and YM1 enzyme expressed in E. coli XL1-Blue (23).
The purified d-AATs from the SK1 and KLS1 strains showed absorption maxima at 330 and 415 nm, corresponding to pyridoxal-5-phosphate bound to the active site lysine residue (23). The molecular masses of the enzymes were also estimated to be around 70 kDa by gel permeation chromatography on a Superose 12 column (Pharmacia), indicating that the enzymes consisted of two identical subunits.
The kinetic characterization of the purified enzymes was performed based on triplicate experiments at 50°C, while changing the concentrations of d-alanine and α-ketoglutarate from 1 to 40 mM. Lineweaver-Burk plots of the bi-reactant reaction showed characteristic parallel lines for a ping-pong-bi-bi reaction. The catalytic rate of the d-AATs from SK1 and KLS1 was determined to be 47 and 55 s−1, respectively, which was close to one-third of the highly active YM1 d-AAT (Table 1).
TABLE 1.
Kinetic parameters of d-AATs
| Enzyme | wXXcXik | Mean ± SD
|
kcat/Km,d-Ala (mM · s)−1 | kcat/Km,α-KG (mM · s)−1 | ||
|---|---|---|---|---|---|---|
| kcat (s−1) | Km,d-Ala (mM) | Km,α-KG (mM) | ||||
| YM1 | 140-LRcD-143 | 134 ± 2 | 2.5 ± 0.2 | 3.1 ± 0.1 | 52 | 42 |
| SK1 | 144-EYcY-147 | 47 ± 2 | 2.5 ± 0.1 | 0.71 ± 0.1 | 18 | 63 |
| KLS1 | 144-EYcY-147 | 55 ± 3 | 1.4 ± 0.2 | 0.26 ± 0.1 | 38 | 203 |
| Mutant SK1 | 144-LRcD-147 | 79 ± 6 | 3.4 ± 0.3 | 1.4 ± 0.3 | 22 | 53 |
The Michaelis constants with d-alanine were all similar for the d-AATs from SK1, KLS1, and YM1, and yet the Km,α -KG values were remarkably smaller for the Geobacillus enzymes. In particular, the Km,α-KG for the KLS1 enzyme was 12 times smaller than that for the YM1 enzyme. Meanwhile, for the Geobacillus enzymes, the Km,d-Ala values were much larger than the Km,α-KG values, in distinct contrast to the same constants for the YM1 enzyme.
Investigations of the substrate preference of the SK1 d-AAT revealed that d-alanine, d-aminobutyrate, and d-aspartate were the best amino donors for α-ketoglutarate (Table 2), whereas d-serine, d-asparagine, and d-norvaline also served as amino donors, but to a lesser extent. Moreover, investigation of the amino acceptors showed that α-ketoglutarate, α-ketobutyrate, and α-ketoisovalrate were the preferred amino acceptors.
TABLE 2.
Substrate specificity of d-AAT from Geobacillus toebii SK1
| Amino donor | Relative activity (%)a | Amino acceptors | Relative activity (%)a |
|---|---|---|---|
| d-Alanine | 100 | α-Ketoglutarate | 100 |
| d-α-Aminobutyrate | 89 | α-Ketobutyrate | 110 |
| d-Aspartate | 70 | α-Ketoisovalerate | 13 |
| d-Serine | 12 | Phenyl pyruvate | 2 |
| d-Asparagine | 11 | ||
| d-Norvaline | 8.2 | ||
| d-Tryptophan | 4.9 | ||
| d-Valine | 1.9 | ||
| d-Histidine | 1.9 | ||
| d-Threonine | 0.9 | ||
| d-Methionine | 0.7 |
The relative activity was calculated by analyzing the formation of d-glutamate from α-ketoglutarate using an automatic amino acid analyzer. The amino acceptor specificity was assayed by measuring the formation of pyruvate from d-alanine as an amino group donor. All reactions were carried out at 50°C in the standard assay mixture containing 10 mM substrates. Inert amino acceptors included benzyl formate, hydroxylphenyl pyruvate, and indole pyruvate.
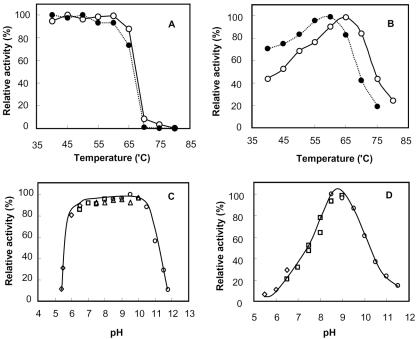
The SK1 d-AAT was further investigated for its stability and the maximum activity at different temperatures. The remaining activity after heat treatment for 20 min confirmed that the enzyme was fully stable up to 65°C (Fig. 3A). The enzyme activity under the assay conditions also increased with temperature up to the stability limit, and the maximum activity was observed at 65°C (Fig. 3B). When incubated in different pH buffers, the SK1 d-AAT remained stable within a pH range of 6.0 to 10.5 (Fig. 3C), and the optimal pH was around pH 8.5 (Fig. 3D).
FIG. 3.
Effect of temperature and pH on SK1 d-AAT. The open symbols indicate the native enzyme, while the closed symbols with dotted lines indicate the mutant enzyme with an LRcD sequence instead of 144-EYcY-147. (A) The native and mutant enzymes were incubated at different temperatures for 20 min in a 0.1 M Tris-HCl buffer (pH 8.5), and the remaining activities determined at 50°C to evaluate the respective thermal stabilities. (B) The native and mutant enzymes were assayed at different reaction temperatures for 20 min. The concentrations of pyruvate from d-alanine were measured by using the salicylaldehyde method. (C) The native enzyme was incubated in different buffers for 1 h at 50°C, and the remaining activities were assayed to evaluate the pH stability. (D) The native enzyme was assayed for 20 min at different pHs with the following 0.1 M buffers: sodium acetate buffer (pH 4.0 to 6.0), potassium phosphate buffer (pH 6.0 to 8.0), Tris-HCl buffer (pH 7.5 to 9.0), and N-cyclohexyl-3-aminopropanesulfonic acid buffer (pH 9.0 to 12.0).
Mutational studies on β5-α5 loop sequence.
As mentioned in the kinetic characterization, the catalytic rate of the SK1 d-AAT was three times slower than that of the YM1 enzyme. To investigate the relationship between the 144-EYcY-147 sequence and the low activity of the SK1 enzyme, the EYcY sequence was mutated to LRcD, making the loop identical to other Bacillus enzymes. Interestingly, the mutated enzyme exhibited a 68% increase in catalytic activity, whereas the Michaelis constant for α-ketoglutarate increased twofold (Table 1). Although the thermal stability of the mutant did not change very much, its maximum activity was identified at 60°C, and the change in activity with temperature was less steep than that for the native enzyme, as represented in Fig. 3A and B (solid symbols for the mutant). The increase in enzyme activity with temperature was interpreted by the Arrhenius equation [ln(k) = ln(A) − (Ea/RT)] to compare the activation energies (Ea) for the native and mutant enzymes (Fig. S3 in the supplemental material). From the slope of ln(k) versus 1/T, the activation energies for the native and mutant enzymes were calculated as 29 and 16 kJ/mol, respectively.
Homology modeling of SK1 d-AAT.
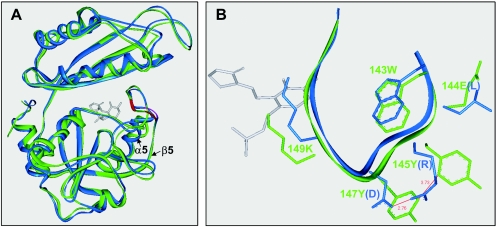
Comparative modeling with DS Modeling 1.1 generated hypothetical structures for the wild and mutant type SK1-DAATs (Fig. 4A), which only differed in the nonaligned loop region. The models displaying the highest stereochemical quality were then determined by using the Protein Health module. The resulting model contained no cis-proline, and the final RMS values for the Cα trace of the wild and mutant enzymes after 200 ps of MD were 1.2 and 1.0 Å, respectively. The estimated structures were very stable during the MD simulations, and at least 95% of the backbone φ and ψ dihedral angles for the residues were located within the allowed regions of the Ramachadran plot (70% core).
FIG. 4.
Homology model for G. toebii SK1 d-AAT. (A) 3-D Align analysis of the monomeric units of the wild and mutant enzymes; (B) magnified structures of the 143-wEYcYik-149 loop. The loop structure bears the cofactor binding lysine (underlined) and is located at the crevice between the N and C domains. The 143W is related to the intersubunit packing to form a catalytic dimer. The capital letters in EYcY indicate the residues substituted for LRcD in the mutant enzyme. The structures of the native and mutant enzymes are represented in green and blue, respectively. The gray molecule shows the structure of d-cycloserine pyridoxalphosphate, adopted from the PDB-entry 2DAA.
In the three-dimensional structure of the YM1 d-AAT, the arginine and aspartate side chains in the LRcD sequence have a short salt-bridge. Meanwhile, the hypothetical model of the SK1 enzyme suggested that the aromatic groups of tyrosine in the EYcY sequence were apart by 3.7 Å (Fig. 4B), allowing a π/π interaction between the aromatic rings (18). Plus, the salt-bridge interaction between the arginine and aspartate was reconstituted at a 2.7-Å distance, using the molecular modeling and dynamics for the LRcD mutant of the SK1 enzyme.
DISCUSSION
The d-AAT from G. toebii SK1 exhibited sequence identities of <45.5% with the enzymes from mesophilic bacilli and thermophilic YM1. However, BLAST searches of genome databases revealed that the SK1 enzyme shared a 71% identity with a hypothetical correspondent of the G. kaustophilus genome. Meanwhile, the enzyme from Geobacillus sp. strain KLS exhibited a 90% identity and close evolutionary relationship with the SK1 enzyme (Fig. S2 in the supplemental material). As such, these thermostable d-AATs with various identity levels can provide a genetic base for evolutionary protein engineering by DNA shuffling methods.
The sequence and structural alignments revealed that most of the residues with key roles in the YM1 enzyme were conserved in the SK1 d-AAT, as described in Results. In previous structural studies of the YM1 d-AAT, the 240-STTS-243 loop has been interpreted as the substrate-binding pocket, and was conserved as 244-STTS-247 in the SK1 d-AAT. An mutational study performed to broaden the substrate specificity by replacing the loop with a SVSS sequence, as found in the B. sphaericus enzyme with a broader substrate range, resulted in a 30% lower activity compared to the original and no significant expansion in the substrate specificity was observed (unpublished data).
In previous crystallographic studies of the YM1 d-AAT, the long loop between β5 and α5 was located in a crevice between the N and C domains and included the cofactor binding lysine residue (26). Plus, mutational and biochemical analyses have indicated that the 139W in the loop plays an important role in subunit packing to form a catalytic dimer, and any mutations replacing the 139W with a nonaromatic residue were detrimental to the enzyme activity (11, 16).
In the primary sequence of the SK1 d-AAT, the EYcY sequence was located between the 143W (139W in YM1 d-AAT) and the cofactor binding lysine residue, 149K (145K). Therefore, substituting LRcD for EYcY was expected to lower the activity and stability of the enzyme by disturbing the active site or subunit interface. However, the mutant enzyme in the present study maintained its stability and even increased its activity up to 68%, while the Michaelis constant for α-ketoglutarate increased twofold (Table 1). Therefore, these results suggest that the LRcD sequence of highly active homologue is recruited effectively and fitted into the structure of the SK1 enzyme. When the profiles in Fig. 3B were replotted based on Arrhenius' equation (Fig. S3 in the supplemental material), the mutant enzyme showed a decreased activation energy (Ea = 16 kJ/mol) compared to that of the wild type (Ea = 29 kJ/mol). The lower activation energy and decreased affinity to substrates as exhibited by the mutant may imply an increased flexibility in the local region adjacent to the active site.
The arginine and aspartate in the LRcD sequence of the YM1 d-AAT were analyzed to form a stable salt bridge within a 3-Å distance (26), whereas homology modeling of the SK1 d-AAT suggested that the two tyrosine residues in the EYcY sequence of the SK1 enzyme were located in a proximate distance to form a π/π interaction (18). Accordingly, the catalytic results and molecular simulation imply that the salt bridge in the YM1 d-AAT and presumed aromatic interaction in the Geobacillus d-AATs are interchangeable with each other, despite the significant difference in molecular size and chemical properties.
Supplementary Material
Acknowledgments
This work was supported by a grant (code no. 20050301034476) from Biogreen 21 Program (Rural Development Administration) and the 2005 research fund of Kookmin University, Republic of Korea.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Ager, D. J., I. G. Fotheringham, S. A. Laneman, D. P. Pantaleone, and P. P. Taylor. 2000. The large scale synthesis of “unnatural” amino acids. Enantiomer 5:235-243. [PubMed] [Google Scholar]
- 2.Ash, C., J. A. E. Farrow, S. Wallbanks, and M. D. Collins. 1991. Phylogenetic heterogeneity of the genus bacillus revealed by comparative analysis of small subunit rRNA sequences. Lett. Appl. Microbiol. 13:202-206. [Google Scholar]
- 3.Bae, H.-S., S.-P. Hong, S.-G. Lee, M.-S. Kwak, N. Esaki, and M.-H. Sung. 2002. Application of a thermostable glutamate racemase from Bacillus sp. SK-1 for the production of d-phenylalanine in a multi-enzyme. Syst. J. Mol. Catal. B. Enzymatic 17:223-233. [Google Scholar]
- 4.Berger, B. J., S. English, G. Chan, and M. H. Knodel. 2003. Methionine regeneration and aminotransferases in Bacillus subtilis, Bacillus cereus, and Bacillus anthracis. J. Bacteriol. 185:2418-2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berntsson, S. 1995. Spectrometric determination of pyruvic acid by the salicylaldehyde method. Anal. Biochem. 27:1659-1660. [Google Scholar]
- 6.Felsenstein, J. 1993. PHYLIP: Phylogenetic Inference Package, version 3.5. University of Washington, Seattle.
- 7.Fiser, A., and A. Sali. 2003. Comparative protein structure modeling with MODELLER: a practical approach. Methods Enzymol. 374:463-493. [Google Scholar]
- 8.Fotheringham, I. G., S. A. Bledig, and P. P. Taylor. 1998. Characterization of the genes encoding d-amino acid transaminase and glutamate racemase, two d-glutamate biosynthetic enzymes of Bacillus sphaericus ATCC 10280. J. Bacteriol. 180:4319-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Galkin, A., L. Kulakova, T. Yoshimura, K. Soda, and N. Esaki. 1997. Synthesis of optically active amino acids from α-keto acids with Escherichia coli cells expressing heterologous genes. Appl. Environ. Microbiol. 61:4651-4656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gutierrez, A., T. Yoshimura, Y. Fuchikami, and N. Esaki. 2000. Modulation of activity and substrate specificity by modifying the backbone length of the distant interdomain loop of d-amino acid aminotransferase. Eur. J. Biochem. 267:7218-7223. [DOI] [PubMed] [Google Scholar]
- 11.Kishimoto, K., C. Yasuda, and J. M. Manning. 2000. Reversible dissociation/ association of d-amino acid transaminase subunits: properties of isolated active dimers and inactive monomers. Biochemistry 39:381-387. [DOI] [PubMed] [Google Scholar]
- 12.Kwak, M.-S., S.-G. Lee, S.-C. Jeung, S.-H. Suh, J.-H. Lee, Y.-J. Jeon, Y.-H. Kim, and M.-H. Sung. 1999. Screening and taxonomic characterization of d-amino acid aminotransferase-producing thermophiles. Kor. J. Microbiol. Biotechnol. 27:184-190. [Google Scholar]
- 13.Lugtenberg, E. J. J., H. J. Wijsman, and D. van Zaane. 1973. Properties of a d-glutamic acid-requiring mutant Escherichia coli. J. Bacteriol. 114:499-506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luthy, R., J. U. Bowie, and D. Eisenberg. 1992. Assessment of protein models with three-dimensional profiles. Nature 356:83-85. [DOI] [PubMed] [Google Scholar]
- 15.MacKerell, A. D., Jr., B. Brooks, C. L. Brooks, III, L. Nilsson, B. Roux, Y. Won, and M. Karplus. 1998. CHARMM: the energy function and its parameterization with an overview of the program, p. 271-277. In P. Schleyer, R. Schleyer, N. L. Allinger, T. Clark, J. Gasteiger, P. Kollman, and H. F. Schaeffer III (ed.), The encyclopedia of computational chemistry 1. John Wiley & Sons, Inc., New York, N.Y.
- 16.Martinez, del Pozo, A., P. W. van Ophem, D. Ringe, G. Petsko, K. Soda, and J. M. Manning. 1996. Interaction of pyridoxal 5′-phosphate with tryptophan-139 at the subunit interface of dimeric d-amino acid transaminase. Biochemistry 35:2112-2116. [DOI] [PubMed] [Google Scholar]
- 17.McMullan, G., J. M. Christie, T. J. Rahman, I. M. Banat, N. G. Ternan, and R. Marchant. 2004. Habitat, applications, and genomics of the aerobic, thermophilic genus Geobacillus. Biochem. Soc. Trans. 32:214-217. [DOI] [PubMed] [Google Scholar]
- 18.Meyer, E. A., R. K. Castellano, and F. Diederich. 2003. Interactions with aromatic rings in chemical and biological recognition. Angew. Chem. Int. Ed. 42:1210-1259. [DOI] [PubMed] [Google Scholar]
- 19.Nazina, T. N., T. P. Tourova, A. B. Poltaraus, E. V. Novikova, A. A. Grigoryan, A. E. Ivanova, A. M. Lysenko, V. V. Petrunyaka, G. A. Osipov, S. S. Belyaev, and M. V. Ivanov. 2001. Taxonomic study of aerobic thermophilic bacilli: descriptions of Geobacillus subterraneus gen. nov., sp. nov. and Geobacillus uzenensis sp. nov. from petroleum reservoirs and transfer of Bacillus stearothermophilus, Bacillus thermocatenulatus, Bacillus thermoleovorans, Bacillus kaustophilus, Bacillus thermoglucosidasius and Bacillus thermodenitrificans to Geobacillus as the new combinations G. stearothermophilus, G. thermocatenulatus, G. thermoleovorans, G. kaustophilus, G. thermoglucosidasius and G. thermodenitrificans. Int. J. Syst. Evol. Microbiol. 51(Pt. 2):433-446. [DOI] [PubMed] [Google Scholar]
- 20.Patel, R. N. 2000. Microbial/enzymatic synthesis of chiral drug intermediates. Adv. Appl. Microbiol. 47:33-78. [DOI] [PubMed] [Google Scholar]
- 21.Patel, R. N. 2001. Enzymatic synthesis of chiral intermediates for Omapatrilat, an antihypertensive drug. Biomol. Eng. 17:167-182. [DOI] [PubMed] [Google Scholar]
- 22.Pucci, M. J., J. A. Thanassi, H.-T. Ho, P. J. Falk, and T. J. Dougherty. 1995. Staphylococcus hemolyticus contains two d-glutamic acid biosynthetic activities, a glutamate racemase and a d-amino acid transaminase. J. Bacteriol. 177:336-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ro, H.-S., S.-P. Hong, H.-J. Seo, T. Yoshimura, N. Esaki, K. Soda, H.-S. Kim, and M.-H. Sung. 1996. Site-directed mutagenesis of the amino acid residues in beta-strand III [Val30-Val36] of d-amino acid aminotransferase of Bacillus sp. YM-1. FEBS Lett. 398:141-145. [DOI] [PubMed] [Google Scholar]
- 24.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 25.Soda, K., and N. Esaki. 1985. d-Amino acid transaminase, p. 463-467. In P. Christen and D. E. Metzler (ed.), Transaminases. John Wiley & Sons, Inc., New York, N.Y.
- 26.Sugio, S., G. A. Petsko, J. M. Manning, K. Soda, and D. Ringe. 1995. Crystal structure of a d-amino acid aminotransferase: how the protein controls stereoselectivity. Biochemistry 34:9661-9669. [DOI] [PubMed] [Google Scholar]
- 27.Sung, M.-H., H. Kim, J.-W. Bae, S.-K. Rhee, C. O. Jeon, K. Kim, J.-J. Kim, S.-P. Hong, S.-G. Lee, J.-H. Yoon, Y.-H. Park, and D.-H. Baek. 2002. Geobacillus toebii sp. nov., a new thermophilic bacterium isolated from hay compost. Int. J. Syst. Evol. Microbiol. 52:2251-2255. [DOI] [PubMed] [Google Scholar]
- 28.Tanizawa, T., Y. Masu, S. Asano, H. Tanaka, and K. Soda. 1989. Thermostable d-amino acid aminotransferase from a thermophilic Bacillus species. J. Biol. Chem. 264:2445-2449. [PubMed] [Google Scholar]
- 29.Taylor, P. P., and I. G. Fotheringham. 1997. Nucleotide sequence of the Bacillus licheniformis ATCC 10716 dat gene and comparison of the predicted amino acid sequence with those of other bacterial species. Biochim. Biophys. Acta 1350:38-40. [DOI] [PubMed] [Google Scholar]
- 30.Taylor, P. P., D. P. Pantaleone, R. F. Senkpeil, and I. G. Fotheringham. 1998. Novel biosynthetic approaches to the production of unnatural amino acids using transaminases. Trends Biotechnol. 16:412-418. [DOI] [PubMed] [Google Scholar]
- 31.Thompson, R. J., H. G. A. Bouwer, D. A. Portnoy, and F. R. Frankel. 1998. Pathogenicity and immunogenicity of a Listeria monocytogenes strain that requires d-alanine for growth. Infect. Immun. 66:3552-3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thomson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoon, J.-H., S. T. Lee, and Y.-H. Park. 1998. Inter- and intraspecific phylogenetic analysis of the genus Nocardioides and related taxa based on 16S rRNA sequences. Int. J. Syst. Bacteriol. 48:187-194. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.