Abstract
Differentiation of the species within the genus Listeria is important for the food industry but only a few reliable methods are available so far. While a number of studies have used Fourier transform infrared (FTIR) spectroscopy to identify bacteria, the extraction of complex pattern information from the infrared spectra remains difficult. Here, we apply artificial neural network technology (ANN), which is an advanced multivariate data-processing method of pattern analysis, to identify Listeria infrared spectra at the species level. A hierarchical classification system based on ANN analysis for Listeria FTIR spectra was created, based on a comprehensive reference spectral database including 243 well-defined reference strains of Listeria monocytogenes, L. innocua, L. ivanovii, L. seeligeri, and L. welshimeri. In parallel, a univariate FTIR identification model was developed. To evaluate the potentials of these models, a set of 277 isolates of diverse geographical origins, but not included in the reference database, were assembled and used as an independent external validation for species discrimination. Univariate FTIR analysis allowed the correct identification of 85.2% of all strains and of 93% of the L. monocytogenes strains. ANN-based analysis enhanced differentiation success to 96% for all Listeria species, including a success rate of 99.2% for correct L. monocytogenes identification. The identity of the 277-strain test set was also determined with the standard phenotypical API Listeria system. This kit was able to identify 88% of the test isolates and 93% of L. monocytogenes strains. These results demonstrate the high reliability and strong potential of ANN-based FTIR spectrum analysis for identification of the five Listeria species under investigation. Starting from a pure culture, this technique allows the cost-efficient and rapid identification of Listeria species within 25 h and is suitable for use in a routine food microbiological laboratory.
The genus Listeria currently embraces six species: L. monocytogenes, L. ivanovii, L. innocua, L. welshimeri, L. seeligeri, and L. grayi, based on DNA homology, 16S rRNA homology, chemotaxonomic properties, and multilocus enzyme analysis (34). All of these species are widespread in the environment, but only L. monocytogenes is considered an opportunistic pathogen for humans and animals. Occasionally, human infections due to L. ivanovii and L. seeligeri have also been reported (8, 25, 35). Since most of the Listeria species are found as food contaminants, which may indicate a potential risk for subsequent contamination by L. monocytogenes, their presence requires immediate action by the food company. For instance, L. innocua has frequently been found as a marker organism of a L. monocytogenes contamination in dairy plants (14, 26, 36). Therefore, a rapid and reliable differentiation of L. monocytogenes from the other species of the genus is particularly important for the food industry with respect to an effective quality assurance strategy.
For the identification of Listeria at the species level in routine laboratories, time-consuming, laborious, and sometimes unreliable biochemical and phenotypical standard methods such as sugar fermentations and the CAMP test are often used (5, 22). Easy and rapid identification systems that are commercially available often fail to accurately identify atypical strains, due to the lack of basic classification marker reactions (3, 13, 21, 33). Therefore, fast molecular methods and immunological procedures have been developed. However, most of them are limited to detecting only the genus Listeria or only L. monocytogenes (1, 6, 9, 29, 30, 32). Some other methods are laborious for species discrimination (7, 11, 20) or fail to identify all Listeria species (19, 27, 39). Recently developed sensitive and specific microarray techniques are still of limited potential for routine laboratories, due to high costs and the requirement of highly skilled personnel (40).
Fourier transform infrared spectroscopy (FTIR) is a vibrational spectroscopic technique with high-resolution power which is able to distinguish microbial cells at different taxonomic levels (16). One attractive application of this inexpensive and rapid technique is the identification of unknown strains using an extensive reference library containing spectra from well-identified microbes (12, 15, 23, 28, 31, 41). The identification is achieved by calculating the overall difference between a test spectrum and all reference spectra. A test strain is assigned to the source of the nearest reference spectrum (15). However, such a procedure is univariate and does not consider patterns of individual differences at different wavelengths, leaving a wealth of information stored in the spectra unused. In case of the differentiation of closely related species within the same genus, advanced multivariate methods for data analysis are therefore required.
Investigation of Listeria species using FTIR spectroscopy has been undertaken previously (18, 24). However, these studies included only a single strain per species, and even in this simple case the unequivocal clustering of different spectra of the same strain was not always possible. It therefore remained unclear whether FTIR spectroscopy would have the capacity to differentiate the Listeria species, especially if the technique was applied to a strain selection covering at least a significant part of the natural microbiodiversity of the species. If many strains from several species are included in the analysis, self-learning systems such as artificial neural networks (ANNs) may be able to extract the information stored in the spectra of such a broad database and greatly enhance the species-specific differentiation of bacterial isolates, when a comprehensive reference data set is used (28, 37, 38). In the present study, ANNs have therefore been applied to establish a classification system for Listeria FTIR spectra and its performance has been compared to univariate FTIR analysis and the standard phenotypical API differentiation of Listeria. We report that the semiautomated, ANN-based FTIR technique allows reliable identification of Listeria species in 25 h and is suitable for use in a routine microbiological laboratory.
MATERIALS AND METHODS
Bacterial strains.
A list of all 520 Listeria strains used in this study can be found in Tables S1 and S2 of the supplementary material.
Reference strain set: sequence analysis of iap and thy genes.
A reference strain set of 243 well-defined strains of Listeria monocytogenes, L. innocua, L. ivanovii, L. seeligeri, and L. welshimeri from international culture collections (American Type Culture Collection and the Special Listeria Culture Collection) and the Weihenstephan Listeria collection served to establish a spectral reference data set. A total of 164 strains of this set have been identified by DNA sequence analysis of the complete iap gene and thymidylate synthase gene (the thy gene), and the remaining strains have been identified by a multiplex PCR system developed by Bubert et al. (4).
For the sequence analysis of the iap and thy genes, 9 ml of liquid culture grown in 10 ml of brain heart infusion broth (Oxoid, England) at 30°C was harvested, resuspended in 2 ml of purified water, and kept at −20°C. Eight microliters of this lysate served as a template for a 100-μl PCR. Thermal cycling was performed with a Techne Cyclone Gradient cycler (Pequlab, Erlangen, Germany). The iap gene was PCR amplified and sequenced with the primers iap-P-V/57 (5′-ATG AAT ATG AAA AAA GCA ACT ATC GC) and iap-P-R/57 (5′-TTA TAC GCG ACC GAA GCC AA). These primers bind at the 5′ and 3′ ends of the iap gene, covering the entire iap sequence, and were designed by ClustalW (http://www.ebi.ac.uk/clustalw/) alignments of Entrez Nucleotides database Listeria sequences (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide). The internal primer iap_F700/58 (5′-GTC ATG GAA TAA TTT ATC T[G/T]C TTC TTC) was used for DNA sequencing. PCR was performed using 50 μl AB-Gene 2× Reddy Mix with 1.5 mM MgCl2 (AB-Gene, Hamburg, Germany), 8 μl of lysate, 1 μl of each primer [50 pmol/μl], and 40 μl of purified water. Thermal cycling conditions were 5 min at 95°C, followed by 29 cycles, each consisting of 20 s at 95°C, 30 s at 50°C, and 1 min 40 s at 72°C.
The thy gene was PCR amplified and sequenced using the primers thy_2_F/62 (5′-GAG GAA ATG ATG GAA CGC TGG GA) and thy_1_R/60 (5′-TAT T[G/C/C C/A]G[G CGC GGT CTT GTG). These primers bind in the noncoding region of the thy gene and were designed based on one L. monocytogenes thy sequence provided by Pascale Cossart (Institute Pasteur, France) and homologous GenBank sequences identified by a BLAST search (http://www.ncbi.nlm.nih.gov/BLAST/). All primers were checked using Netprimer (http://www.premierbiosoft.com/netprimer/netprlaunch/netprlaunch.html). PCR was performed as described for the iap gene, using the following thermal cycling conditions: 5 min at 95°C, followed by 27 cycles, each consisting of 20 s at 95°C, 30 s at 54°C, and 1 min 50 s at 72°C. PCR products were purified using the QIAquick 96 PCR Purification kit (QIAGEN, Hilden, Germany). Sequencing was performed at Sequiserve (Willi Metzger, Vaterstetten, Germany). The complete coding region of the thy gene was sequenced. Sequences were aligned using ClustalW (http://genius.embnet.dkfz-heidelberg.de/menu/w2h/w2hdkfz/ or http://www.ebi.ac.uk/clustalw/) and edited using Jalview (http://genius.@embnet.dkfz-heidelberg.de/menu/w2h/w2hdkfz/ or http://www.ebi.ac.uk/∼michele/jalview/). Dendrograms were constructed using Jalview.
Identification of the validation strain set isolates.
A set of 277 strains representing five species of Listeria of diverse habitats (foods, environment, and animals and humans from South and North America and central, northern, and southern Europe) were used for the external validation of the ANN model. This strain set will be referred to in the rest of this paper as the external validation strain set. These strains have been identified by a phenotypic API Listeria test (bioMérieux, Marcy-l'Etoile, France) according to the manufacturer's instructions. Additionally, these strains were examined for the presence of hemolysis activity by a tube test with an erythrocyte suspension. Briefly, the serum was carefully removed from fresh, sterile, and defibrinated sheep blood (Oxoid, England), and a 2% (vol/vol) suspension of the erythrocytes was made in phosphate-buffered saline (pH 7.2). A total of 1 ml of Listeria culture grown overnight at 37°C in brain heart infusion broth was mixed with 1 ml of the erythrocyte suspension. After incubation for 24 h at 37°C, the presence or absence of hemolysis was observed.
The Multiplex PCR system developed by Bubert et al. (4) was used for the differentiation of Listeria spp. belonging to the external validation strain set when API and FTIR identification was discordant. DNA was prepared by using one loop of bacterial cells, which were homogenized in 200 μl Mili-Q water. Cells were disrupted with 0.5 g zirconia-silica beads (0.1-mm diameter; Roth, Karlsruhe, Germany) by being shaken two times at 6.5 m s−1 for 45 s in a Hybaid RiboLyser (Middlesex, United Kingdom) cell disrupter. Afterwards, the lysate was separated by centrifugation at 13,000 × g for 3.5 min. DNA amplification reactions were carried out in a 50-μl final volume containing 25 μl Reddy Mix [75 mM Tris-HCl (pH 8.8), 20 mM (NH4)2SO4, 1.5 mM MgCl2, 0.01% (vol/vol) Tween 20, 0.2 mM each deoxyribonucleoside triphosphate, 1.25 U of Taq polymerase] from PCR Master Mix, ABgene (Surrey, United Kingdom), 8 μl of lysate, and 1 mM each primer. The primer combination and PCR conditions were as described previously (4).
Measurement of FTIR spectra.
The strains stored at −80°C were streaked and subcultured on tryptone soy agar plates (Oxoid, Basingstoke, Hampshire, England) for 24 h. The growth temperature was 37 ± 2°C. The sample preparation for the infrared (IR) absorbance measurements was performed as described by Oberreuter et al. (31). Prior to the spectral measurements, the sample holder was sealed with a KBr cover plate to prevent contamination of the spectrometer. The spectra were recorded and evaluated according to Oberreuter et al. (31). For data processing, such as calculation of derivatives and normalization, OPUS software, version 4.2 for Windows NT (Bruker, Germany), was used. First derivatives of the original IR spectra were calculated with a 9-point Savitzky-Golay filter to minimize problems from unavoidable baseline shifts. It was observed that the identification quality increases with the inclusion of additional repetitive measurements per strain in the reference data set. Best identification was achieved with 10 independent measurements (from independent bacterial cultures) of each strain being included in the reference database to ensure a sufficient coverage of biological variance of growth and sampling procedures, although a standard and strict operation procedure was established (data not shown). The success of ANN modeling strongly depends on the quality of the spectra (37). Therefore, the thresholds for minimum absorbance (0.25) and maximum absorbance (1.20) for detector linearity, signal-noise ratios with a noise maximum of 1.5 × 10−4 units, and a water vapor content of spectral measurements of <3 ×10−4 U were predefined, and this quality test procedure was applied to each spectrum.
Univariate FTIR analysis.
The selection of relevant spectral ranges and establishment of the cutoff values of spectral distance (SD) for the identification of Listeria at the species level were done. To establish the SD value, three independent measurements of five strains of each Listeria species were used. The calibration of their SD threshold value for a correct identification of an isolate at species level was done using a procedure based on Oberreuter et al. (31). In our case, the windows from 900 to 1,200, 1,250 to 1,650, and 2,830 to 3,030 cm−1 (all weight factors were 30) and a cutoff value for the SD of 0.5 were used. This implies that the spectral distance between an isolate and the first hit of the identification hit list must be <0.5 to yield a valid identification at the species level. Then, 10 repetitive measurements from independent sample preparations of all reference strains were performed, resulting in 2,430 spectra, which were added to the reference spectral library.
Artificial neural network-based FTIR identification.
Before artificial neural network analysis, hierarchical cluster analysis (HCA) of the spectra in the reference library was used as a first step in developing the Listeria species identification scheme based on ANN. The HCA was performed using the first derivative of the original spectra as input in the regions ranging from 700 to 1,200, 1,500 to 1,800, and 2,800 to 3,100 cm−1, correlation with scaling to first range and Ward's algorithm according to the OPUS software (Bruker). The two major groupings resulting from this HCA were used to establish the first layer of the two-layered neural network. Afterwards, the subsequent subnets were optimized for respective classification at the species level.
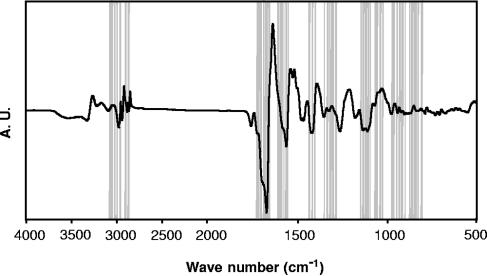
For the ANN analysis, 2,430 spectra of the reference data set were randomly distributed into a training set (8 spectra of each strain), a prevalidation set (1 spectrum of each strain), and a test set (1 spectrum of each strain). Prior to the artificial neural network analysis, the spectral windows between 700 and 1,800 cm−1, and 2,800 and 3,100 cm−1 were predefined in a data preprocessing step. For spectral feature selection, the most discriminative 61 wavelengths (Fig. 1) were selected based on the calculation of the covariance of the spectra data points (37). NeuroDeveloper software (Synthon, Heidelberg, Germany) was used to perform feature selection and to establish a two-layer neural network with 61 input neurons, one hidden unit, and two output units. For each classification level, a fully connected feed-forward neural network was trained with the Rprop algorithm (37).
FIG. 1.
First derivative of a Listeria FTIR spectrum. The regions of the infrared spectra contributing most significantly to the differentiation of the five Listeria species are highlighted. A.U., arbitrary units.
Validation of FTIR identification procedures.
To test both FTIR univariate and FTIR artificial neural network identification models, an internal validation was performed. One randomly selected spectrum of each strain in the database was excluded and used to construct a test set. This test set, containing independent spectra of each reference strain, was used to test the reference data set, and the results were determined at species level.
As a final test of performance, the validation strain set of 277 independent Listeria isolates, whose spectra were not included in the reference database, was identified by both univariate and ANN methods in an external validation.
RESULTS AND DISCUSSION
Modular architecture of the artificial neural network.
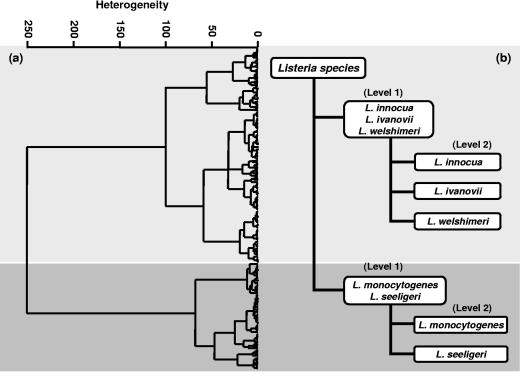
A modular ANN model was constructed for species identification at the basis of HCA groupings. HCA is a technique that groups IR spectra based on the overall similarity to other spectra. This technique can be applied “unsupervised,” due to its ability to perform the comparisons mathematically without predetermined information. In contrast, the ANN model was used as a “supervised” method of analysis, based on a learning procedure, which can classify unknown samples into predetermined groups. The similarity between the species observed in the HCA analysis, representing the Listeria reference spectra data set, provided information to develop modules of ANN with optimized classification performance through individual feature selection and network architecture. The modules are later integrated in one ANN classification system (37). Figure 2a shows the two major groups resulting from the cluster analysis, which were used to establish the first level of the ANN architecture comprising the L. innocua-L. ivanovii-L. welshimeri net and the L. monocytogenes-L. seeligeri net. Then, according to the outputs of this first level in the ANN classification scheme, specialized networks were activated at a second level, determining the species-specific subnetworks (Fig. 2b). Based on this classification scheme, the discrimination of Listeria down to the species level resulted from the projection of an unknown Listeria spectrum from the first level to the second level. When, in the first level, this spectrum was predicted to belong to one of the nets at this level, the output from this first level was projected to the second level to distinguish between the respective species available in the respective subnet.
FIG. 2.
(a) Hierarchical cluster analysis of the first derivative of 243 Listeria spectra included in the reference data set. It was performed by using the regions from 700 to 1,200, 1,500 to 1,800, and 2,800 to 3,100 cm−1, correlation with scaling to first range, and Ward's algorithm. (b) The two major groups resulting from the cluster analysis (a) were used to establish the first level of the architecture of the neural network for the identification of Listeria species. In the first level, the L. innocua-L. ivanovii-L. welshimeri net and the L. monocytogenes-L. seeligeri net were established. In the second level of this classification scheme, the species-specific subnetworks (L. innocua, L. ivanovii, L. monocytogenes, L. seeligeri, and L. welshimeri) were activated.
Validation of the spectral reference databases.
Univariate-based FTIR and ANN-based FTIR identification procedures were internally validated, based on 243 reference strains contained in the database (Table 1). The overall correctness of identification using the spectral window combination described in Materials and Methods was 88.9% at the species level for the univariate method. Less-satisfactory results were obtained for L. seeligeri (77.6%), due to the relatively high degree of misidentification of this species as L. monocytogenes. The overall performance of the ANN model was a rate of correct identification of 96.3%. The worst performance for both methods was observed with L. welshimeri, which showed the same misidentification results for the same two strains as L. ivanovii and L. innocua for the univariate and multivariate methods, respectively. This fact reveals that for these two particular strains, the intraspecific biodiversity represented in the database with only 19 L. welshimeri strains is limited.
TABLE 1.
Internal validation of the Listeria infrared spectral reference database
| Species | No. of strains tested | Univariate FTIR analysis
|
ANN identification
|
||||
|---|---|---|---|---|---|---|---|
| Correct identification % (no.) | Misidentificationa % (no.) | No identificationb % (no.) | Correct identification % (no.) | Misidentificationc % (no.) | No identificationd % (no.) | ||
| L. innocua | 65 | 95.4 (62) | 3.1 (2) | 1.5 (1) | 98.5 (64) | 1.5 (1) | |
| L. ivanovii | 41 | 87.8 (36) | 12.2 (5) | 95.1 (39) | 4.9 (2) | ||
| L. monocytogenes | 69 | 91.3 (63) | 8.7 (6) | 97.1 (67) | 2.9 (2) | ||
| L. seeligeri | 49 | 77.6 (38) | 22.4 (11) | 96.0 (47) | 4.0 (2) | ||
| L. welshimeri | 19 | 89.5 (17) | 10.5 (2) | 90.0 (17) | 10.0 (2) | ||
| Total | 243 | 88.9 (216) | 11.0 (26) | 0.1 (1) | 96.3 (234) | 3.7 (9) | |
Strains yielding an SD value below or equal to the threshold value of 0.5 used for correct identification of Listeria species, but their identification corresponded to a different species.
Strains yielding an SD value higher than the threshold value of 0.5 used for correct identification of Listeria species.
Strains yielding identification results corresponding to a different species.
Strains not yielding identification.
Once the FTIR univariate and the ANN models using a 243-reference-strain data set were established, external validation comprising 277 isolates not included in the reference database was used to challenge both FTIR models. According to Table 2, 129 of 130 strains of L. monocytogenes were correctly identified by the neural network method, whereas the univariate approach identified only 121 strains correctly. Similarly, L. innocua, L. ivanovii, and L. seeligeri reached better identification results by ANN than by the univariate model. Only L. welshimeri showed the same poor rate of accuracy of identification by both methods. This indicates that the low number of L. welshimeri strains included in the database limited their identification. While the univariate FTIR analysis procedure allowed the correct identification of 85.2% (236 of 277) of all test strains, the ANN method was able to identify 96.0% (266 of 277) of strains correctly at the species level. Furthermore, comparable results of the prediction accuracy in the internal (96.3%) and external (96.0%) validation of the ANN reveals the stability of this model, indicating that a significant part of the microbiodiversity of the Listeria species was covered by the reference database and represented by the ANN classification system.
TABLE 2.
External validation of the identification potential of univariate FTIR, ANN, and API using 277 Listeria strains not included in the reference data set
| Species | No. of strains tested | Univariate FTIR analysis
|
ANN identification
|
API Listeria
|
||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Correct identification % (no.) | Misidentificationa % (no.) | No identificationb % (no.) | Correct identification % (no.) | Misidentificationc % (no.) | No identificationd % (no.) | Correct identification % (no.) | Misidentificatione % (no.) | No identificationf % (no.) | ||
| L. innocua | 60 | 91.7 (55) | 8.3 (5) | 93.3 (56) | 6.7 (4) | 90.0 (54) | 8.3 (5) | 1.7 (1) | ||
| L. ivanovii | 28 | 71.4 (20) | 28.6 (8) | 96.4 (27) | 3.6 (1) | 89.3 (25) | 3.6 (1) | 7.1 (2) | ||
| L. monocytogenes | 130 | 93.0 (121) | 7.0 (9) | 99.2 (129) | 0.8 (1) | 93.1 (121) | 6.9 (7) | 1.5 (2) | ||
| L. seeligeri | 48 | 64.6 (31) | 35.4 (17) | 93.8 (45) | 6.2 (3) | 75.0 (36) | 2.1 (1) | 22.9 (11) | ||
| L. welshimeri | 11 | 81.8 (9) | 18.2 (2) | 81.8 (9) | 18.2 (2) | 72.7 (8) | 27.3 (3) | |||
| Total | 277 | 85.2 (236) | 14.8 (41) | 96.0 (266) | 4.0 (11) | 88.0 (244) | 5.1 (14) | 6.9 (19) | ||
Strains yielding an SD value below or equal to the threshold value of 0.5 used for correct identification of Listeria species, but their identification corresponded to an incorrect species.
Strains yielding an SD value higher than the threshold value of 0.5 used for correct identification of Listeria species.
Strains yielding identification results corresponding to an incorrect species.
Strains not yielding identification results.
Strains yielding identification results corresponding to an incorrect species.
Strains yielding no or multiple results.
Influence of the number of the reference strains on the identification success.
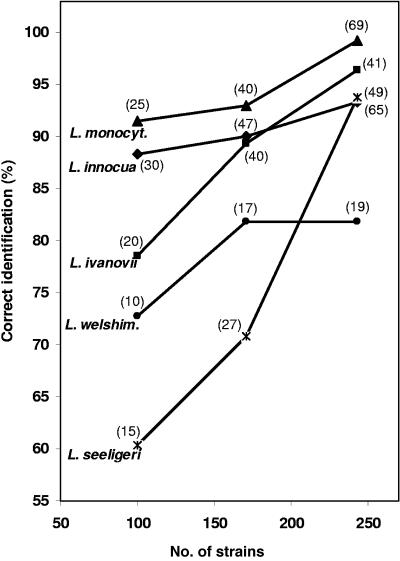
Three ANN reference databases with 100, 171, and 243 randomly chosen strains, respectively, were compiled. The same 277-validation-strain set described above was identified by all three databases (Fig. 3). In general, as expected, the inclusion of more biological intraspecies variability leads to an improvement of interspecies differentiation. This is in accordance with previous studies for other microbes (28, 31). Oberreuter et al. (31) reported for the coryneform bacteria that, on average, 5 to 10 strains of a species are needed to achieve an identification success of approximately 90%. We have observed that the species of the genus Listeria require more strains per species (around 20 to 25) to better cover the natural intraspecies variability range (data not shown). Hence, a good representation of the species biodiversity in the reference database will allow a reasonable identification capacity. We have observed that the addition of more L. innocua strains not only improved the identification results of L. innocua, but also improved those of L. ivanovii. This is in agreement with the observation that L. ivanovii was mostly misidentified as L. innocua by the ANN models, based on analysis of 100 and 171 strains (data not shown). This was supported by the noticeable increase of correct L. ivanovii identification from 89.3% to 96.4% when only a single strain of L. ivanovii was added to the ANN model with 171 strains. It was also noted that inclusion of more L. monocytogenes and L. seeligeri strains yielded a large improvement of their correct identifications. While the rate of identification success for L. monocytogenes was nearly perfect (99.2%), L. welshimeri identification results did not improve when ANN based on 171 strains was compared to ANN comprising all 243 strains. This led us to speculate that the species L. welshimeri contains strains whose FTIR absorption differences are not mainly due to specific cellular structures. Therefore, the database must include more strains for L. welshimeri to cover the entire biological variance of this species.
FIG. 3.
Comparison of the external validation of the ANN model using three different reference data sets including 100, 171, and 243 strains. The number of strains per species included in each data set is indicated in parentheses.
Comparison of API- and FTIR-ANN-based Listeria identification.
Considering that the API Listeria system has been listed as one of the preferred rapid methods for the biochemical identification of Listeria species in the routine microbiology food laboratory (17), we applied this technique as a second identification method to the 277 isolates of the validation strain set. This system allows a 24-h identification of all Listeria species, based on 10 sugar fermentation reactions and enzymatic reactions in microtubes, usually without the need for additional tests (3). Discordances between the FTIR-ANN method and API analysis were found for 39 out of the 277 strains. To resolve this conflicting data, multiplex PCR of the iap gene was performed (4). This method confirmed the FTIR-ANN results for 28 of the 39 discordances. On the other side, the multiplex PCR confirmed the API test for six strains only. The remaining 5 of the 39 discordant strains were unidentified by the API kit and were misidentified by the ANN method. The API Listeria test kit therefore provided a correct identification for 244 of the 277 isolates (88.0%), while the FTIR-artificial neural network correctly identified 266 of the 277 isolates (96.0%) (Table 2). In this study, the API Listeria system misidentified 14 (5.1%) of the isolates. Most important, seven strains of the pathogenic species L. monocytogenes were misidentified as the nonpathogenic L. innocua, due to ambiguous results from the DIM reaction of the API test system. Additionally, for 10 strains the hemolysis test has been used as a supplementary test when the API Listeria system indicated inconclusive results. Several publications on the identification capacity of the API for Listeria species reported similar limitations (3, 21, 33).
Based on the identification of the 277-validation-strain set, the sensitivity, specificity, and accuracy of the two methods in terms of their reliability to detect the human pathogen L. monocytogenes were evaluated. Sensitivity is defined as the ability of a test to detect a true L. monocytogenes sample when it is truly present. Specificity is defined as the ability of the test to detect the presence of L. monocytogenes in the sample when it is truly not present; accuracy relates to the closeness of the results to the true identification (2, 10). The data in Table 3 clearly demonstrate for all three parameters that ANN-based FTIR identification is the superior method. No other phenotypical method so far described in the literature provides an overall correct identification of 96% for all Listeria species and a success rate of 99.2% for correct L. monocytogenes identification. ANN-based FTIR identification therefore appears to be a promising technique for the semiautomated and rapid identification of Listeria species in 25 h in a routine food microbiological laboratory.
TABLE 3.
Comparison of the sensitivity, specificity, and accuracy of ANN and API identification proceduresa
| Method | No. of strainsb | Result
|
||||||
|---|---|---|---|---|---|---|---|---|
| TP | TN | FP | FN | Sensitivity (%) | Specificity (%) | Accuracy (%) | ||
| ANN | 277 | 129 | 142 | 5 | 1 | 99.2 | 96.6 | 97.8 |
| API Listeria | 277 | 121 | 144 | 3 | 9 | 94.6 | 96.0 | 95.7 |
Abbreviations: TP, true positives; TN, true negatives; FP, false positives; FN, false negatives. Sensitivity is calculated as TP/(TP + FN); specificity is TN/ (TN + FP); and accuracy is (TN + TP)/(TN + TP + FN + FP).
These strains were not included in the reference database.
Supplementary Material
Acknowledgments
This work was supported in part by the FEI (Forschungskreis der Ernährungsindustrie e.V., Bonn), the AiF (Arbeitskreis für industrielle Forschung), and the Ministry of Economics and Technology, project no. 14126N.
We thank H. Hof, Mannheim, Germany; M. Wagner, Vienna, Austria; J. Buck, Wangen, Germany; C. Montel, Aurillac, France; K. Pellicer and M. G. Echeverría, La Plata, Argentina; B. Becker and W. H. Holzapfel, Karlsruhe, Germany; M. Schmidt, Germany; F. Eliskases-Lechner, Rotholz, Austria; and A. Bubert, Germany, for kindly providing Listeria strains. We thank Pascale Cossart, Paris, France, for providing us with the thymidylate synthase gene sequence of L. monocytogenes. Special thanks to Jochen Dietrich and Stefanie Rubenwolf for practical support in our laboratory.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Allerberger, F., M. Dierich, G. Petranyi, M. Lalic, and A. Bubert. 1997. Nonhemolytic strains of Listeria monocytogenes detected in milk products using VIDAS immunoassay kit. Zentralbl. Hyg. Umweltmed. 200:189-195. [PubMed] [Google Scholar]
- 2.American College of Physicians. 2005. Sensitivity, specificity definition. [Online.] http://www.acponline.org.
- 3.Bille, J., B. Catimel, E. Bannerman, C. Jacquet, M.-N. Yersin, I. Caniaux, D. Monget, and J. Rocourt. 1992. API Listeria, a new and promising one-day system to identify Listeria isolates. Appl. Environ. Microbiol. 58:1857-1860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bubert, A., I. Hein, M. Rauch, A. Lehner, B. Yoon, W. Goebel, and M. Wagner. 1999. Detection and differentiation of Listeria spp. by a single reaction based on multiplex PCR. Appl. Environ. Microbiol. 65:4688-4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bubert, A., J. Riebe, N. Schnitzler, A. Schonberg, W. Goebel, and P. Schubert. 1997. Isolation of catalase-negative Listeria monocytogenes strains from listeriosis patients and their rapid identification by anti-p60 antibodies and/or PCR. J. Clin. Microbiol. 35:179-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bubert, A., P. Schubert, S. Kohler, R. Frank, and W. Goebel. 1994. Synthetic peptides derived from the Listeria monocytogenes p60 protein as antigens for the generation of polyclonal antibodies specific for secreted cell-free L. monocytogenes p60 proteins. Appl. Environ. Microbiol. 60:3120-3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cocolin, L., K. Rantsiou, L. Iacumin, C. Cantoni, and G. Comi. 2002. Direct identification in food samples of Listeria spp. and Listeria monocytogenes by molecular methods. Appl. Environ. Microbiol. 68:6273-6282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cummins, A. J., A. K. Fielding, and J. McLauchlin. 1994. Listeria ivanovii infection in a patient with AIDS. J. Infect. 28:89-91. [DOI] [PubMed] [Google Scholar]
- 9.Deneer, H. G., and I. Boychuk. 1991. Species-specific detection of Listeria monocytogenes by DNA amplification. Appl. Environ. Microbiol. 57:606-609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dytham, C. 1999. Choosing and using statistics, p. 30-42. Blackwell Science, Ltd., Berlin, Germany.
- 11.Farber, J. M., and C. J. Addison. 1994. RAPD typing for distinguishing species and strains in the genus Listeria. J. Appl. Bacteriol. 77:242-250. [DOI] [PubMed] [Google Scholar]
- 12.Goodacre, R., E. M. Timmins, P. J. Rooney, J. J. Rowland, and D. B. Kell. 1996. Rapid identification of Streptococcus and Enterococcus species using diffuse reflectance-absorbance Fourier transform infrared spectroscopy and artificial neural networks. FEMS Microbiol. Lett. 140:233-239. [DOI] [PubMed] [Google Scholar]
- 13.Gracieux, P., S. M. Roche, P. Pardon, and P. Velge. 2003. Hypovirulent Listeria monocytogenes strains are less frequently recovered than virulent strains on PALCAM and Rapid′ L. mono media. Int. J. Food Microbiol. 83:133-145. [DOI] [PubMed] [Google Scholar]
- 14.Hahn, G., and P. Hammer. 1990. Listerien-freie Käse—höhere Sicherheit für den Verbraucher. Dtsch. Milchwirtsch. 33:1120-1125. [Google Scholar]
- 15.Helm, D., H. Labischinski, and D. Naumann. 1991. Elaboration of a procedure for identification of bacteria using Fourier-transform IR spectral libraries: a stepwise correlation approach. J. Microbiol. Methods 14:127-142. [Google Scholar]
- 16.Helm, D., H. Labischinski, G. Schallehn, and D. Naumann. 1991. Classification and identification of bacteria by Fourier-transform infrared spectroscopy. J. Gen. Microbiol. 137:69-79. [DOI] [PubMed] [Google Scholar]
- 17.Hitchins, A. D. 2003. Detection and enumeration of Listeria monocytogenes in foods. In George J. Jackson, Robert I. Merker, and Ruth Bandler (ed.), Bacteriological analytical manual online, 8th ed. U.S. Department of Health and Human Services, U.S. Food and Drug Administration Center for Food Safety and Applied Nutrition. [Online.] http://www.cfsan.fda.gov/∼ebam/bam-10.html.
- 18.Holt, C., D. Hirst, A. Sutherland, and F. MacDonald. 1995. Discrimination of species in the genus Listeria by Fourier transform infrared spectroscopy and canonical variate analysis. Appl. Environ. Microbiol. 61:377-378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Howard, P. J., K. D. Harsono, and J. B. Luchansky. 1992. Differentiation of Listeria monocytogenes, Listeria innocua, Listeria ivanovii, and Listeria seeligeri by pulsed-field gel electrophoresis. Appl. Environ. Microbiol. 58:709-712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jersek, B., E. Tcherneva, N. Rijpens, and L. Herman. 1996. Repetitive element sequence-based PCR for species and strain discrimination in the genus Listeria. Lett. Appl. Microbiol. 23:55-60. [DOI] [PubMed] [Google Scholar]
- 21.Johnson, J. L., and C. P. Lattuada. 1993. Comparison of nucleic acid hybridization assays and biochemical characterization tests for the confirmation of Listeria monocytogenes. J. Food Prot. 56:834-840. [DOI] [PubMed] [Google Scholar]
- 22.Jones, D., and H. P. R. Seeliger. 1992. The genus Listeria, p. 1595-1616. In A. Balows, H. G. Trüper, M. Dworkin, W. Harder, and K.-H. Schleifer (ed.), The prokaryotes, 2nd ed., vol. 2. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 23.Kümmerle, M., S. Scherer, and H. Seiler. 1998. Rapid and reliable identification of food-borne yeasts by Fourier-transform infrared spectroscopy. Appl. Environ. Microbiol. 64:2207-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lefier, D., D. Hirst, C. Holt, and A. G. Williams. 1997. Effect of sampling procedure and strain variation in Listeria monocytogenes on the discrimination of species in the genus Listeria by Fourier transform infrared spectroscopy and canonical variates analysis. FEMS Microbiol. Lett. 147:45-50. [DOI] [PubMed] [Google Scholar]
- 25.Lessing, M. P., G. D. Curtis, and I. C. Bowler. 1994. Listeria ivanovii infection. J. Infect. 29:230-231. [DOI] [PubMed] [Google Scholar]
- 26.Loessner, M., and S. Scherer. 2002. Listerien-Aktuelle Lage und Entwicklung neuer Schutz- und Reifungskulturen. Dtsch Molkerei-Z. 126:217-221.
- 27.Manzano, M., L. Cocolin, C. Cantoni, and G. Comi. 2000. Temperature gradient gel electrophoresis of the amplified product of a small 16S rRNA gene fragment for the identification of Listeria species isolated from food. J. Food Prot. 63:659-661. [DOI] [PubMed] [Google Scholar]
- 28.Maquelin, K., C. Kirschner, L. P. Choo-Smith, N. A. Ngo-Thi, T. van Vreeswijk, M. Stammler, H. P. Endtz, H. A. Bruining, D. Naumann, and G. J. Puppels. 2003. Prospective study of the performance of vibrational spectroscopies for rapid identification of bacterial and fungal pathogens recovered from blood cultures. J. Clin. Microbiol. 41:324-329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ninet, B., E. Bannerman, and J. Bille. 1992. Assessment of the Accuprobe Listeria monocytogenes culture identification reagent kit for rapid colony confirmation and its application in various enrichment broths. Appl. Environ. Microbiol. 58:4055-4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Norton, D. M. 2002. Polymerase chain reaction-based methods for detection of Listeria monocytogenes: toward real-time screening for food and environmental samples. J. AOAC Int. 85:505-515. [PubMed] [Google Scholar]
- 31.Oberreuter, H., H. Seiler, and S. Scherer. 2002. Identification of coryneform bacteria and related taxa by Fourier-transform infrared (FT-IR) spectroscopy. Int. J. Syst. Evol. Microbiol. 52:91-100. [DOI] [PubMed] [Google Scholar]
- 32.Olsen, J. E., S. Aabo, W. Hill, S. Notermans, K. Wernars, P. E. Granum, T. Popovic, H. N. Rasmussen, and O. Olsvik. 1995. Probes and polymerase chain reaction for detection of food-borne bacterial pathogens. Int. J. Food Microbiol. 28:1-78. [DOI] [PubMed] [Google Scholar]
- 33.Paillard, D., V. Dubois, R. Duran, F. Nathier, C. Guittet, P. Caumette, and C. Quentin. 2003. Rapid identification of Listeria species by using restriction fragment length polymorphism of PCR-amplified 23S rRNA gene fragments. Appl. Environ. Microbiol. 69:6386-6392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rocourt, J. 1999. The genus Listeria and Listeria monocytogenes: phylogenetic position, taxonomy, and identification, p. 1-20. In E. T. Ryser and E. H. Marth (ed.), Listeria, listeriosis, and food safety, 2nd ed. Marcel Dekker, New York, N.Y.
- 35.Rocourt, J., H. Hof, A. Schrettenbrunner, R. Malinverni, and J. Bille. 1986. Acute purulent Listeria seeligeri meningitis in an immunocompetent adult. Schweiz. Med. Wochenschr. 116:248-251. (In French.) [PubMed] [Google Scholar]
- 36.Rudolf, M., and S. Scherer. 2001. High incidence of Listeria monocytogenes in European red smear cheese. Int. J. Food Microbiol. 63:91-98. [DOI] [PubMed] [Google Scholar]
- 37.Schmitt, J., and T. Udelhoven. 2001. Use of artificial neural networks in biomedical diagnosis, p. 379-419. In H. U. Gremlich and B. Yan (ed.), Infrared and Raman spectroscopy of biological materials. Marcel Dekker, New York, N.Y.
- 38.Udelhoven, T., D. Naumann, and J. Schmitt. 2000. Development of a hierarchical classification system with artificial neural networks and FT-IR spectra for the identification of bacteria. Appl. Spectrosc. 54:1471-1479. [Google Scholar]
- 39.Vaneechoutte, M., P. Boerlin, H. V. Tichy, E. Bannerman, B. Jager, and J. Bille. 1998. Comparison of PCR-based DNA fingerprinting techniques for the identification of Listeria species and their use for atypical Listeria isolates. Int. J. Syst. Bacteriol. 48:127-139. [DOI] [PubMed] [Google Scholar]
- 40.Volokhov, D., A. Rasooly, K. Chumakov, and V. Chizhikov. 2002. Identification of Listeria species by microarray-based assay. J. Clin. Microbiol. 40:4720-4728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wenning, M., H. Seiler, and S. Scherer. 2002. Fourier-transform infrared microspectroscopy, a novel and rapid tool for identification of yeasts. Appl. Environ. Microbiol. 68:4717-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.