Abstract
Disruption of genes by homologous recombination occurs at a low frequency in the basidiomycete Schizophyllum commune. For instance, the SC3 and SC15 genes were inactivated at frequencies of 1 and 5%, respectively. As an alternative to disruption, we used gene silencing through the introduction of a hairpin construct. The SC15 gene, which encodes an abundantly secreted structural protein, was silenced at a frequency of 80% in monokaryons of S. commune after introduction of a hairpin construct of the gene. Silencing also occurred in dikaryons in which one of the partners was not a silenced strain. The silencing mechanism resembles RNAi in other filamentous fungi and is a powerful tool for the functional analysis of genes expressed in monokaryons or dikaryons.
RNA interference (RNAi) can be used for the functional analysis of genes. In this process double-stranded RNA (dsRNA) molecules induce the degradation of transcripts that are homologous in sequence. RNAi can result from the direct administration of dsRNA to cells or by inducing dsRNA formation through the expression of complementary sequences (10). Sequences similar to the dcl-2, qde-2, and qde-3 genes of Neurospora crassa, which are required for RNAi in N. crassa, are present in a partial genomic sequence of the homobasidiomycete Schizophyllum commune that represents ∼40% of the single-copy DNA (GenBank DQ275629, DQ275628, and DQ275630). dcl-2 (GenBank NCU06766.1) is an ortholog of Dicer (3), which encodes an endonuclease that processes dsRNA in small interfering RNAs (siRNAs) (1). qde-2 (GenBank AF217760) encodes a component of the RNA-induced silencing complex (RISC), which is guided by a siRNA to mRNA with a homologous sequence (2). Finally, qde-3 (GenBank AF205407) encodes a putative RecQ DNA helicase (4). The presence of sequences in S. commune homologous to dcl-2, qde-2, and qde-3 suggests that the proteins required for RNAi are present and that this mechanism of gene silencing might be functional in this basidiomycete.
Our objective here was to determine whether a mechanism similar to RNAi is operating in S. commune. To this end, a hairpin construct of SC15 was made. This gene encodes an abundantly secreted 17-kDa protein of S. commune, which is involved in aerial hyphae formation and attachment in the absence of the SC3 protein (9). This is the first report of RNA-mediated gene silencing in a filamentous homobasidiomycete.
MATERIALS AND METHODS
Strains and growth conditions.
The isogenic Schizophyllum commune strains 4-39 (MATA41 MATB41, CBS 341.81), 4-40 (MATA43 MATB43, CBS 340.81), and FL1 (MATA43 MATB43) were used. The latter strain contains the SC15::GFP fusion construct pSC15gfp (see DNA constructs). Strains were grown at 25°C in the light or at 30°C in the dark on minimal medium (6) solidified with 1.5% agar. For microscopy, strains were grown in a thin layer of medium between a cover glass and a porous polycarbonate (PC) membrane (diameter, 76 mm; pore size, 0.1 μm; Osmonics; GE Water Technologies, Trevose, PA).
DNA constructs.
A fragment of SC15 (GenBank AJ007503) cDNA encompassing the region between the start of the 5′ untranslated region and the 93rd codon was amplified by PCR with primers sc15kpnfw (5′-GGTACCAGTCGAACCCACACCGACTACC-3′) and sc15kpnrv (5′-GGTACCTGAGCTCCTCAATGCC-GTCGTTGG-3′), which both introduce KpnI recognition sites. The fragment was cloned in inverse orientation in the KpnI site just after the stop codon of genomic SC15 in plasmid pSC15gspz (Fig. 1). The resulting hairpin construct, pSC15hp, encodes a SC15 mRNA with a stem of 334 nucleotides and with a loop of 333 bp. Plasmid pSC15gspz consists of a pUC20 backbone containing a phleomycin resistance cassette (11) and a 4.1-kb SalI genomic fragment encompassing the SC15 gene. pSC15gfp is a derivative of pSC15gspz in which a green fluorescent protein (GFP) cDNA (GenBank AF188479) has been cloned in frame with the SC15 coding region (A. Vinck and L. G. Lugones, in preparation).
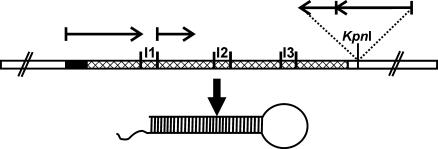
FIG. 1.
Silencing construct pSC15hp and resulting hairpin mRNA. The SC15 promoter and terminator (white boxes), 5′ untranslated region (black box), and coding sequence (cross-hatched box) are indicated, as well as the SC15 introns (I1, I2, and I3).
Transformation procedure.
S. commune was transformed essentially as previously described (11), except that protoplast generation was done in 1 M MgSO4 containing 1 mg of lysing enzymes ml−1 from Trichoderma harzianum (Applied Plant Research, Horst, The Netherlands). A total of 5 to 10 μg of DNA was added to 3 × 107 protoplasts in 100 μl of 1 M sorbitol. Transformants were selected on minimal medium containing 25 μg of phleomycin (Cayla, Toulouse, France) ml−1.
RNA and protein analysis.
RNA was isolated from mycelium that had been ground in liquid nitrogen using TRIzol reagent (Invitrogen, Carlsbad, CA). Electrophoresis and hybridization of RNA were performed as previously described (5), except that the filters were washed at 65°C. For analysis of SC3 and SC15 secretion, colonies were grown for 3 to 4 days on the surface of a PC membrane overlaying solidified minimal medium. The membrane supporting the colonies was transferred to a fresh plate topped with a polyvinylidene difluoride (PVDF) membrane. SC15 and SC3 secretion was monitored by immunodetection after incubation of the colonies on PVDF membrane for 2 and 17 h, respectively. The SC3 and SC15 antisera (9, 17) were diluted 10,000- and 5,000-fold, respectively. Goat anti-rabbit-conjugated alkaline phosphatase was used as a second antibody with BCIP (5-bromo-4-chloro-3-indolylphosphate) and nitroblue tetrazolium as substrates (7).
Fluorescence microscopy.
GFP fluorescence was monitored with an Axioskop 2 Plus microscope (Zeiss, Jena, Germany) equipped with an HBO 100-W mercury lamp and a Photometrics Cool SNAP camera (1,392 × 1,024 pixels) using standard fluorescein isothiocyanate filters.
RESULTS
The homokaryotic wild-type S. commune strain 4-39 was transformed with pSC15hp. This construct encodes an SC15 hairpin mRNA with a stem of 334 nucleotides and a loop of 333 bp. Of 34 transformants, 27 produced less SC15 protein than did the 4-39 wild-type strain. The intensity of the immunolocalization signal of the other seven colonies was similar to that of the wild type. Of the transformants showing no secretion of SC15, three (S1, S3, and S5) were randomly selected for further study. These three strains and the wild-type strain were grown on minimal medium on a porous PC membrane. Immunodetection on PVDF membranes that had been placed underneath the colonies in between the medium and the PC membrane revealed that, in contrast to SC15, secretion of SC3 was unaffected (Fig. 2). This shows that the reduction in SC15 secretion was not due to a thn mutation. (The thn mutation occurs frequently spontaneously and has a pleiotropic phenotype that includes downregulation of SC3 and SC15 and a reduction in the formation of aerial hyphae [16].) The low amount of SC15 secreted by transformants S1, S3, and S5 was accompanied by greatly reduced accumulation of SC15 mRNA (Fig. 3).
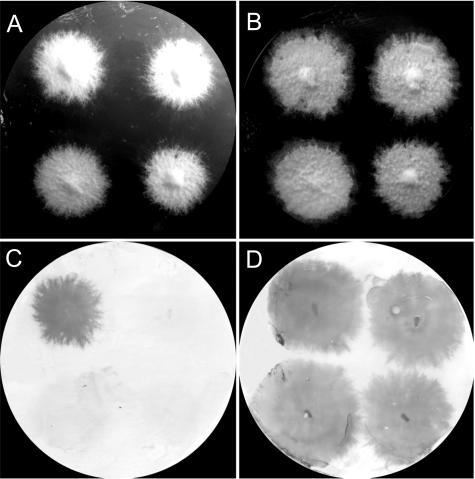
FIG. 2.
Secretion of SC15 is severely affected in strains transformed with the SC15 hairpin construct. (A and B) A PVDF membrane was placed underneath colonies grown on top of a PC membrane. This was followed by immunodetection for SC15 (C) and SC3 (D), respectively. The colonies on each plate are arranged in the following order: wild-type strain 4-40 (top, left), and recombinant strains S1 (top, right), S3 (bottom, left), and S5 (bottom, right).
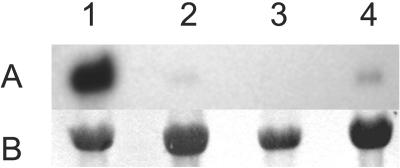
FIG. 3.
SC15 mRNA level is severely reduced in strains transformed with the SC15 hairpin construct. (A) Accumulation of SC15 mRNA in 4-day-old colonies of the wild-type strain 4-40 (lane 1) and transformed strains S1 (lane 2), S3 (lane 3), and S5 (lane 4). (B) Methylene blue staining of 18S rRNA (loading control).
The silencing of SC15 was not related to the introduction of extra copies of the gene rather than due to the hairpin mRNA, since when S. commune strain 4-39 was transformed with pSC15gspz containing SC15 there was no change in SC15 secretion. Silencing also was not due to cytosine methylation, since when colonies of transformants S1, S3, and S5 were grown on solid minimal medium containing 10 μM 5-azacytidine (AZC), there was no change in the amount of detectable SC15 protein. (AZC inhibits DNA-methylation by incorporating into the DNA in place of cytidine and/or by direct inhibition of the activity of methylases [8].)
Transformants S1, S3, and S5 were crossed with FL1, which expresses the endogenous copy of SC15 and a translational fusion of SC15 and GFP. GFP fluorescence in the dikaryons was significantly reduced relative to a dikaryon composed of nuclei from FL1 and 4-39 (Fig. 4). Fluorescence intensity corresponded with mRNA levels of the SC15::GFP fusion, as shown by hybridization with probes against SC15 and GFP (Fig. 5). SC15 mRNA was not detected in dikaryons that expressed pSC15hp in only one of the parental nuclei (Fig. 5). Accumulation of SC15::GFP mRNA was also reduced. Thus, the hairpin RNA not only leads to degradation of SC15 mRNA originating from the parental nucleus but also of comparable mRNA produced by the partner nucleus in the dikaryotic cell.
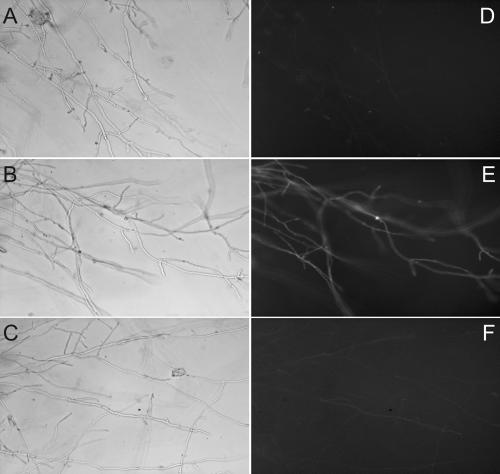
FIG. 4.
Bright-field (A, B, and C) and fluorescence (D, E, and F) images of 4-day-old colonies of wild-type strains 4-39 × 4-40 (A and D) and strains 4-39 × FL1 (B and E) and S1 × FL1 (C and F).
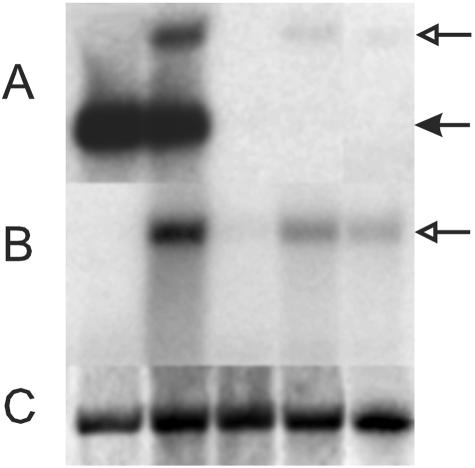
FIG. 5.
The expression level of SC15 and SC15::GFP is reduced in dikaryons that express the SC15 hairpin construct in one of the parental nuclei. (A) Hybridization of an SC15 probe with RNA from 4-day-old colonies of crosses of the wild-type strains 4-39 × 4-40 (lane 1), 4-39 × FL1 (lane 2), and FL1 crossed with, respectively, transformants S1 (lane 3), S3 (lane 4), and S5 (lane 5). SC15 mRNA is indicated by a filled arrowhead, and SC15::GFP is indicated by an open arrowhead. (B) Same as in panel A but probed with GFP. (C) Methylene blue staining of 18S rRNA (loading control).
DISCUSSION
The introduction of extra copies of the SC3 hydrophobin gene into S. commune resulted in silencing of both the introduced and endogenous SC3 genes in 90% of the transformants (13). In this case, the silencing was triggered by SC3 mRNA through a cytosine methylation mechanism, and the gene in the wild-type nucleus of the dikaryon was not silenced (12). SC15 was not silenced by the introduction of multiple copies of the gene. These results distinguish silencing of SC3 (13) and the silencing of SC15 we describe here and suggest that two different silencing mechanisms are operative in S. commune.
We anticipate that the RNA-based silencing mechanism described here also operates in other homobasidiomycetes, e.g., the commercially important species Agaricus bisporus and Pleurotus ostreatus. Orthologs of the N. crassa genes dcl-2, qde-2, and qde-3 are also present in the genomic sequences of Coprinus cinereus (http://www.broad.mit.edu/cgi-in/annotation/fungi/coprinus_cinereus/blast_page.cgi) and Phanerochaete chrysosporium (http://genome.jgi-psf.org/Phchr1/Phchr1.home.html), which is consistent with this hypothesis. In the homobasidiomycetes, RNA-based gene silencing is a powerful alternative to gene inactivation by homologous recombination, which occurs at low frequency in this group of fungi. For instance, gene inactivation of SC3, SC4, and SC15 in S. commune occurs at a frequency of 1 to 5% (9, 14, 15). More importantly, functional analysis of genes that are active in the heterokaryotic phase, e.g., in mushroom formation and sporulation, requires inactivation of the gene in both nuclei of the dikaryon. RNA-based gene silencing occurs at a much higher frequency than homologous recombination and enables silencing of functional genes in both nuclei of a dikaryon by the expression of a hairpin construct in only one of the nuclei.
Acknowledgments
We thank Ischa Lamot for technical support.
REFERENCES
- 1.Bernstein, E., A. A. Caudy, S. M. Hammond, and G. J. Hannon. 2001. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409:363-366. [DOI] [PubMed] [Google Scholar]
- 2.Catalanotto, C., G. Azzalin, G. Macino, and C. Cogoni. 2000. Gene silencing in worms and fungi. Nature 404:245-254. [DOI] [PubMed] [Google Scholar]
- 3.Catalanotto, C., M. Pallotta, P. ReFalo, M. S. Sachs, L. Vayssie, G. Macino, and C. Cogoni. 2004. Redundancy of the two dicer genes in transgene-induced posttranscriptional gene silencing in Neurospora crassa. Mol. Cell. Biol. 24:2536-2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cogoni, C., and G. Macino. 1999. Posttranscriptional gene silencing in Neurospora by a RecQ DNA helicase. Science 286:2342-2344. [DOI] [PubMed] [Google Scholar]
- 5.de Vries, R. P., P. J. I. van de Vondervoort, L. Hendriks, M. van de Belt, and J. Visser. 2002. Regulation of the a-glucoronidase-encoding gene (aguA) from Aspergillus niger. Mol. Genet. Genomics 268:96-102. [DOI] [PubMed] [Google Scholar]
- 6.Dons, J. J. M., O. M. H. de Vries, and J. G. H. Wessels. 1979. Characterization of the genome of the basidiomycete Schizophyllum commune. Biochim. Biophys. Acta 563:100-112. [DOI] [PubMed] [Google Scholar]
- 7.Harlow, E., and D. Lane. 1988. Antibodies: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 8.Jones, P. A. 1985. Altering gene expression with 5-azacytidin. Cell 40:485-486. [DOI] [PubMed] [Google Scholar]
- 9.Lugones, L. G., J. F. de Jong, O. M. H. de Vries, R. Jalving, J. Dijksterhuis, and H. A. B. Wösten. 2004. The SC15 protein of Schizophyllum commune mediates formation of aerial hyphae and attachment in the absence of the SC3 hydrophobin. Mol. Microbiol. 53:707-716. [DOI] [PubMed] [Google Scholar]
- 10.Mello, C. C., and D. Conte, Jr. 2004. Revealing the world of RNA interference. Nature 431:338-342. [DOI] [PubMed] [Google Scholar]
- 11.Schuren, F. H. J., and J. G. H. Wessels. 1994. Highly-efficient transformation of the homobasidiomycete Schizophyllum commune to phleomycin resistance. Curr. Gen. 26:179-183. [DOI] [PubMed] [Google Scholar]
- 12.Schuurs, T. A. 1998. Regulation of hydrophobin genes in Schizophyllum commune. University of Groningen, Groningen, The Netherlands.
- 13.Schuurs, T. A., E. A. Schaeffer, and J. G. Wessels. 1997. Homology-dependent silencing of the SC3 gene in Schizophyllum commune. Genetics 147:589-596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Wetter, M.-A., F. H. J. Schuren, T. A. Schuurs, and J. G. H. Wessels. 1996. Targeted mutation of the SC3 hydrophobin gene of Schizophyllum commune affects formation of aerial hyphae. FEMS Microbiol. Lett. 140:265-269. [Google Scholar]
- 15.van Wetter, M.-A., H. A. B. Wösten, and J. G. H. Wessels. 2000. SC3 and SC4 hydrophobins have distinct roles in formation of aerial structures in dikaryons of Schizophyllum commune. Mol. Microbiol. 36:201-210. [DOI] [PubMed] [Google Scholar]
- 16.Wessels, J. G. H., O. M. H. de Vries, S. A. Ásgeirsdóttir, and J. Springer. 1991. The thn mutation of Schizophyllum commune, which suppresses formation of aerial hyphae, affects expression of the Sc3 hydrophobin gene. J. Gen. Microbiol. 137:2439-2445. [DOI] [PubMed] [Google Scholar]
- 17.Wösten, H. A. B., S. A. Ásgeirsdóttir, J. H. Krook, J. H. H. Drenth, and J. G. H. Wessels. 1994. The fungal hydrophobin SC3p self-assembles at the surface of aerial hyphae as a protein membrane constituting the hydrophobic rodlet layer. Eur. J. Cell Biol. 63:122-129. [PubMed] [Google Scholar]