Abstract
Like most gram-positive oral bacteria, Actinomyces naeslundii is resistant to salivary lysozyme and to most other lytic enzymes. We are interested in studying the lysins of phages of this important oral bacterium as potential diagnostic and therapeutic agents. To identify the Actinomyces phage genes encoding these species-specific enzymes in Escherichia coli, we constructed a new cloning vector, pAD330, that can be used to enrich for and isolate phage holin genes, which are located adjacent to the lysin genes in most phage genomes. Cloned holin insert sequences were used to design sequencing primers to identify nearby lysin genes by using whole phage DNA as the template. From partial digestions of A. naeslundii phage Av-1 genomic DNA we were able to clone, in independent experiments, inserts that complemented the defective λ holin in pAD330, as evidenced by extensive lysis after thermal induction. The DNA sequence of the inserts in these plasmids revealed that both contained the complete lysis region of Av-1, which is comprised of two holin-like genes, designated holA and holB, and an endolysin gene, designated lysA. We were able to subclone and express these genes and determine some of the functional properties of their gene products.
Phages of oral bacteria have been studied in this laboratory for a number of years since we believe they are likely to play a significant ecological role in regulating the oral microflora. We are also interested in phage-encoded lytic enzymes as potential diagnostic tools (10, 19, 23) and as therapeutic agents (1, 6, 7, 13) to control specific oral pathogens. Actinomyces naeslundii is a gram-positive, facultative anaerobe that is found in high numbers in dental plaque and is believed to be causally involved in gingivitis and root surface caries in the oral cavity, in addition to actinomycosis in other regions of the body (22). The cell wall of this organism is resistant to salivary lysozyme and most other peptidoglycan-hydrolyzing enzymes, even though it is chemically similar to many other gram-positive cell walls that are lysozyme sensitive (18). Phage Av-1, originally isolated in this laboratory (3), is a small group I phage that is lytic for several human strains of A. naeslundii (2). This implies that its genome encodes an endolysin that can degrade the cell wall of this species. Preliminary experiments in this laboratory demonstrated that phage lysates of Av-1 contained a lytic enzyme that is specific for A. naeslundii. To clone the Av-1 lysin gene in Escherichia coli, which is also resistant to the enzyme, we devised an indirect strategy in which the functional activity of a plasmid-encoded λ endolysin is restored via complementation of a defective λ holin. Since holins are not species specific, we constructed a cloning vector to isolate plasmids containing phage inserts that express lethal, holin-like activity in E. coli. By sequencing these inserts, identifying potential open reading frames (ORFs), and analyzing their deduced amino acid sequences, putative holins could be tentatively identified on the basis of size, possession of transmembrane regions, and charge distribution, even though holins frequently cannot be identified by sequence similarities to known phage holins (25-27). Since holin and lysin genes are located in the same region in most phage genomes, we reasoned that by using DNA sequencing primers originating from putative holin sequences, it should be possible to identify nearby endolysin genes, based on amino acid similarities to known phage lysins. The lysin gene(s) could then be PCR amplified from whole phage DNA and cloned into a suitable expression vector to allow production and characterization of the encoded enzyme. Here we describe the construction and use of a new cloning vector that we used to enrich for phage genome fragments that express holin-like genes. In the case of phage Av-1, the use of this vector enabled us to clone the phage's holin and lysin genes. These genes were subcloned and expressed in E. coli and the unusual and potentially useful functional properties of the proteins encoded by these genes were partially characterized.
MATERIALS AND METHODS
Bacterial strains, phages, and plasmids.
The organisms and plasmids used in the present study are listed in Table 1. A. naeslundii strain MG-1S is a spontaneous streptomycin-resistant mutant of MG-1, the host strain that was used to originally isolate phage Av-1 (3); it was isolated on Trypticase soy broth (BBL Laboratories) agar plates containing 1 g of streptomycin/liter. Phage Av-1 and its host strain MG-1 (formerly classified as A. viscosus) are available from the American Type Culture Collection (Manassas, VA) and the Felix d'Herelle Reference Center for Bacterial Viruses (Laval University, Quebec, Canada).
TABLE 1.
Bacterial strains, phages, and plasmids used in this study
| Strain, phage, or plasmid | Relevant genotype or phenotypea | Source and/or reference |
|---|---|---|
| Strains | ||
| Actinomyces naeslundii MG-1S | Host for phage Av-1; Strr | Lab stock (3) |
| Escherichia coli K-12 | ||
| DH5α | Cloning host for blue-white screening | Invitrogen |
| MC4100 | General host for plasmids and prophages | R. Young |
| LE392 | supE supF; suppresses λSam7 | Lab stock |
| Rosetta | LacY cloning host carrying pRARE(camr) | Novagen |
| Phages | ||
| Av-1 | Group I phage of A. naeslundii | Lab stock (3) |
| λcI857 Sam7 | Amber mutation in S gene | Lab stock |
| λcI857 ΔSR kan | Lysis genes deleted, Q+; Kanr | R. Young (15) |
| Plasmids | ||
| pS105 | λQ−S+R+Rz+; Ampr | R. Young (21) |
| pAD330 | λQ−S−R+Rz+; Ampr | This study |
| pQE81L | Cloning vector for expressing N-terminal His6 tag fusions | QIAGEN |
| pRARE | Provides tRNAs that recognize the rare codons AGG, AGA, AUA, CUA, CCC, and GGA | Novagen |
Strr, streptomycin resistance; Kanr, kanamycin resistance; camr, chloramphenicol resistance.
Media and growth conditions.
MG-1S was grown in TSYPS broth, which is composed of 30 g of Trypticase soy broth, 5 g of yeast extract, 5 g of K2HPO4, and 0.5 g of streptomycin sulfate/liter, at 34 or 37°C under static conditions, in air, or on plates containing TSYPS solidified with 15 g of agar/liter. E. coli strains were routinely grown in LB broth or on LB agar at 32°C, 37°C or 42°C (depending on the experiment). Broth cultures were incubated under vigorous aeration (250 to 300 rpm). To maintain plasmids, select transformants or isolate lysogens, antibiotics were added to the following final concentrations as appropriate: ampicillin (100 μg/ml), kanamycin (50 μg/ml), and chloramphenicol (35 μg/ml). SOC broth (Invitrogen) was used to recover transformants after electroporation before plating on antibiotic-containing medium.
Growth of phage Av-1 and purification of Av-1 DNA.
Phage Av-1 was grown by infecting log-phase TSYPS broth cultures of MG-1S at a multiplicity of infection of 0.1 and incubating 16 h at 34°C. DNase and RNase were added to the crude phage lysates (1 U/ml each) and, after 30 min at 37°C, the lysates were chilled on ice and bacterial debris was removed by two centrifugations of 30 min at 10,000 × g. Phage were sedimented from the supernatant by centrifugation for 2 h at 280,000 × g (Beckman SW 70.1 rotor, 65K rpm). Phage DNA was extracted and purified by a standard proteinase K-CTAB procedure (4), modified as follows. The phage pellets from two to four tubes were combined by resuspending in 900 μl of a solution containing 690 μl of water, 100 μl of 0.5 M EDTA, 100 μl of 1 M Tris (pH 8.0), and 10 μl of proteinase K (10 mg/ml). After incubation for 30 min at 37°C, the phage DNA was isolated by adding 100 μl of CTAB (cetyltrimethylammonium bromide; 0.5% in 0.5 M NaCl) and heating the mixture 5 min in a water bath at 65°C. After the mixture cooled to room temperature, a sterile bent glass rod was used to remove the DNA-detergent precipitate to a screw-cap 15-ml tube containing 2 ml of 1.2 M NaCl, and the DNA was allowed to slowly redissolve overnight by placing the tube on a vertical rotating apparatus. The DNA was then precipitated by adding 4 ml of 95% ethanol, collected by centrifugation for 10 min at 10,000 × g, washed in 70% ethanol, and then resuspended in 1 to 2 ml of water.
Growth of lambda phages.
The lambda Sam7 and ΔSR phages, both of which carry the cI857 temperature-sensitive repressor allele, were grown by heating log-phase lysogenic cultures for 15 min at 42°C, incubating them 4 h at 37°C, and then lysing the cultures with CHCl4 (0.1% [vol/vol]). Debris and unlysed cells were removed by low-speed centrifugation, and the supernatants were sterile filtered (0.45-μm-pore-size Millipore filter). LE392 was used as a host to titer the Sam7 mutant, using TCMB bottom agar (10 g of Trypticase/liter, 5 g of NaCl/liter, 10 mM MgSO4, 1 ml of 0.1% sterile filtered vitamin B1/liter, and 11 g of agar/liter) and TCMB top agar (7 g of agar/liter) to enhance plaque size. To obtain λΔSR lysogens, host strains were plated in LB top agar on kanamycin LB agar plates, spotted with phage suspension, and incubated at 32°C. Kanamycin-resistant (Kanr) colonies were picked and purified by restreaking, and the presence of the prophage was confirmed by testing for absence of growth at 42°C, resistance to kanamycin, and immunity to a wild-type λ phage in spot tests on seeded top agar overlays; λvir was also used in spot tests to verify that apparent immunity was not due to phage resistance.
PCR and sequencing primers.
PCR primers used for vector construction, sequencing cloned inserts and cloning holin and lysin genes are listed in Table 2 All primers were synthesized by the University of Maryland, Baltimore, Biopolymer Laboratory. This core facility also carried out all sequencing reactions, utilizing dye terminator chemistry and automated ABI sequencers.
TABLE 2.
PCR and sequencing primers used in this study
| Primer pair | Sequence (5′-3′)a | Use |
|---|---|---|
| P136 | CGCGAATTCACCCGGGAGATCTA-CGGAGTAGA AGATGGTAGAAATCAATAATC EcoRI SmaI BglII | Construction of pAD330; replace ClaI-EcoRI region of pS105, |
| P137 | TGACAGCTTATCATCGATATGGGCCAACTC | create ′S and add SmaI and |
| ClaI | BglII sites | |
| P368holA | GCGGATCCAGGAGG-TTCGTCCGATGATACACATTAACCCTAC | Clone Av-1 holA gene |
| BamHI | ||
| P371holA | GCGAGATCT-GGACTCGTGCTCACCCATTG | |
| BglII | ||
| P341holB | GCCGGATCC-CAGAACCGTCAGCTAAGAGAAATCATCACCTACC | Clone Av-1 holB gene |
| BamHI | ||
| P340holB | GCGAGATCT-GGCAATGATGTCTGCGCGAGTTGCCACTTCTAC | |
| BglII | ||
| P368holA | Same as for P341holB | Clone Av-1 holA and holB together |
| P340holB | Same as for P340holB | |
| P295lys | GCCGGATCC-GCAACTCGCGCAGACATCATT | Clone Av-1 lysA gene |
| BamHI | ||
| P296lys | GCGAGATCT-AACTGTTTATGGGCGCTCTGC | |
| BglII | ||
| P231F | CGGAAGCAGAACCGGATCAC | Sequence inserts in pAD330 |
| P232R | CCACGCCAGCATATCGAGGAAC |
Restriction sites are underlined; the RBS and start codon are in italics.
Vector construction and experimental design.
We utilized the pS105 plasmid of Smith et al. (21) to construct a holin-selective cloning vector. This plasmid is a pBR-based medium-copy-number plasmid that carries a modified λ lysis region. The modifications consist of a truncated Q gene (eliminating antiterminator activity) and the single, but functional, S105 allele of the dual-start holin S gene, which normally encodes two proteins, the S105 holin and the S107 antiholin. pS105 has wild-type late promoter (pR′) and terminator (tR′) sites and functional R and Rz lysin genes. Using primers P136 and P137 (Table 2), we amplified the region of pS105 spanning the ClaI site to the C-terminal region of the S gene, which overlaps the ribosome-binding and start sites of the downstream R gene, and in the process added BglII, SmaI, and EcoRI restriction sites at the end of the truncated S gene. The resulting amplicon was purified and cut with ClaI and EcoRI before it was ligated into the large fragment of pS105, which was purified from a ClaI-EcoRI digest and dephosphorylated. The ligation products were electroporated into MC4100, and recombinants were selected on LB-ampicillin plates. The resulting vector, pAD330 (Fig. 1), was sequenced from upstream of the EcoRI site through the ClaI site to confirm that the sequence in this region was not altered during amplification or cloning. Inserts cloned in or downstream of the EcoRI site in pAD330 cannot be expressed since this vector, like pS105, is “Q-less” and transcription, which initiates at the late promoter pR′, terminates at tR′. Expression can only proceed in the presence of a functional Q antiterminator, which in our case is provided in trans in the cloning host from the resident λ cI857 ΔSR prophage upon thermal induction. If a protein expressed from the insert complements the defective ′S in the vector, the downstream lysin gene products can gain access to and degrade the peptidoglycan of E. coli, which results in cell lysis. This releases the plasmid into the medium, from which it can be concentrated and purified, as described below.
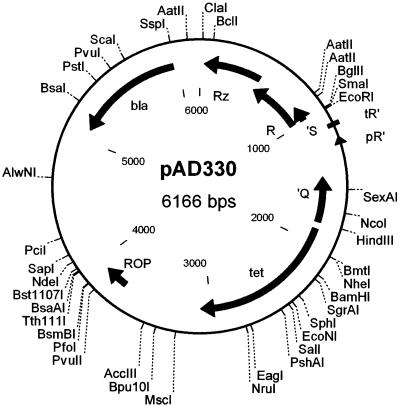
FIG. 1.
Holin-selective cloning vector pAD330. bla and tet regions encode resistance to ampicillin and tetracycline, respectively. Deletions in Q and S inactivate antitermination and holin activities of these proteins. ROP indicates region involved in regulation of plasmid replication in E. coli. The R and Rz genes are wild-type λ endolysin alleles.
Cloning Av-1 genome fragments.
Standard procedures were used to perform restriction enzyme digestions, agarose gel electrophoresis, dephosphorylation of vectors and plasmids, and ligation reactions (16), using enzyme buffers and reaction conditions recommended by suppliers. Partial Sau3AI restriction digests of Av-1 DNA were electrophoresed in 1% agarose gels, 2- to 3-kb fragments were excised from the gel, and the DNA was extracted and purified with a QIAquick gel extraction kit (QIAGEN); ligated into BglII-cut, dephosphorylated pAD330; and electroporated into E. coli lysogenized with λΔSR. After a 1-h growth period in SOC broth, transformants were selected by plating them on ampicillin LB agar plates and incubation at 32°C. The resulting clones were pooled by adding 5 ml of LB broth to each plate and scraping all of the colonies into the broth with a sterile glass rod. This suspension was then used in the “plasmid release” protocol, described below, to enrich for plasmids carrying holin genes.
Electroporation.
A BTX ECM399 electroporator (Genetronics, Inc.) was used to introduce (transform) plasmids into E. coli strains. Cells were made electrocompetent by the following procedure. One-liter LB broth cultures were grown to mid to late log phase (optical density at 600 nm of 0.5 to 1.0) at 32 or 37°C and chilled on ice. The cells were collected by low-speed centrifugation (15 min at 4,000 × g) and resuspended in 0.5 volume of sterile, cold water. The cells were washed twice more in the same manner, and the final pellets were resuspended in a 2 ml of sterile, cold 10% (vol/vol) glycerol. Cells were held on ice and used immediately, or aliquots were stored at −70°C for up to 2 weeks before use. Electroporation was carried out by mixing 2 μl of plasmid or ligation product with 98 μl of cold cells in a 1-mm sterile electroporation cuvette and then pulsing the mixture at 1,500 V; 900 μl of SOC medium was immediately added, and the cells were incubated 1 h at 37°C or 32°C on a shaker. Aliquots of 50 to 100 μl were then plated on LB plates containing the appropriate antibiotic(s) and incubated overnight to select recombinants or plasmid-carrying cells.
Enrichment of holin-complementing inserts.
We utilized a “plasmid release” protocol (provided by W. Roof, Texas A&M University) to enrich for plasmids expressing holin-like genes. Initial lysogenic transformant clones were pooled, as described above, diluted 1/100 into 25 ml of antibiotic-supplemented LB broth, grown to mid-log phase at 32°C, and then heat induced at 42°C for 15 min. The culture was then incubated for 15 min to 2 h at 37°C, chilled on ice, and centrifuged 10 min at 4,000 × g to remove unlysed cells and debris. The supernatant was then filtered through a Millex GV filter (0.2-μm pore size). Plasmids in the filtrate were concentrated and purified by adding EDTA to 10 mM, mixing with an equal volume of 2.5 M sodium acetate, loading the mixture onto a QIAGEN QIAPrep spin column, washing with PB and PE buffers, spinning to remove residual buffer, and then elution with 50 μl of water. This concentrate was then used to transform another culture of E. coli by electroporation, and the treated cells were plated to recover a second round of recombinants. This process was repeated three times to enrich for rare plasmids expressing holin-complementing activity (i.e., lysis). Plasmids from 20 to 30 clones from the last cycle of each experiment were extracted, digested with ClaI and EcoRI, and analyzed by agarose gel electrophoresis to determine approximate the sizes of their inserts. The plasmid inserts in selected clones were then sequenced, and their deduced amino acid sequences were determined. The amino acid sequences were then used to identify plasmids encoding putative holin-like proteins, based on size, charge distribution, and possession of one or more transmembrane regions.
Plasmid isolation and purification.
Plasmids were extracted and purified from 10-ml overnight LB cultures by using QIAprep Spin Miniprep kits (QIAGEN), which were also used in subsequent cloning procedures to remove restriction enzymes and alkaline phosphatase prior to ligations. For larger cultures, QIAfilter Plasmid Midi Kits (QIAGEN) were used. All plasmid sizes were checked by agarose gel electrophoresis, and DNA concentrations were measured by UV absorption at 260 nm.
PCR methods and PCR cloning.
Conventional PCR protocols were used for vector construction and to amplify specific genes from the Av-1 genome, using the primers listed in Table 2. Platinum Taq High Fidelity polymerase, buffer, and deoxynucleoside triphosphates from Invitrogen were used in a Hybaid PCRExpress thermocycler. Each 50-μl reaction, carried out in thin-walled tubes, contained 5 μl of 10× buffer, 1 μl of 10 mM deoxynucleoside triphosphate mix, 2 μl of 50 mM MgSO4, 1 μl of primer mix (10 mM each), 1 μl of template DNA (200 μg/ml), 0.5 μl of Taq enzyme (2.5 U), and 39.5 μl of autoclaved deionized water. Initial denaturation was carried out for 1 min at 94°C, and then 30 cycles of the following program was used: 94°C for 30 s, 55°C for 30 s, and 68°C for 1 min. The resulting amplicons (checked for size by agarose gel electrophoresis) were purified with a QIAGEN QIAquick PCR Purification Kit, restricted with the appropriate enzymes, and then repurified before being added to ligation mixtures. Insert orientation in the resulting plasmid recombinants was checked by restriction digest analysis, and their sequences were determined on both strands to confirm that no alterations occurred during amplification or cloning.
Expression and detection of cloned phage proteins.
A 10-ml portion of LB broth cultures of cells carrying phage Av-1 genes cloned in pAD330 or pQE81L was grown to mid-log phase (optical density 0.5 at 550 nm), induced (by adding 1 mM IPTG [isopropyl-β-d-thiogalactopyranoside] or by heating 15 min at 42°C), and then incubated 4 h at 37°C. Cells were collected by centrifugation (3,000 × g), washed two times by resuspension in phosphate-buffered saline (PBS; pH 7.2), and then resuspended in 0.5 ml of PBS containing a 1 μM concentration of the protease inhibitor phenylmethylsulfonyl fluoride. The cells were broken by sonication for 30 s, in an ice bath, with a Branson 250 Sonifier, using a microtip and a power setting of 2. The resulting extract was centrifuged 10 min at 18,000 × g, and proteins in both supernatant and pellet fractions were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). Detection of proteins produced in E. coli utilized conventional Laemmli denaturing polyacrylamide gels (4% stacking gel, 12% resolving gel), in a Bio-Rad Mini Protean III system, followed by Coomassie blue staining to visualize the protein bands.
Detection of lysin activity.
Lysoplate assays were used to detect phage endolysin activity. Water-washed, stationary-phase cells of MG-1S were concentrated 100-fold by centrifugation, and 2 ml was added to 7 ml of 1% molten agarose (in PBS) before the mixture was poured into 50-mm petri plates. After solidification, 5-mm wells were made with a no. 2 cork borer and filled with 50 μl of cell extract (adjusted to contain a total of 40 μg of total protein), and the plates were then incubated and examined for clear zones surrounding the wells.
Holin assays.
Holin-like activity was evaluated by three different methods. (i) E. coli λΔSR lysogens carrying holin genes cloned in pAD330 were grown to early log phase at 32°C and heated 15 min at 42°C to induce gene expression, and lysis was then monitored turbidimetrically during subsequent incubation at 37°C by measuring the absorbance at 550 nm (Fig. 6). (ii) A 2-μl portion of overnight 32°C broth cultures Av-1 holin plasmid-carrying Rosetta strains (LacZ+ LacY−) was spotted onto LB agar plates containing X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) and IPTG. The plates were incubated overnight at 32°C, placed in a 42°C incubator for 2 to 3 h, reincubated overnight at 37°C, and then examined for blue zones surrounding the colonies, which resulted from holin-like damage to the cytoplasmic membrane and leakage of β-galactosidase into the surrounding agar (8). (iii) Serial dilutions of λSam7 phage were plated on prophage-free, holin plasmid-carrying Rosetta cells, and the resulting titers were used to quantitate the ability of the plasmid-carried genes to complement the defective S gene in this phage by calculating the efficiency-of-plating on each strain compared to the titer obtained with the suppressor strain LE392. In these experiments the infecting phage provides a functional Q gene product in trans, which allows expression of the holin gene inserted in pAD330.
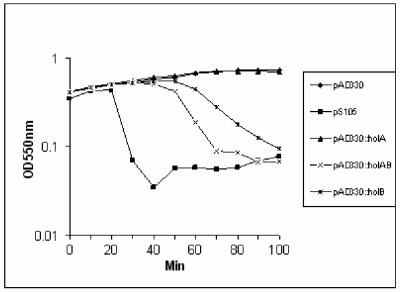
FIG. 6.
Cell lysis after heat induction of holin expression. E. coli Rosetta strains lysogenized with different holA, holB, and holAB plasmids were induced, and lysis was followed by periodically measuring the optical density (OD) at 550 nm as described in Materials and Methods.
Bioinformatic analyses.
Putative ORFs were identified and analyzed by the Vector NTI Suite 8 (InforMax, Inc.; Invitrogen) and Clone Manager Suite 7 (SciEd) programs, using the start codons of ATG, GTG, and TTG (including nested starts), the three conventional stop codons, and a minimal length of 40 amino acids. Potential ORFs were then examined manually to ensure that they had a potential Shine-Dalgarno sequence located 5 to 10 nucleotides upstream of the start codon or that its start codon was very close to or overlapped the stop codon of an upstream ORF. NCBI programs (BLAST 2.0) were then used for similarity searches of nucleotide sequences (blastx) and translated sequences (blastp) in the nonredundant GenBank database. The PSI-BLAST program was used to identify conserved protein domains (12). Evaluation of transmembrane regions in deduced protein sequences were determined by using the TMpred program (http://www.ch.embnet.org).
Nucleotide sequence accession number.
The nucleotide sequence of the 1,768-bp holin-complementing fragment of Av-1 DNA has been deposited in GenBank under accession number DQ123818.
RESULTS
Isolation of holin-complementing DNA fragments from the Av-1 genome.
Initial plates of transformed cells containing pAD330-Av-1 recombinants generally resulted in 400 to 1,000 colonies per plate. When pools of these cells were diluted in LB broth, grown to log phase, heat induced, and then incubated for 15 to 30 min, no holin-complementing plasmids were apparently released into the supernatant since no recombinants were obtained in subsequent enrichment cycles by using DNA concentrated from the supernatant. This was in contrast to control experiments with heat-induced lysogens carrying pS105, which released this plasmid within 15 min (see below). By extending the postinduction incubation period to 1 h or longer, holin-complementing plasmids were released into the supernatant as concentrates and then resulted in recovery of 10 to 100 recombinant clones per plate in the second enrichment cycle. After three cycles, the plasmids in 20 to 30 clones were extracted, and their insert sizes were determined by agarose gel electrophoresis after digestion with ClaI and EcoRI. Clones which carried plasmid inserts of 1 to 3 kb were then grown, heat induced, and observed for lysis during subsequent incubation. Two of five recombinants (obtained from separate, independent enrichments) that showed extensive lysis within 1 to 2 h were selected for sequencing in order to identify putative holins. These two plasmids were found to be carrying an identical 1,768-bp insert, the sequence of which was found by in silico analysis to include three genes, two putative holin genes, designated holA and holB, and an adjacent putative endolysin gene, designated lysA (Fig. 2). Both holin genes are predicted to utilize an AUG start codon, whereas the endolysin gene likely utilizes GUG as its start codon. The deduced amino acid sequences of the three proteins encoded by these genes are presented in Fig. 3. The other three recombinant plasmids, which caused significantly less lysis (data not shown), were not sequenced or examined further.
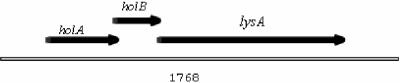
FIG. 2.
Putative genes encoded on the Av-1 holin-complementing fragment. The holA gene (in frame 1) overlaps the start of the holB gene (frame 2) by five nucleotides. The 5′ end of the lysA gene (frame 1) starts six nucleotides downstream of the end of the holB gene.
FIG. 3.
Amino acid sequences of HolA, HolB and LysA. Charged residues of the holins are indicated above the respective amino acids, and the predicted transmembrane regions are indicated by a double underline. The peptidoglycan-binding domain of LysA is indicated in boldface italics.
Subcloning and expression of the lysA gene.
The lysA gene was PCR amplified (by using the relevant PCR primers in Table 2), cloned into the N-terminal His6 tag vector pQE81L and introduced into DH5α. The product of the resulting gene fusion was expressed at high levels after IPTG induction (Fig. 4), and cell extracts containing it displayed strong lytic activity against washed, stationary-phase cells of host MG-1S (Fig. 5), confirming that lysA encodes an Av-1 endolysin. Interestingly, lysoplate assay plates prepared with conventional agar in place of agarose prevented lysin activity, suggesting the presence of some inhibitory substances in the former. Significantly, the N-terminal His tag did not prevent enzymatic activity and therefore can be utilized in future experiments to purify the enzyme for characterization studies.
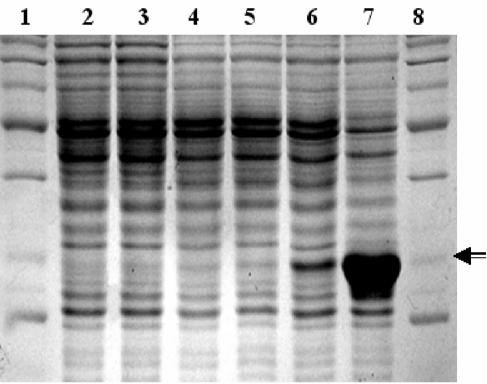
FIG. 4.
SDS-PAGE gel of N-terminal His6-tagged Av-1 lysin expressed in E. coli DH5α. Lanes 1 and 8, protein MW standards; lane 2, uninduced DH5α cells; lane 3, DH5α cells induced with IPTG; lane 4, uninduced cells containing pQE81L vector; lane 5, pQE81L-containing cells induced with IPTG; lane 6, uninduced cells containing Av-1 lysA-pQE81L construct; lane 7, Av-1 lysA-pQE81L-containing cells induced with IPTG. An arrow indicates the 30-kDa standard.
FIG. 5.
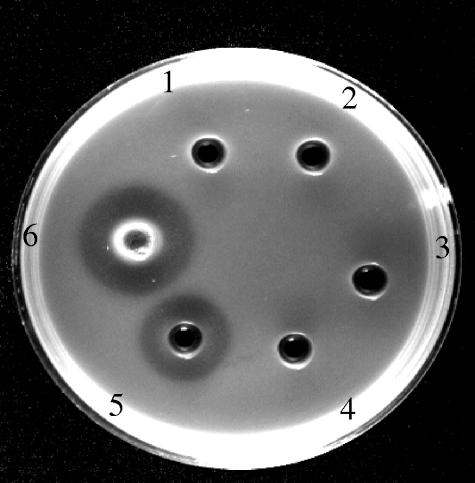
Lysoplate detection of lytic activity of His6-tagged Av-1 lysin. Wells contained extracts of DH5α cells with or without plasmids, as indicated: 1, uninduced plasmid-free cells; 2, IPTG-induced plasmid-free cells; 3, uninduced cells containing pQE81L; 4, pQE81L-containing cells induced with IPTG; 5, uninduced cells containing the Av-1 lysA-pQE81L construct; and 6, Av-1 lysA-pQE81L-containing cells induced with IPTG. All wells contained 40 μg of protein. The plate was photographed after 5 h at 37°C. The clear zones surrounding wells 5 and 6 are due to lysis of A. naeslundii cells embedded in the agarose; the enzymatic activity of uninduced cells carrying the lysA plasmid (well 5) is due to leaky expression from the pQE81L Lac promoter in the absence of inducer, which wasalso observed in SDS-PAGE gels (lane 6 in Fig. 4).
Subcloning and expression of holin genes.
The holA and holB genes were cloned separately and together into pAD330 (using the PCR primers indicated in Table 2), and the three plasmids were introduced into DH5α and MC4100 cells lysogenized with λcI857ΔSRkanr. No lysis of these cells was observed after thermal induction of the prophage, and no production of the holin proteins was detectable in SDS-PAGE gels (data not shown). Examination of the codon usage of these two genes revealed that both utilized several codons that are rarely used in E. coli. We therefore introduced these plasmids into the Novagen (EMD Biosciences) Rosetta strain, whose pRARE plasmid provides six rarely used tRNAs. These plasmid-carrying strains were lysogenized with the λΔSR phage. Thermal induction of these lysogens resulted in various degrees of cell lysis, as shown in Fig. 6. The holA holB plasmid caused more rapid and extensive lysis than did the holB plasmid, whereas the holA plasmid did not cause detectable lysis after induction. This result suggests that HolA and HolB proteins may interact in some cooperative fashion to effect lysis. When produced together the two holins caused more leakage of β-galactosidase than did either holin alone (Fig. 7), which is consistent with this interpretation. Each of the holin gene products by themselves was able to complement the defective S gene in λSam7 to a limited extent, whereas the pair together had a much larger effect on the efficiency of plating of this phage (Table 3). In these experiments, the infecting phage provides a functional Q gene product, which allows expression of insert genes but visible plaques cannot be formed on nonsuppressor strains unless the genes expressed can complement the defective Sam7 gene product. Plaque formation thus indicates that the produced protein(s) has holin-like activity. Taken together, our data suggest that both HolA and HolB proteins are required for optimal complementation of the Sam7 allele to wild-type activity.
FIG. 7.
β-Galactosidase leakage from plasmid-containing cells after heat induction of holin gene expression. Colonies on LB-X-Gal-IPTG agar, printed in grayscale, contain the following plasmids: 1, pAD330; 2, pS105; 3, pAD330-holA; 4, pAD330-holAB; and 5, pAD330-holB.
TABLE 3.
Complementation of lambda Sam7 mutation by HolA and HolB
| Host strain (plasmid) | Efficiency of platinga |
|---|---|
| LE392b | 1 |
| Rosettac | 5.3 × 10−7 |
| Rosetta (pS105) | 0.8 |
| Rosetta (pAD330) | 2.4 × 10−5 |
| Rosetta (pAD330::holA) | 3.5 × 10−2 |
| Rosetta (pAD330::holB) | 0.2 |
| Rosetta (pAD330::holAB) | 2 |
Efficiency of plating = titer of λSam7 phage on indicated host/titer on LE392. Results are averages of two experiments, performed in triplicate, in which plaque counts varied <±4%.
supE and supF suppress λSam7.
All Rosetta strains carry pRARE, which provides six tRNAs for codons rarely used in E. coli.
DISCUSSION
The plasmid release protocol required a relatively lengthy postinduction incubation period (30 to 60 min) before holin-complementing plasmids were released. This unexpectedly long delay is most likely due to the presence of rare codons in the holin genes. The combination of rare codon usage and the utilization of a GUG start codon for the endolysin gene could be a mechanism for regulating the timing of lysis of A. naeslundii by phage Av-1.
Expression of genes cloned into the BglII site in pAD330 is very tightly regulated, based on our observation that cells carrying the functional, highly lethal holin gene holB in this plasmid grow normally at low temperature (data not shown); however, when required, expression can be turned on by brief heating at 42°C to induce the Q-supplying prophage, which allows transcription to proceed beyond the λ terminator, tR′. When we utilized the plasmid enrichment protocol, we successfully used pAD330 to enrich for and isolate holin-complementing genes. By sequencing the two inserts that contained putative holin genes, we found that they fortuitously also contained an adjacent endolysin gene. We were subsequently able to also identify the endolysin gene in the Av-1 genome by using a holB-specific primer to sequence downstream of this gene on a sample of purified, whole-phage DNA (data not shown). This approach should therefore be generally useful in identifying and isolating new phage lysin genes, even if they do not degrade the peptidoglycan of E. coli.
Analysis of the region upstream of the three lysis genes failed to reveal the presence of a recognizable sigma 70-like promoter or a Shine-Dalgarno sequence, and no promoters were found for the holB or lysA genes. The start codons of the latter two genes, however, were located an appropriate distance from putative Shine-Dalgarno sequences. We tentatively postulate that the three lysis genes are expressed as part of a late gene operon. Subsequent sequencing of the genome of Av-1 has in fact revealed that this operon potentially consists of five genes, with the lysA gene being the last one expressed, and that the start of the holA gene overlaps the stop codon of an upstream gene (unpublished data).
Neither of the holin genes has a dual-start motif, indicating that the timing of A. naeslundii lysis by Av-1 may not be an antiholin-holin interaction, such as occurs in the λ S107-S105 system (26). There is no detectable sequence homology between either of the two putative holins in the present study and any other known or unknown phage proteins in the nonredundant GenBank database. The HolB protein (64 amino acids) appears to be a typical type II holin, with two transmembrane regions and a charged C terminus (25, 26). We are confident that, based on its structure and functional properties, the HolB protein is the phage Av-1 holin.
The HolA protein (101 amino acids) may have a single transmembrane region, although this region has only 15 amino acids between charged groups (Fig. 3). If this region does insert into the membrane, it may have a lysis function similar to the phage T4 t holin (25), the temperate S. mitis phage SM1 holin (20) and the temperate S. pneumoniae phage MM1 Hol2 protein (14), all of which have single transmembrane regions. Its structure was found to be distantly related to the BvgS sensor protein of Bordetella pertussis, a membrane-embedded protein, which suggests that the predicted transmembrane region of Av-1 HolA localizes it to the cytoplasmic membrane. We considered whether HolA acts as an antiholin, but we could not establish this from our experiments. Antiholins generally delay phage lysis in order to optimize progeny phage production. Loessner et al. (8) and Vukov et al. (24) used complementation tests to demonstrate that Staphylococcus aureus phage antiholins delayed the activity of their cognate holins in E. coli. In our λSam7 complementation tests we observed no difference in plaque size when this phage was plated on E. coli strains expressing either or both holins. This should have been observed if HolA acts as an antiholin, and its absence resulted in smaller plaques due to premature lysis. Not all double-stranded DNA phages utilize an antiholin to regulate lysis timing since some apparently simply rely on delaying expression of their holin genes (5, 9), which may be the case with phage Av-1. An additional difficulty in interpreting our results is the possibility that the two Av-1 holin proteins may behave differently in the heterologous host E. coli (a gram-negative organism having a G+C composition of 50%) than in their normal host, A. naeslundii, which is a gram-positive organism with a DNA composition of 68% G+C. The actual function of HolA in A. naeslundii therefore remains to be determined.
The LysA protein (272 amino acids) is the phage Av-1 endolysin. It does not contain a signal sequence, like the unusual lysin of Oenococcus oeni phage fOg44 (17), so its function is strictly dependent on holin-mediated membrane permeabilization to gain access to the peptidoglycan wall. Its amino acid sequence is unique and unrelated to other known phage lysins, although we found a distant relationship to a muramidase autolysin of Clostridium acetobutylicum (E = 0.09). LysA also contains a putative peptidoglycan-binding domain in its C-terminal region (Fig. 3). The catalytic domain of the enzyme was not identifiable by BLAST searches, although a portion of the C-terminal region showed a very distant relationship to an l-alanine amidase of Bacillus subtilis. The catalytic site may thus not be in the N-terminal region like most other phage lysins. In terms of its lytic activity, we observed that the lysin is able to lyse even aged, stationary-phase cells of MG-1S. The ability of LysA to lyse the thick cell wall of A. naeslundii could be used to develop a fast, gentle procedure for extracting mRNA from this organism, thus facilitating transcription studies (11). The successful production of an enzymatically active N-terminal His-tagged LysA fusion protein will greatly facilitate purification and further characterization of the enzyme, including determining its spectrum of activity, potential ability to detect and quantitate A. naeslundii in oral samples, and potential use as a therapeutic agent. Determining the specific peptidoglycan bonds that are cleaved by the lysin will also help us to ultimately explain why A. naeslundii is resistant to conventional lysozyme-like enzymes.
Acknowledgments
This study was supported by NIH/NIDCR grant DE13181.
We thank Ry Young for helpful discussions, advice, and encouragement; for providing several strains and plasmids (along with I.-N. Wang of the Young lab); and for originally suggesting using pS105 as a starting point from which to develop a holin-selective vector. We also thank W. Roof for providing details of his plasmid release protocol prior to publication.
REFERENCES
- 1.Cheng, Q., D. Nelson, S. Zhu, and V. A. Fischetti. 2005. Removal of group B streptococci colonizing the vagina and oropharynx of mice with a bacteriophage lytic enzyme. Antimicrob. Agents Chemother. 49:111-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Delisle, A. L., and J. A. Donkersloot. 1995. Relationships among Actinomyces naeslundii (A. viscosus) bacteriophages isolated from sewage and the oral cavity. Microb. Ecol. Health Dis. 8:121-127. [Google Scholar]
- 3.Delisle, A. L., R. K. Nauman, and G. E. Minah. 1978. Isolation of a bacteriophage for Actinomyces viscosus. Infect. Immun. 20:303-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kricker, M., and K. Carlson. 1994. Isolation of T4 phage DNA, p. 455-456. In J. D. Karam (ed.), Molecular biology of bacteriophage T4. ASM Press, Washington, D.C.
- 5.Labrie, S., N. Vukov, M. J. Loessner, and S. Moineau. 2004. Distribution and composition of the lysis cassette of Lactococcus lactis phages and functional analysis of bacteriophage ul36 holin. FEMS Microbiol. Lett. 233:37-43. [DOI] [PubMed] [Google Scholar]
- 6.Loeffler, J. M., S. Djurkovic, and V. A. Fischetti. 2003. Phage lytic enzyme Cpl-1 as a novel antimicrobial for pneumococcal bacteremia. Infect. Immun. 71:6199-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loeffler, J. M., D. Nelson, and V. A. Fischetti. 2001. Rapid killing of Streptococcus pneumoniae with a bacteriophage cell wall hydrolase. Science 294:2170-2172. [DOI] [PubMed] [Google Scholar]
- 8.Loessner, M. J., S. Gaeng, and S. Scherer. 1999. Evidence for a holin-like protein gene fully embedded out of frame in the endolysin gene of Staphylococcus aureus bacteriophage 187. J. Bacteriol. 181:4452-4460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loessner, M. J., S. Gaeng, G. Wendlinger, S. K. Maier, and S. Scherer. 1998. The two-component lysis system of Staphylococcus aureus bacteriophage Twort: a large TTG-start holin and an associated amidase endolysin. FEMS Microbiol. Lett. 162:265-274. [DOI] [PubMed] [Google Scholar]
- 10.Loessner, M. J., K. Kramer, F. Ebel, and S. Scherer. 2002. C-terminal domains of Listeria monocytogenes bacteriophage murein hydrolases determine specific recognition and high-affinity binding to bacterial cell wall carbohydrates. Mol. Microbiol. 44:335-349. [DOI] [PubMed] [Google Scholar]
- 11.Loessner, M. J., A. Schneider, and S. Scherer. 1995. A new procedure for efficient recovery of DNA, RNA, and proteins from Listeria cells by rapid lysis with a recombinant bacteriophage endolysin. Appl. Environ. Microbiol. 61:1150-1152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Marchler-Bauer, A., J. B. Anderson, P. F. Cherukuri, C. Weese-Scott, L. Y. Geer, M. Gwadz, S. He, D. I. Hurwitz, J. D. Jackson, Z. Ke, C. J. Lanczycki, C. A. Liebert, C. Liu, F. Lu, G. H. Marchler, M. Mullokandov, B. A. Shoemaker, V. Simonyan, J. S. Song, P. A. Thiessen, R. A. Yamashita, J. J. Yin, D. Zhang, and S. H. Bryant. 2005. CDD: a conserved domain database for protein classification. Nucleic Acids Res. 33:D192-D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nelson, D., L. Loomis, and V. A. Fischetti. 2001. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. USA 98:4107-4112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Obregon, V., J. L. Garcia, E. Garcia, R. Lopez, and P. Garcia. 2003. Genome organization and molecular analysis of the temperate bacteriophage MM1 of Streptococcus pneumoniae. J. Bacteriol. 185:2362-2368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Raab, R., G. Neal, C. Sohaskey, J. Smith, and R. Young. 1988. Dominance in lambda S mutations and evidence for translational control. J. Mol. Biol. 199:95-105. [DOI] [PubMed] [Google Scholar]
- 16.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 17.Sao-Jose, C., R. Parreira, G. Vieira, and M. A. Santos. 2000. The N-terminal region of the Oenococcus oeni bacteriophage fOg44 lysin behaves as a bona fide signal peptide in Escherichia coli and as a cis-inhibitory element, preventing lytic activity on oenococcal cells. J. Bacteriol. 182:5823-5831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schleifer, K. H., and O. Kandler. 1972. Peptidoglycan types of bacterial cell walls and their taxonomic implications. Bacteriol. Rev. 36:407-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schuch, R., D. Nelson, and V. A. Fischetti. 2002. A bacteriolytic agent that detects and kills Bacillus anthracis. Nature 418:884-889. [DOI] [PubMed] [Google Scholar]
- 20.Siboo, I. R., B. A. Bensing, and P. M. Sullam. 2003. Genomic organization and molecular characterization of SM1, a temperate bacteriophage of Streptococcus mitis. J. Bacteriol. 185:6968-6975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smith, D. L., D. K. Struck, J. M. Scholtz, and R. Young. 1998. Purification and biochemical characterization of the lambda holin. J. Bacteriol. 180:2531-2540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Suzuki, J. B., and A. L. Delisle. 1984. Pulmonary actinomycosis of periodontal origin. J. Periodontol. 55:581-584. [DOI] [PubMed] [Google Scholar]
- 23.Trudil, D. 28 May 2002. Use of phage associated lytic enzymes for the rapid detection of bacterial contaminants. U.S. patent 6,395,504.
- 24.Vukov, N., S. Scherer, E. Hibbert, and M. J. Loessner. 2000. Functional analysis of heterologous holin proteins in a lambda DeltaS genetic background. FEMS Microbiol. Lett. 184:179-186. [DOI] [PubMed] [Google Scholar]
- 25.Wang, I. N., D. L. Smith, and R. Young. 2000. Holins: the protein clocks of bacteriophage infections. Annu. Rev. Microbiol. 54:799-825. [DOI] [PubMed] [Google Scholar]
- 26.Young, R. 1992. Bacteriophage lysis: mechanism and regulation. Microbiol. Rev. 56:430-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Young, R., and U. Blasi. 1995. Holins: form and function in bacteriophage lysis. FEMS Microbiol. Rev. 17:191-205. [DOI] [PubMed] [Google Scholar]