Abstract
Overexpression of a class 1 Hb (GLB1) protects Arabidopsis thaliana plants from the effects of severe hypoxia. Overexpression of the bifunctional symbiotic Hb (GLB1S) from Parasponia andersonii in A. thaliana also increases survival after hypoxia. Plants overexpressing the Hb 1 protein, mutated to have a low oxygen affinity, are as susceptible to hypoxia as WT plants, suggesting that the protection against hypoxia depends on the ability of the Hb to bind ligands, such as oxygen, with high affinity. A mild hypoxia pretreatment (5%) induces the Hb gene and increases the survival of plants after severe hypoxic treatment (0.1%). These results with Hb 1 show that plant Hbs have a role other than in nitrogen-fixing root nodules. Plants overexpressing the GLB1 protein show early vigorous growth in nonhypoxic conditions and are 50% larger in weight than the controls at 14 days. The constitutive expression of GLB1 also resulted in a reduced number of root hairs and increased number of laterals in the root system.
Hemoglobin is an important oxygen binding protein, occurring in most living organisms. Hbs were described originally in animals, and in vertebrates they have been shown to facilitate the transport of oxygen in blood. However, animals, plants, fungi, and bacteria make a number of different Hbs (1–3), many of which have functions other than the transport of oxygen (1–5). Only a few amino acid residues are conserved in all Hbs; these are involved in heme and ligand binding and maintaining the tertiary arrangement of the protein's α-helices. One of the conserved amino acids is a histidine or glutamine in the E helix (E7). The class 1 and 2 Hbs (3) of plants are similar to vertebrate Hbs in that all of these molecules have an E7 histidine residue. The E7 amino acid is necessary for binding oxygen and other ligands in the distal pocket of the molecule. Replacement of the E7 residue with a hydrophobic amino acid destroys the capacity of the Hb to bind oxygen and other ligands (6, 7).
The cellular functions of most plant Hbs are unknown (3). Many of these Hbs have higher oxygen binding affinities than cytochrome c oxidase, so they are unlikely to be involved in oxygen transport to the mitochondria (8–10). The symbiotic Hbs found in legumes and other nitrogen-fixing plants have a lower oxygen binding affinity and function to transport oxygen during nitrogen fixation in symbiotic root nodules (11).
We have found that induction of hypoxic-response genes is crucial for the survival of plants after a severe hypoxic challenge (12). Hypoxia induces expression of the class 1 Hb gene, GLB1, but not the class 2 or 3 genes in either the roots or shoots of Arabidopsis thaliana (9). During exposure to hypoxic conditions, GLB1 mRNA and protein accumulate. In this article we show that overexpression of GLB1 increases survival of plants under hypoxic stress. By replacing the E7 histidine residue with a hydrophobic leucine residue, we have shown that the ligand binding capacity of GLB1 is essential for increased tolerance of plants to hypoxic stress.
One other remarkable property of GLB1 overexpression is that it results in increased early vigor of plants under normal oxygen concentrations. In the first 2 weeks of plant growth, plants overexpressing GLB1 have considerably faster growth of both roots and shoots compared with control plants.
Methods
Plant Growth Conditions.
The C24 ecotype of A. thaliana was used for all experiments. Plants were grown on Murashige–Skoog (MS) media-agar under 18-h days with fluorescent lighting at intensities between 50 and 100 μmol photons⋅m−2·s−1. Agar (0.8%) was used for the majority of experiments, but for growth before and after hypoxia assays 0.5% agar was used to facilitate removal of roots from the medium. For microscopy, plants were grown on vertical plates containing 0.8% or 1.6% agar; the data reported in Fig. 4 are for plants grown on 0.8% agar with half-strength MS medium. These plants were grown aerobically on vertically orientated media, and root morphology and growth were monitored through a 10-day period starting at 13 days after imbibition.
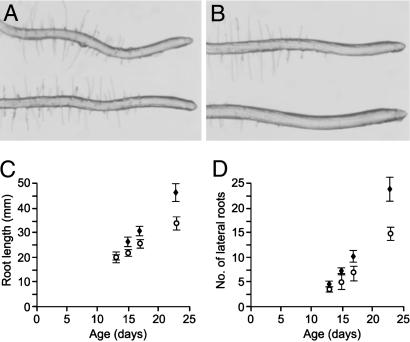
Fig 4.
Differential interference contrast photomicrographs of root tips from WT (A) and 35S:GLB1(D4) (B) plants. The zones of elongation and root hair differentiation are longer in 35S:GLB1(D4) plants. (C) Root growth in WT (○) and 35S:GLB1 (♦) plants. 35S:GLB1 plants have a greater growth rate than WT controls. (D) Production of lateral roots from WT (○) and 35S:GLB1 (♦) plants. As root development progresses, 35S:GLB1 plants produce significantly more lateral roots. Error bars are standard deviations in C and D.
Western Blots.
Polyclonal rabbit antisera against purified recombinant GLB1 protein (9) and Parasponia andersonii nodule Hb (GLB1S) (13) were used for protein detection.
A. thaliana protein extracts were obtained by grinding plant material in 0.01 M NaPO4 (pH 7.0), 1 mM EDTA, centrifuging (10,000 × g for 5 min), and collecting the supernatants. Fifty micrograms of total protein (as measured with the BioRad protein detection reagent) was used per lane for SDS/PAGE separation (15% acrylamide), and Western blots were prepared and probed as described (9). All Western blots were repeated a minimum of three times. The D1, D11, and D4 35S:GLB1 strains were compared seven times on separate Western blots. Quantitation of the protein levels was undertaken on a representative blot by using the imagequant program (Molecular Dynamics).
Production of 35S:GLB1, 35S:GLB1(HE7L), and 35S:GLB1S Plants.
The ORFs of A. thaliana GLB1, GLB1(HE7L), or P. andersonii GLB1S were removed from a bacterial expression vector (9) by digestion with BamHI and ligated to BamHI cut pJ35SN (GLB1) or pART7 [GLB1(HE7L), GLB1S] containing the cauliflower mosaic virus 35S promoter. Subsequently, the 35S:GLB1 fragment was removed by digesting with SalI/ScaI and ligated into SalI/SmaI-digested pBIN19 (GLB1) or by digesting with NotI and ligated into pART27 [GLB1(HE7L), GLB1S]. After sequencing to confirm insert sequences, vectors were introduced into Agrobacterium tumefaciens AGL1 by electroporation and then into A. thaliana plants by immersion of influorescences (14). Control plant lines CON3 and CON5 were generated by using the same protocol, using the empty vector pBI101.1 (15). Other control plants contained the β-glucuronidase (GUS) reporter transgenes GLB2:GUS or GLB1:GUS (3).
Plants from each of five lines (D1, D4, D5, D7, and D11) with increased GLB1 protein levels, three lines expressing the GLB1(HE7L) protein (E74, E75, and E76), and two lines expressing the GLB1S protein (Pa4 and Pa5) were used for subsequent experiments. Homozygous T3 selections from the D4, Pa4, Pa5, and GUS-reporter control lines were used in the experiments detailed in Fig. 3 B–D; all other work was conducted with kanamycin-selected plants from the T2 generation.
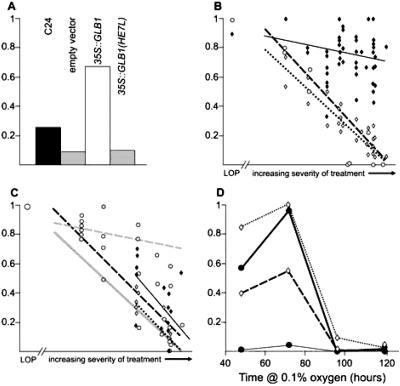
Fig 3.
All four graphs have the proportion of plants surviving after hypoxia indicated by the y axis. 1 indicates that all plants survived and 0 indicates that none survived. (A) Survival of WT (black bar), empty vector controls (gray bar), 35S:GLB1 (empty bar), and 35S:GLB1(HE7L) (striped bar) plants after hypoxia (average of four separate experiments). Overexpression of GLB1, as well as increasing survival of plants after hypoxia, may also have overcome a penalty imposed by the presence of the npt II transgene; compare the transgenic controls to WT. (B and C) Exposure to hypoxia had a variable effect on survival of WT plants in separate experiments. In these graphs the severity of the experiment as indicated by survival of WT plants is plotted on the x axis. A low oxygen pretreatment (LOP) is shown to the left of the broken axis for comparison. The y axis represents posthypoxia survival, so that the line for WT has a slope of 1. Regression lines are plotted in B and C. (B) Response of WT (dotted line, ◊), 35S:GLB1(D4) (black line, ⧫), and GUS reporter controls (dashed line, ○) to increasing severity of hypoxia. (C) Response of 35S:GLB1(D1) (black line, ⧫), 35S:GLB1(D11) (dotted line, ◊), and two 35S:GLB1S lines (dashed line, ○) to increasing severity of hypoxia. The data from the two GLB1S lines were combined because they were not significantly different. (C) The WT and 35S:GLB1(D4) regression lines from B are shown in pale gray for comparison. (D) Survival of plants (y axis) after differing periods of exposure to 0.1% oxygen (x axis) after 5% oxygen pretreatment (dashed lines) or no pretreatment (solid lines). 35S:GLB1(D4) plants (crosses and gray lines) had a higher survival rate than WT plants (ellipses and black lines) in the absence of pretreatment for the first two time points. When pretreatment was provided, both genotypes had high survival rates at 48 and 72 h, but most plants died after 96 h. The small difference at 96 h between 35S:GLB1(D4) and WT plants with pretreatment is statistically significant.
Hypoxic Stress Response Assay.
Response to hypoxia was assessed by measuring plant growth parameters subsequent to stress in low oxygen conditions as described (12). Two low oxygen regimes were used; in some assays a pretreatment of 5% oxygen (mild hypoxia) preceded exposure to 0.1% oxygen (24 or 48 h), whereas in a more severe treatment plants were exposed to 0.1% oxygen without pretreatment for up to 96 h (severe hypoxia). Bacterial anaerobic growth chambers (Oxo) were used to expose plants to the different gas mixtures. Root growth was determined by measuring the difference in length of the total root system immediately after hypoxia and the 3-week recovery period. Either the primary or one of the secondary roots was measured to avoid a false negative result caused by accidental damage of the primary root. Second, for comparisons of transgenic lines containing 35S:GLB1, 35S:GLB1(HE7L), and 35S:GLB1S the plants were exposed to low oxygen conditions in 25-mm Petri dishes. In these experiments, shoot survival, shoot weight, and root weight were the only measurements used to assess plant responses to hypoxia.
Because of differences between anaerobic growth chambers and experiments conducted at different times, the survival of WT plants after hypoxia varied between experiments. Experiments were compared by using percentage survival of WT plants as a measure of severity. Data for multiple experiments are presented in Fig. 2 B and C, with the x axis indicating the severity of treatment as measured by survival of WT plants.
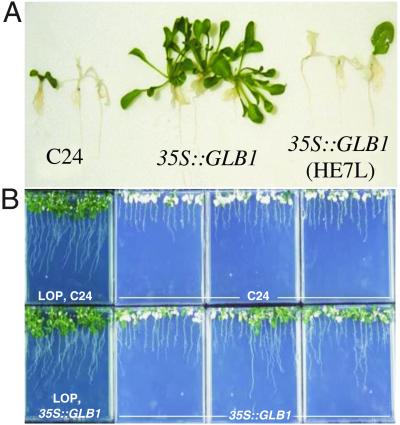
Fig 2.
(A) Plants 17 days after hypoxia. 35S:GLB1(D4) plants survived and grew, whereas most WT and 35S:GLB1(HE7L)(E76) plants died. (B) WT and 35S:GLB1(D4) plants 21 days after hypoxia treatments. Plants were exposed to hypoxia either with no pretreatment or after low oxygen pretreatment (LOP). In the absence of pretreatment, 31% of 35S:GLB1 plants survived and grew, but no WT plants survived in this experiment. All plants of both genotypes survived hypoxia after pretreatment.
Empty vector controls (selected with kanamycin, Table 1, see Fig. 3A) and GUS-reporter transgenic plants (kanamycin selection not used, see Fig. 3B) used as controls did not survive hypoxia as well as WT plants and survival was less responsive to treatment severity. Because of these differences, WT was used as a benchmark for comparing experiments rather than the transgenic control plants.
Table 1.
Survival and growth after hypoxic stress of plants containing different transgenes expressing WT GLB1 or mutant GLB1 compared with control lines
| Plant type | Shoot survival, % | Shoot weight, mg per 10 plants | Root weight, mg per 10 plants |
|---|---|---|---|
| WT | 27 (107)†‡ | 100 (8)†‡ | 8 (8)† |
| CON3 | 8 (91)* | 70 (8)* | 5 (8)‡ |
| CON5 | 10 (94)* | 90 (8)† | 6 (8)† |
| 35S:GLB1 (D4) | 77 (117)*†‡ | 230 (12)*†‡ | 20 (12)*†‡ |
| 35S:GLB1 (D5) | 63 (104)*†‡ | 200 (12)*†‡ | 19 (12)*†‡ |
| 35S:GLB1 (D7) | 63 (115)*†‡ | 220 (12)*†‡ | 24 (12)*†‡ |
| 35S:GLB1(HE7L) (4) | 7 (112)* | 80 (12)† | 7 (12)† |
| 35S:GLB1(HE7L) (5) | 11 (111)* | 110 (12)†‡ | 8 (12)†‡ |
| 35S:GLB1(HE7L) (6) | 12 (134)* | 120 (12)†‡ | 9 (12)†‡ |
The number of plants (shoot survival) or number of replicate samples (weight measures) are given in parentheses in each cell. Symbols represent statistical differences (P < 0.05) between data of transgenic plant groups and WT plants (*), empty vector control line CON3 (†), or empty vector control line CON5 (‡).
Statistical Analysis.
Proportions of plants surviving after hypoxia were analyzed by z tests. The remaining data for these experiments were analyzed by ANOVA using genstat or pair-wise Student's t tests using Microsoft excel. Data were also compared across a range of experiments with differing severity by regression analysis using excel, and slopes of regression lines were compared by using James' second-order approximation and Alexander's normalized-t approximation tests (16, 17) as incorporated into the altmmr program (http://members.aol.com/IMSAP/api.html). Root morphology and plant growth data were analyzed by Student's t tests.
Microarray Analysis of Transcription.
The gene expression profiles of 35S:GLB1 and C24 plants were compared by using microarrays. An experiment was conducted in which RNAs from the roots or shoots of plants grown for 3 weeks in tissue culture were compared. Data for fermentative and glycolytic pathway genes, which normally are induced by hypoxia, are listed in Table 2. The remainder of these data and additional analysis of GLB1 overexpression will be published elsewhere. The array contained cDNA clones from roots subjected to a mild hypoxia treatment (5% O2). Microarray hybridization experiments and subsequent analysis were conducted as described (18).
Table 2.
Genes from the fermentative and glycolytic pathways are not induced by GLB1 overexpression
| Gene name | Shoots | Roots |
|---|---|---|
| ADH1 | −0.56 | −2.78 |
| AlaAT1 | −0.12 | −1.53 |
| AlaAT2 | −0.31 | −1.89 |
| ASUS1 | 0.21 | −4.10 |
| LDH1 | −0.13 | −0.86 |
| PDC1 | −3.10 | −6.36 |
| PDC3 | −1.04 | −1.06 |
| GLB1 | 14.74 | −1.16 |
Values shown are log2 transformed and normalized ratios of expression for 35S:GLB1 plants compared to WT controls. All plants were grown under aerobic conditions to 3 weeks of age. RNA from root and shoot tissues were analyzed separately. The values that are outside of the −3.20 to +3.20 range are significant. GLB1 is not overexpressed in the roots as expression in the roots of WT plants is also high under these growth conditions.
Oxygen Binding Kinetics of GLB1(HE7L).
Oxygen binding to the GLB1(HE7L) protein was measured by laser flash photolysis (19) and stopped-flow rapid mixing (20). Rate constants were calculated by using published procedures (21).
Results
Plants Overexpressing GLB1 Survive Hypoxic Treatments That Kill WT Plants.
Hypoxia induces expression of A. thaliana GLB1 in both roots and shoots, which is shown by both mRNA level and promoter-reporter gene expression (GUS) (3, 9). GLB1 mRNA levels increase with a moderately low oxygen treatment (5% oxygen) and are enhanced further by a subsequent severe hypoxia treatment (0.1% oxygen) (9). GLB1 protein is induced in roots by a 16-h treatment with 5% oxygen, and this induction is increased in 0.1% oxygen (Fig. 1A).
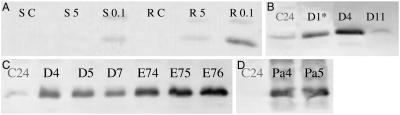
Fig 1.
Western blots showing levels of Hb proteins in transgenic and WT (C24) plants. (A) GLB1 in shoot or root tissues from control plants or plants subjected to 5% or 0.1% oxygen. Lanes contain protein extracted from shoot tissue control (SC), shoot 5% oxygen (S5), shoot 0.1% oxygen (S0.1), root tissue control (RC), root 5% oxygen (L5), and root 0.1% oxygen (R0.1) plants. GLB1 bands migrated at an apparent size of 18 kDa. (B) GLB1 overexpression in the 35S:GLB1 plant lines D1, D4, and D11 compared with WT control (the picture has been split to remove lanes). (C) GLB1 overexpression in the 35S:GLB1 plant lines D4, D5, and D7 and in plant lines overexpressing GLB1(HE7L) (lines E74, E75, and E76) are shown compared with WT. (D) P. andersonii GLB1S levels for 35S:GLB1S(Pa4) and 35S:GLB1S(Pa5) plants and WT controls. High levels of P. andersonii GLB1S were produced in the transgenic plants, but these levels cannot be compared with the levels of A. thaliana GLB1 shown in A–C, because these Western blots use different antisera.
To test whether an increased level of GLB1 enhances the ability of plants to survive hypoxic stress, we generated transgenic plants containing the GLB1 ORF fused to a cauliflower mosaic virus 35S promoter (35S:GLB1), which drives high levels of gene expression throughout the plant. Three lines showed substantially increased levels of GLB1 under nonhypoxic conditions (D4, D5, and D7; Fig. 1C). After hypoxic stress, in which plants were exposed to 0.1% oxygen for 48 h, these three lines had high survival rates (63–77% compared with 8–27% for WT or transgenic controls) (Figs. 2A and 3A). The increased survival was paralleled by increases in the weights of roots and shoots in the plants grown during the recovery period (Table 1).
The hypoxia treatment we used varied in severity, and we monitored survival of WT plants in each experiment (see Methods). The difference in survival between 35S:GLB1 (D4) and control plants increased with severity of the hypoxia treatment (Fig. 3B). In a severe hypoxia treatment, in which no WT plants survived without a pretreatment, 31% of the 35S:GLB1 (D4) plants survived (Table 3, Fig. 2B). Growth of 35S:GLB1 (D4) plants subsequent to hypoxic stress was greater than WT, as shown by root lengths, shoot weights, and chlorophyll content (Table 3).
Table 3.
Survival and growth of 35S:GLB1 and WT plants after two different hypoxic stress treatments
| Survival/growth
|
Hypoxia, no pretreatment | Hypoxia after pretreatment with 5% O2 | ||
|---|---|---|---|---|
| 35S:GLB1 | WT | 35S:GLB1 | WT | |
| Shoot survival, % | 31.3 | 0.0 | 100 | 100 |
| Primary root tip survival, % | 69.0 | 55.7 | 100 | 100 |
| Chlorophyll conc., μg/g | 61.7 | 25.9 | 71.1 | 78.4 |
| Chlorophyll content, μg per plant | 0.7 | 0.2 | 1.5 | 1.5 |
| Root growth, mm | 56.9 | 33.3 | 56.6 | 53.6 |
| Root weight, mg per 10 plants | 131 | 119 | 129 | 254 |
| Shoot weight, mg per 10 plants | 375 | 329 | 407 | 565 |
Hypoxia treatments were 48 h of 0.1% oxygen in nitrogen. Pretreatments were also 48 h in duration.
Statistically significant difference (P < 0.05) between 35S:GLB1 and WT plants.
This treatment involved a small number of plants that exhibited highly variable root systems.
A 35S:GLB1 line (D1) with a GLB1 level intermediate between WT and the three high expression lines (4.7-fold induction, compared with 8.8-fold for D4) (Fig. 1B) had an intermediate hypoxia survival response (Fig. 3C, Table 5, which is published as supporting information on the PNAS web site, www.pnas.org); D11, a kanamycin-resistant line with no overexpression, had a hypoxia response equivalent to WT (Fig. 3C, Table 5). The D1 and D11 lines were examined in the more severe hypoxia stress treatments where a significant increase in survival and growth compared with controls was observed with D1 but not D11 (statistics in Table 5).
High-Affinity Ligand Binding Is Essential for GLB1 to Improve Survival After Hypoxic Stress.
Ligands bind to the ferrous iron of the heme prosthetic group of Hbs and the binding is made reversible via interactions with amino acid side chains that surround the iron atom on the distal side of the planar heme molecule. In many Hbs, including GLB1, a histidine residue at position E7 (α-helix E, residue 7) is essential for binding ligands in the distal pocket. To determine whether the ligand binding capacity of GLB1 was the critical factor in promoting survival after hypoxic stress, we replaced the E7 histidine with leucine. The mutated protein (GLB1 HE7L) had a much lower oxygen affinity than the WT molecule, with a calculated P50 for oxygen of 350 nM compared with 9.7 nM for the WT protein (22).
Plants [35S:GLB1(HE7L) lines E74, E75, and E76] expressing the leucine-substituted Hb protein at a level slightly higher than that of WT GLB1 in the 35S:GLB1 lines (D4, D5, and D7) (Fig. 1C) did not show enhanced survival after hypoxic stress, and shoot survival was similar to WT and transgenic control plants (Figs. 2A and 3A, Table 1). 35S:GLB1(HE7L) plants, grown for 17 days after hypoxic stress, have root and shoot weights similar to control plants and much lower root and shoot weights than those of the 35S:GLB1 plants (Table 1).
Overexpression of GLB1S also Enhances Survival After Hypoxic Stress.
In testing whether Hbs other than A. thaliana GLB1 could confer increased survival after hypoxia we chose the bifunctional symbiotic Hb (GLB1S) (Fig. 1D) from P. andersonii (13). This is a class 1 Hb [other symbiotic Hbs are class 2 (3)], which has a low oxygen affinity similar to that of other symbiotic Hbs [P50 O2 = 100 nM (22, 23)]. Two lines expressing the 35S:GLB1S transgene survived hypoxic stress better than WT, but were not as vigorous as the 35S:GLB1(D4) line (Fig. 3C). The data from the two lines were similar and are combined.
Low Oxygen Pretreatment Increases Survival After Hypoxia in 35S:GLB1 Plants.
We found that survival and growth parameters of 35S:GLB1 and control plants were identical after a severe hypoxic stress (48 h, 0.1% oxygen), which was preceded by a low oxygen pretreatment (48 h, 5% oxygen) (Table 3). The protection provided by the adaptive pretreatment was greater than that provided by GLB1 overexpression alone.
To test whether longer periods of hypoxic stress would differentiate the 35S:GLB1 and control plants, low oxygen pretreated plants were exposed to increasing periods of 0.1% oxygen. Survival of WT plants declined to zero after 72–96 h of 0.1% oxygen treatment, but 9% of the 35S:GLB1 plants survived at 96 h (Fig. 3D). This difference is small but statistically significant (P = 0.04) and suggests that GLB1 overexpression may add to the level of hypoxia tolerance imparted by low oxygen pretreatment.
In control plants where the 5% oxygen pretreatment permitted survival in 0.1% O2 the pretreatment induced a set of genes coding for proteins involved in the fermentation or glycolytic pathways. We had shown that ADH1 activation is critical for survival (12). Overexpression of GLB1 promotes survival without a low oxygen treatment but the fermentation and glycolytic pathway genes are not induced by GLB1 overexpression in the absence of hypoxia (Table 2).
35S:GLB1 Plants Have Enhanced Early Growth Rates.
35S:GLB1(D4) plants grew to a greater size than WT controls during the first 2 weeks after germination under normal atmospheric oxygen concentrations (Table 4). 35S:GLB1 plant wet weights were ≈1.5-fold greater than the controls at 14 days postimbibition. The increased growth was not a result of faster development. The rate of development in 35S:GLB1(D4) plants was unaltered relative to the controls as determined by leaf numbers, which were identical at all of the measurement times.
Table 4.
Increased early growth of 35S:GLB1 over WT control plants
| Time, days
|
No sucrose | Sucrose supplemented | ||
|---|---|---|---|---|
| 35S:GLB1 | WT | 35S:GLB1 | WT | |
| 14 | 0.0636 ± 0.0030 | 0.0434 ± 0.0021 | 0.0347 ± 0.0008 | 0.0249 ± 0.0022 |
| 21 | 0.1101 ± 0.0022 | 0.0976 ± 0.0332 | 0.0870 ± 0.0064 | 0.0703 ± 0.0018 |
| 28 | 0.1762 ± 0.0148 | 0.1780 ± 0.0059 | 0.1311 ± 0.0100 | 0.1130 ± 0.0120 |
| 32 | 0.3076 ± 0.0409 | 0.3871 ± 0.0492 | 0.1954 ± 0.0026 | 0.1819 ± 0.0086 |
Plant wet weights shown as g per 10 plants, n = 2; errors shown are SEM. There was no difference (P = 0.548) in the weight of samples of 50 seeds between 35S:GLB1 (0.0010 g, n = 3) and WT (0.0011 g, n = 3) plants. 35S:GLB1 plants at 14 days have significantly higher weights (P < 0.01) than controls in either sucrose-supplemented or nonsupplemented media.
Plants grown in sucrose-supplemented media have lower weights than those grown with sucrose at these time points (P < 0.01).
We measured a number of root growth parameters in the GLB1 overexpression and control plants (Fig. 4). 35S:GLB1(D4) roots grew more during the 9-day observational period to a final mean length of 46 mm (n = 5) compared with 34 mm for the controls (n = 6) (Fig. 4C). 35S:GLB1(D4) plants also had 40% lower root hair density and 60% more lateral roots compared with the controls (Fig. 4D). Over the 9 days, the average growth rate for the primary roots of the control plants was 2.1 mm per day, compared with 3.2 mm per day for the 35S:GLB1(D4) plants (P = 0.02). Roots of 35S:GLB1(D4) plants had a different appearance to control plants (Fig. 4 A and B), with sparser root hairs (Table 6, which is published as supporting information on the PNAS web site) and slightly longer root elongation zones (Table 6).
Discussion
Overexpression of GLB1 Enhances Survival After Severe Hypoxia.
We had shown that treatment of A. thaliana plants with low oxygen increases the expression of both GLB1 mRNA and protein in roots and shoots, and that hypoxia after low oxygen pretreatment induces even greater GLB1 expression. Class 1 Hbs, such as GLB1, are found throughout the families of flowering plants (3) and may have a universal role during hypoxic stress. For example, the GLB1 Hbs from barley, maize, and rice accumulate in response to low oxygen treatment (24) and cotton GLB1 was detected in a cDNA library from anaerobic-treated roots (3). In addition, barley GLB1 expression can be induced by treatments other than hypoxia, which reduce ATP levels, suggesting that GLB1 might have a central role in responding to lowered cellular energy reserves (25).
In the present experiment overexpression of GLB1 in A. thaliana increased the survival of plants exposed to severe hypoxia, the enhanced survival being proportional to the level of GLB1 protein. We showed that Hb-mediated hypoxia tolerance depends on high-affinity ligand binding, where the ligand may be oxygen or another small polar molecule. A mutant GLB1 protein (HE7L), which has a 10-fold reduction in oxygen binding affinity (22), provided no protection against hypoxia, even when present at high concentration. A symbiotic plant Hb from P. andersonii (GLB1S) with a lower affinity for oxygen than GLB1, but a higher affinity than GLB1(HE7L), provided an intermediate level of protection against hypoxia (Fig. 3C). Because of the demonstrated dependence on high-affinity binding, if the ligand is oxygen then it seems unlikely that oxygen transport is the mechanism by which increased GLB1 expression results in hypoxia tolerance. The oxygen binding affinity of GLB1 is too high for it to assist oxygen delivery to mitochondria (9).
Similar results have been reported for barley GLB1 overexpressed in a maize cell line (26). In these cells, the detrimental impacts of low oxygen were reduced and the energy status, as defined by ATP and NADH levels, was maintained in the cell lines overexpressing GLB1, but depleted in control cell lines. When cells were treated with antimycin A, an inhibitor of mitochondrial metabolic function, the GLB1-overexpressing cells were less affected than controls. Our results show that GLB1 overexpression provides protection against hypoxia in an entire plant. It is likely that the biochemical effects observed in the maize cell culture experiment are also occurring in the 35S:GLB1 plants, with some resistance to lowered cellular energy “charge” also providing an increased capacity to survive severe hypoxia.
Sowa and colleagues (26, 27) have postulated that barley GLB1 is able to generate NAD+ from NADH and increase flux through the glycolytic pathway, increasing ATP production in the absence of mitochondrial respiration. Our results using A. thaliana GLB1 do not conflict with this hypothesis. Changes in glycolytic flux may not be the main consequence of GLB1 overexpression as indicated by our finding that genes for glycolytic enzymes do not show enhanced expression in 35S:GLB1 relative to WT plants (Table 2).
GLB1 Overexpression Phenocopies Low Oxygen Pretreatment.
We have observed a dramatic increase in plant survival after severe hypoxia, when GLB1 protein levels were increased, whereas overexpression of either of the hypoxia response genes ADH1 or LDH1 did not provide any protection from hypoxic stress (28). Low oxygen pretreatment enhances hypoxia survival in both 35S:GLB1 and control plants. In the controls, GLB1 is induced to a high level by hypoxia, and this response is enhanced further by low oxygen pretreatment before hypoxia. Our results show it is likely that induction of GLB1 during pretreatment is important for increased hypoxia tolerance and that constitutive expression of GLB1 can partially replace a low oxygen pretreatment. GLB1 overexpression and low oxygen pretreatment can have additive effects on hypoxia survival. A greater proportion of low oxygen pretreated 35S:GLB1 plants survived 96 h of hypoxia compared with identically treated control plants.
The enhanced hypoxia survival of by 35S:GLB1 plants is not caused by increased expression of glycolytic and anaerobic metabolism genes before hypoxia. The Hb gene cannot be involved in signaling the expression of the suite of anaerobic response genes (e.g., ADH, LDH, and PDC1). Apparently GLB1 has a metabolic role in the cell that enables enhanced survival of hypoxia independent of changes in the expression levels of the glycolytic and anaerobic metabolism genes.
GLB1 Overexpression Increases Early Growth.
The mechanism by which GLB1 enhances hypoxia survival may also be responsible for enhanced early growth of 35S:GLB1 plants in normal atmospheric conditions. These plants have greater root and shoot weights at 14 days. The roots also produce more laterals and have altered morphology at the primary root tip. We do not understand how GLB1 overexpression changes plant growth, but similar claims have been made for overexpression of the Hb from the bacterium Vitreoscilla aquorea (VITaq GLB) in tobacco plants. These plants have increased growth, speed of germination, and production of secondary metabolites (reviewed in ref. 29). Our results show that GLB1 reproduces some of the effects of VITaq GLB; however, the two proteins have quite different biochemical properties. The oxygen affinity of VITaq GLB is low, ≈6,000 nM, compared with 9.7 nM for GLB1 (22). Overexpression of a mutant GLB1(HE7L), which has an oxygen affinity of 350 nM, did not generate hypoxia tolerance.
The increased growth of 35S:GLB1 plants is not caused by an altered developmental rate of leaf production so it may be caused by greater leaf cell number or size. Possibly 35S:GLB1 plants have an increased ability to grow during this phase because of an enhanced tolerance of a localized, transient hypoxia. A hypoxic phase is normally experienced during germination, causing a lag in respiration rate shortly after imbibition (30), which induces anaerobic response proteins such as ADH (31, 32). If 35S:GLB1 plants have a shorter lag phase or retain more energy reserves after the lag phase, they may have greater early growth. In a study of barley GLB1, the viability of stored seed was found to be correlated with GLB1 levels, and hypoxia during germination was closely matched by GLB1 expression (33). Root meristems can also be oxygen deficient even under normal growth conditions (34). The changes we found in root growth, and morphology in 35S:GLB1 plants may be indicative of an increased ability of root meristems to withstand this hypoxia.
Supplementary Material
Acknowledgments
We acknowledge the assistance of Mark Hargrove in analyzing oxygen binding to the GLB1(HE7L) protein. We thank Celia Miller, Rosemary White, and Tracey Mitchell for assistance with microscopy and photography; Cyril Appleby and John Jacobsen for helpful discussions; and The Ken and Yasuko Myer Plant Science Research Fund for providing a postdoctoral fellowship to P.W.H.
Abbreviations
GUS, β-glucuronidase
References
- 1.Blaxter M. L. (1993) Parasitol. Today 9, 353-360. [DOI] [PubMed] [Google Scholar]
- 2.Doyle J. J. (1998) Trends Plant Sci. 3, 473-478. [Google Scholar]
- 3.Hunt P. W., Watts, R. A., Trevaskis, B., Llewellyn, D. J., Burnell, J., Dennis, E. S. & Peacock, W. J. (2001) Plant Mol. Biol. 47, 677-692. [DOI] [PubMed] [Google Scholar]
- 4.Burr A. H. J., Hunt, P., Wagar, D. R., Dewilde, S., Blaxter, M. L., Vanfleteren, J. R. & Moens, L. (2000) J. Biol. Chem. 275, 4810-4815. [DOI] [PubMed] [Google Scholar]
- 5.Burmester T., Weich, B., Reinhardt, S. & Hankeln, T. (2000) Nature 407, 520-523. [DOI] [PubMed] [Google Scholar]
- 6.Rohlfs R. J., Mathews, A. J., Carver, T. E., Olson, J. S., Springer, B. A., Egeberg, K. D. & Sligar, S. G. (1990) J. Biol. Chem. 265, 3168-3176. [PubMed] [Google Scholar]
- 7.Mathews A. J., Rohlfs, R. J., Olson, J. S., Tame, J., Renaud, J. P. & Nagai, K. (1989) J. Biol. Chem. 264, 16573-16583. [PubMed] [Google Scholar]
- 8.Duff S. M. G., Wittenberg, J. B. & Hill, R. D. (1997) J. Biol. Chem. 272, 16746-16752. [DOI] [PubMed] [Google Scholar]
- 9.Trevaskis B., Watts, R. A., Andersson, C. R., Llewellyn, D. J., Hargrove, M. S., Olson, J. S., Dennis, E. S. & Peacock, W. J. (1997) Proc. Natl. Acad. Sci. USA 94, 12230-12234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arredondo P. R., Hargrove, M. S., Sarath, G., Moran, J. F., Lohrman, J., Olson, J. S. & Klucas, R. V. (1997) Plant Physiol. 115, 1259-1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bergersen F. J., Turner, G. L. & Appleby, C. A. (1973) Biochim. Biophys. Acta 291, 271-282. [DOI] [PubMed] [Google Scholar]
- 12.Ellis M. H., Dennis, E. S. & Peacock, W. J. (1999) Plant Physiol. 119, 57-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Appleby C. A., Tjepkema, J. D. & Trinick, M. J. (1983) Science 220, 951-953. [DOI] [PubMed] [Google Scholar]
- 14.Clough S. J. & Bent, A. F. (1998) Plant J. 16, 735-743. [DOI] [PubMed] [Google Scholar]
- 15.Jefferson R. A. (1987) Plant Mol. Biol. Rep. 5, 387-405. [Google Scholar]
- 16.DeShon R. P. & Alexander, R. A. (1994) Educ. Psychol. Meas. 54, 328-335. [Google Scholar]
- 17.Alexander R. A. & Govern, D. M. (1994) J. Educ. Stat. 19, 91-101. [Google Scholar]
- 18.Klok E. J., Wilson, I. W., Wilson, D., Chapman, S. C., Ewing, R. M., Somerville, S. C., Peacock, W. J., Dolferus, R. & Dennis, E. S. (2002) Plant Cell 14, 2481-2494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hargrove M. S. (2000) Biophys. J. 79, 2733-2738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hargrove M. S., Barry, J. K., Brucker, E. A., Berry, M. B., Phillips, G. N., Olson, J. S., Arredondo-Peter, R., Dean, J. M., Klucas, R. V. & Sarath, G. (1997) J. Mol. Biol. 266, 1032-1042. [DOI] [PubMed] [Google Scholar]
- 21.Trent J., Hvitved, A. N. & Hargrove, M. S. (2001) Biochemistry 40, 6155-6163. [DOI] [PubMed] [Google Scholar]
- 22.Watts R. A., (1999) Ph.D. dissertation (Australian National University, Canberra).
- 23.Wittenberg J. B., Wittenberg, B. A., Gibson, Q. H., Trinick, M. J. & Appleby, C. A. (1986) J. Biol. Chem. 261, 13624-13631. [PubMed] [Google Scholar]
- 24.Taylor E. R., Nie, X. Z., MacGregor, A. W. & Hill, R. D. (1994) Plant Mol. Biol. 24, 853-862. [DOI] [PubMed] [Google Scholar]
- 25.Nie X. I. H., Hill, R. D. & Nie, X. Z. (1997) Plant Physiol. 114, 835-840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sowa A. W., Duff, S. M. G., Guy, P. A. & Hill, R. D. (1998) Proc. Natl. Acad. Sci. USA 95, 10317-10321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sowa A. W., Guy, P. A., Sowa, S. & Hill, R. D. (1999) Acta Biochim. Polonica 46, 431-445. [PubMed] [Google Scholar]
- 28.Dolferus R., Klok, E. J., Ismond, K., Delessert, C., Wilson, S., Good, A., Peacock, J. & Dennis, L. (2001) IUBMB Life 51, 79-82. [DOI] [PubMed] [Google Scholar]
- 29.Bulow L., Holmberg, N., Lilius, G. & Bailey, J. E. (1999) Trends Biotechnol. 17, 21-24. [DOI] [PubMed] [Google Scholar]
- 30.Bewley J. D. & Black, M., (1994) Seeds: Physiology of Development and Germination (Plenum, London).
- 31.Duffus J. H. (1968) Phytochemistry 7, 1135-1137. [Google Scholar]
- 32.Gambhir P. N., Pande, P. C. & Ratcliffe, R. G. (1997) Magn. Reson. Chem. 35, S125-S132. [Google Scholar]
- 33.Maia I. G., Benedetti, C. E., Leite, A., Turcinelli, S. R., Vercesi, A. E. & Arruda, P. (1998) FEBS Lett. 429, 403-406. [DOI] [PubMed] [Google Scholar]
- 34.Koch K. E., Ying, Z., Wu, Y. & Avigne, W. T. (2000) J. Exp. Bot. 51, 417-427. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.