Abstract
Biosynthesis of the toxic and carcinogenic aflatoxins by the fungus Aspergillus flavus is a complicated process involving more that 27 enzymes and regulatory factors encoded by a clustered group of genes. Previous studies found that three enzymes, encoded by verA, ver-1, and aflY, are required for conversion of versicolorin A (VA), to demethylsterigmatocystin. We now show that a fourth enzyme, encoded by the previously uncharacterized gene, aflX (ordB), is also required for this conversion. A homolog of this gene, stcQ, is present in the A. nidulans sterigmatocystin (ST) biosynthesis cluster. Disruption of aflX in Aspergillus flavus gave transformants that accumulated ∼4-fold more VA and fourfold less aflatoxin than the untransformed strain. Southern and Northern blot analyses confirmed that aflX was the only gene disrupted in these transformants. Feeding ST or O-methylsterigmatocystin, but not VA or earlier precursor metabolites, restored normal levels of AF production. The protein encoded by aflX is predicted to have domains typical of an NADH-dependent oxidoreductase. It has 27% amino acid identity to a protein encoded by the aflatoxin cluster gene, aflO (avfA). Some of domains in the protein are similar to those of epoxide hydrolases.
The aflatoxins (AF), produced by Aspergillus species, are perhaps the most intensively studied polyketide metabolites (19-21). They are among the most potently toxic and carcinogenic compounds in nature (3). Biosynthesis of AF involves a set of coregulated genes that encode at least 27 proteins, including a Cys6Zn2-type pathway-specific transcription factor, a polyketide synthase, two dedicated fatty acid synthases, six cytochrome P450 monooxygenases, one esterase, two O-methyltransferases, two nonoxidative proteins, ten oxidoreductases, and two proteins for which there is little homology to known proteins in sequence databases (8, 20).
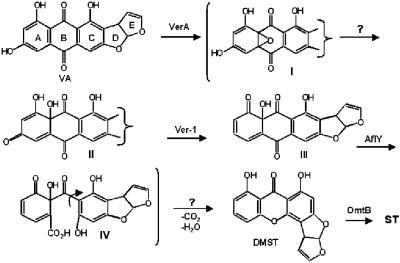
The roles of the proteins encoded by most of these genes in AF biosynthesis have been confirmed by either gene knockout or gene complementation studies (22). Conversion of versicolorin A (VA) to demethylsterigmatocystin (DMST) is predicted to involve multiple steps (Fig. 1). The probable sequence of steps has recently been clarified by analysis of precursor incorporation results (9, 10). The predicted steps involve oxidation of the anthraquinone moiety of VA, reductive deoxygenation of the A-ring, Baeyer-Villiger oxidation and rearrangement, and dehydration and decarboxylation. Enzymes previously shown to be involved in these steps are: a cytochrome P450 monooxygenase (VerA/StcS) (13, 14), an NADH-dependent deoxygenase (Ver-1/StcU) (16) with similarity to the melanin biosynthesis enzyme, tetrahydroxynaphthalene reductase (18), and a novel predicted metallo-oxidase, AflY (HypA/StcR) (6). The hypothetical pathway shown in Fig. 1 suggests that additional enzymes may be required to catalyze two additional steps in the conversion process: the epoxide ring-opening after VA oxidation and the dehydration/decarboxylation that follows the Baeyer-Villiger oxidation. Four genes with unknown function in the AF cluster are predicted to encode oxidoreductases. One of these genes, aflX, is located near the proximal end of the AF cluster. We now report that the protein encoded by aflX is involved in the conversion of VA to DMST. Based on its structure, this protein is most likely the enzyme responsible for catalysis of the epoxide ring-opening step in the conversion scheme shown in Fig. 1.
FIG. 1.
Putative enzymatic steps for conversion of VA to DMST. The suggested intermediates shown in brackets are hypothetical. Arrows shown with a question mark are steps that are expected to require enzymatic catalysis.
MATERIALS AND METHODS
Microorganisms and growth conditions.
The niaD mutant of A. flavus AF13 (ATCC 96044) was obtained from Peter Cotty, Southern Regional Research Center, New Orleans, LA. A. parasiticus ver-1 (aflM) disruptant VAD102 (15) was obtained from John Linz, Michigan State University, East Lansing, MI. A cypX (aflV) gene disruptant (CYPX33) in A. parasiticus BN009-E (7) that accumulates averufin (AVF) and does not produce aflatoxin (K. C. Ehrlich and B. G. Montalbano, unpublished results) was also used in the present study. Cultures were maintained on 5/2 agar (5% V-8 vegetable juice, 2% agar [pH 5.2]) in the dark at 31°C and, for feeding experiments, were grown on yeast extract-sucrose medium (YES; 60 g of sucrose and 2 g of yeast extract per liter).
Nucleic acid isolation and analysis.
DNA was purified from A. flavus by the method of Horng et al. (11). DNAs from A. flavus AF13 and the AF13 aflX knockout isolates were subjected to EcoRI, HindIII, or XhoI restriction enzyme digestion and separated by agarose gel electrophoresis, followed by vacuum blotting to Nytran Plus nylon membranes (Schleicher & Schuell, Inc., Keene, NH). The filters were hybridized with an [α-32P]dCTP-labeled fragment corresponding to the aflX open reading frame. Hybridization was carried out overnight in ULTRAHyb buffer (Ambion, Austin, TX) at 42°C. Membranes were washed once for 15 min in 2× SSPE (20× SSPE per liter: 3.6 M NaCl, 0.2 M NaPO4 [pH 7.7], 20 mM EDTA)-0.1% sodium dodecyl sulfate at 42°C. Filters were then washed an additional three times in 0.1× SSPE-0.1% sodium dodecyl sulfate at 42, 50, and 60°C. Damp filters were placed on Kodak X-Omat AR autoradiography film (Eastman Kodak, Rochester, NY) and allowed to expose for the desired time with intensification at −80°C.
For measurement of RNA levels total fungal RNA was purified by using the QIAGEN RNeasy Plant Minikit (QIAGEN) and reverse transcribed by using the iScript cDNA Synthesis kit (Bio-Rad). The resulting cDNAs were treated with RNase-Free DNase (QIAGEN) to ensure complete removal of DNA prior to PCR. To confirm that knockout transformants lacked aflX expression, cDNA was PCR amplified with primers to internal regions of aflX. Primer pairs designed to internal regions of the two genes flanking aflX in the cluster, moxY and aflY, were used as control reactions for PCR amplification to show that only aflX was inactivated in the A. flavus aflX knockout isolates. The locations of PCR primers within their respective gene is based on nucleotide sequence data for the A. flavus AF13 aflatoxin gene cluster (GenBank accession no. AY510451) and are shown in parentheses following the primer sequence: 5′ aflX, 5′-GCTGAACCGATTGCATCCTGAGATCAC-3′ (nucleotide [nt] 75289); 3′ aflX, 5′-GTCGTATCTCTTATCGTCCACGTTAGC-3′ (nt 74784); 5′ aflY, 5′-CCACTCTAAGTTTCACACCTTTCATGACG-3′ (nt 77212); 3′ aflY, 5′-CTTAAGGGCTCGTTTCAGCAGCGCATCAG-3′ (nt 76482); 5′ moxY, 5′-GAAGGCCCAGAATGAGAATCTCCGTAGCT-3′ (nt 73908); and 3′ moxY, 5′-CTAGCGGTTACTGTCAGAAACTCCATTGG-3′ (nt 74361). Neither aflX nor moxY has introns, so the aflY primer set was designed to amplify a region that includes two introns of 51 and 37 bp in order to confirm that the cDNA was not contaminated with genomic DNA. PCRs were performed with ExTaq polymerase (Takara) and 1 μl of cDNA template according to the manufacturer's specifications. The template cDNA was amplified by using the following thermocycler parameters: 94°C for 2 min; followed by 35 cycles of 94°C for 30 s, 65°C for 30 s, and 72°C for 30 s, with a final extension at 72°C for 7 min. The annealing temperature was lowered to 60°C for the aflX amplification. The PCRs were separated on a 1% agarose gel. An aliquot of each reaction was subcloned into a pCR2.1-TOPO vector (Invitrogen) and sequenced. DNA sequencing was performed on a CEQ8000 Automated DNA Sequencer (Beckman Coulter, Fullerton, California), and the sequence data were analyzed by using DNAMAN DNA analysis software (Lynon Biosoft, Quebec, Canada).
aflX disruption vector construction and fungal transformation.
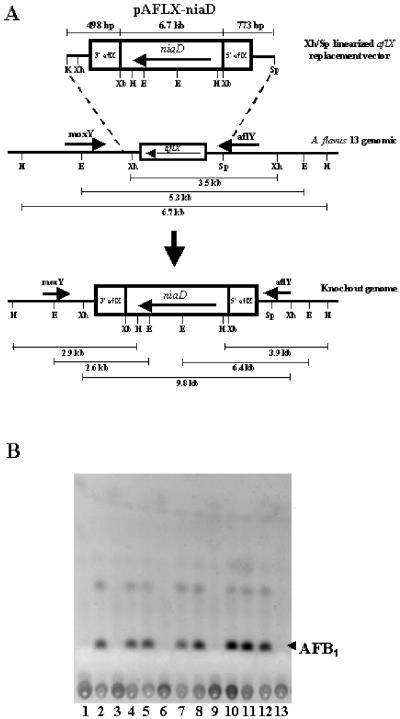
A PCR-based method was used to construct the A. flavus AF13 aflX-niaD knockout plasmid in which a central 469-bp region of the aflX coding region is replaced by the A. parasiticus niaD selectable marker gene (Fig. 2). Briefly, 5′ and 3′ regions the aflX gene were amplified by using oligonucleotide primers. The location of the primers (in parentheses following the primer sequence) within and flanking aflX is based on the GenBank nucleotide sequence data for the A. flavus AF13 aflatoxin gene cluster. Primers used were as follows: 5′ aflX KpnI, 5′-TATGGTACCCGCTAGGTACGAAGAGGTGG-3′ (nt 74356) and 5′ aflX XbaI, 5′-ATTCTAGACGCCTGTTTCCTTCCTCGAC-3′ (nt 74854). The 3′ end of aflX was amplified with the following primers: 3′ aflX XbaI, 5′-TATATCTAGACAGAACGGCAGTAGGCGTGG-3′ (nt 75323) and 3′ aflX SphI, 5′-ATACCGGGTACCGACCCAA-3′ (nt 76181). The 5′ end of the aflX gene was amplified by using primers engineered with KpnI and XbaI sites (underlined), and the 3′ aflX XbaI primer also was designed with an XbaI site (underlined) to assist in subcloning into pUC18 plasmid vector. The 3′ aflX SphI primer was designed to hybridize to a region just downstream (nt 76181) of a SphI site (nt 76096) so that an XbaI-SphI digest of the 3′ aflX PCR product would release a fragment of 773 bp. After PCR amplification of A. flavus AF13 genomic DNA with ExTaq polymerase an aliquot of the PCRs was separated on a 1% agarose gel to confirm the correct size of the products. The expected products of 498 bp for the 5′ aflX amplification and 858 bp for the 3′ aflX amplification were obtained. The 498-bp PCR product was digested with KpnI and XbaI and subcloned into KpnI/XbaI-digested pUC18. Subsequently, pUC18-5′-498-bp KpnI-XbaI plasmid DNA was XbaI/SphI digested and ligated to the XbaI/SphI-digested 773-bp 3′ aflX PCR product. The resulting pUC18-aflX 5′-498/3′-773-bp plasmid DNA was isolated and digested with XbaI. Plasmid pSL82 (4) harboring the A. parasiticus niaD gene was also digested with XbaI to release a fragment of about 6.7 kb containing the niaD gene and flanking regions. The 6.7-kb niaD gene was then ligated to XbaI digested, calf intestinal alkaline phosphatase-treated pUC18-aflX 5′-498/3′-773-bp plasmid DNA to generate the final pAFLX-niaD knockout vector (Fig. 2).
FIG. 2.
Preparation of aflX knockout mutants. (A) Schematic diagram of the knockout vector used to prepare the aflX knockout mutants. The dashed lines show the results expected by replacement of the wild-type DNA with DNA containing the niaD transformant selection cassette. Direction of transcription is indicated by horizontal arrows. The lengths of expected DNA fragments upon restriction enzyme digestion of either the wild-type A. flavus or the transformant DNA are shown under the horizontal lines. (B) TLC profiles of putative aflX knockout and nonknockout niaD+ transformants produced by transformation of A. flavus AF13 with the aflX knockout cassette shown in panel A. For TLC, 10 μl of culture medium was spotted on silica plates and developed with toluene-ethyl acetate-acetic acid (80:10:10 [vol/vol/vol]). The position of the aflatoxin B1 (AFB1) standard is shown on the right. Lanes are designations of individual transformant colonies. The plates were visualized under UV light at 312 nm.
Transformation was performed as follows. Protoplasts were obtained from about 106 conidia of the A. flavus AF13 niaD mutant grown on 250 ml of potato dextrose broth (Difco) for 12 h at 30°C with shaking (150 rpm). Mycelia were vacuum filtered through a sterile piece of Whatman filter paper and washed once with sterile water. One gram of mycelia was transferred into 20 ml of filter-sterilized enzyme solution (per 20 ml: 17 ml of H2O, 2 ml of 0.2 M NaPO4 [pH 5.8], 0.4 ml of 1.0 M CaCl2, 1.4 g of NaCl, 0.2 ml of β-glucuronidase [105 U/ml; Sigma], 200 mg of lysing enzyme [Sigma catalog no. L-1412], and 50 mg of driselase [Sigma catalog no. D-9515]). Mycelia were incubated at 30°C with shaking (80 rpm) for 3 h. Protoplasts were separated from intact mycelia by passage through Miracloth into a sterile 50-ml conical tube, and 20 ml of sterile STC buffer (1.2 M sorbitol, 10 mM CaCl2, 10 mM Tris-HCl [pH 7.5]) was added. Protoplasts were pelleted by low-speed centrifugation (1,000 rpm) at room temperature for 5 min. The supernatant was carefully removed, and the protoplasts were washed once more in 20 ml of STC and pelleted by centrifugation as described above. The protoplast pellet was resuspended in 1.0 ml of STC buffer, and the protoplast concentration was determined by using a hemacytometer. Protoplasts (about 105 in 0.1 ml of STC) were transferred to a chilled 0.2-ml microfuge tube, and 10 μg of XhoI-SphI-linearized pAFLX-niaD plasmid DNA and 0.05 ml of cold 25% PEG 4000 solution (0.6 M KCl, 50 mM CaCl2, 10 mM Tris-HCl [pH 8.0], 25% PEG 4000) was added, followed by gentle mixing and incubation on ice for 15 min. Then, 1 ml of 50% PEG 4000 solution (same as 25% PEG 4000 solution except 50% PEG 4000) was added, followed by gentle mixing and incubation for 15 min at room temperature. The protoplast solution was gently pipetted into a petri plate (100 by 15 mm), and 15 ml of selection-regeneration medium (Czapek-Dox broth [sodium nitrate as the sole nitrogen source for selection; Difco], 3.5%; 1.0 M sucrose; 1% agar) was added, followed by gentle swirling to disperse the protoplasts. Solidified agar plates were incubated at 30°C in the dark until colonies grew to the desired size for further analysis.
TLC and aflatoxin precursor feeding experiments.
Production of metabolites by putative aflX knockout transformants was assessed by Silica Gel TLC of 10-μl aliquots of 5-ml cultures grown from spores in YES medium for 5 days. On some of the transformants the culture was extracted with 5 ml of acetone for 1 h and filtered, and the acetone extract diluted with approximately 2 volumes of water and 1 volume of methylene chloride. The methylene chloride layer was collected and evaporated to dryness. The metabolite mixture was redissolved in 100 μl of acetone and a 10-μl aliquot was subjected to thin-layer chromatography (TLC) on Si250 silica gel plates (Baker, Phillipsburg, NJ) or analyzed by liquid chromatography-mass spectrometry (LC-MS) (see below). To separate metabolites, plates were developed with either toluene-ethyl acetate-acetic acid (8:1:1 [vol/vol/vol]), toluene-acetone (2:1 [vol/vol]) or ethyl ether-methanol-water (97:3:1 [vol/vol/vol]) and examined under 312-nm-wavelength light on a UV transilluminator. In feeding studies, 2-day cultures of transformant O1 in YES medium were separately incubated for 24 h with 1 μg each of the following AF precursor metabolites: averantin (AVN), averufin (AVF), VA, ST, and OMST. Following incubation, the metabolites were extracted and separated by TLC as described above. Digital images were made with a GelDoc System using Molecular Analyst Software (Bio-Rad, Hercules, CA) or a Canon Powershot S200 camera.
LC-MS analysis.
A 20-μl aliquot of the metabolite mixtures in acetone (described above) was concentrated to dryness under a flow of nitrogen. The residue was dissolved in 17 μl of acetonitrile and diluted with 3 μl of 0.1% trifluoroacetic acid (TFA) (aq). The 20-μl sample was injected onto two connected LUNA C18 (2 by 50 mm, 3 μ) columns (Phenomenex, Torrance, CA), kept at 30°C with a column heater. The chromatographic separation was performed on an Alliance HPLC (Waters Corp., Milford, MA) with a solvent gradient of 40 to 60% acetonitrile-0.1% TFA in 20 min, followed by a 5-min gradient to 70% acetonitrile-0.1% TFA at 0.3 ml/min. Detection was accomplished with a 996 photodiode array (210 to 320 nm) and Micromass ZMD mass spectrometer (Waters Corp., Milford, MA) equipped with an atmospheric pressure chemical ionization source (corona, 3.5 kV; cone, 30 V; source block temperature, 140°C; atmospheric pressure chemical ionization heater, 500°C; desolvation gas, 500 liters/min; cone gas, 100 liters/min). A positive and negative scan from m/z 300 to 400 was performed every 0.4 s. Aflatoxins were best detected in positive ionization mode, eluting near the solvent front (2.7 min). Versicolorin-type compounds were best detected in negative ionization mode with VB, VA, AVN, and AVF eluting at 10.7, 12.4, 21.1, and 22.2 min with (M-H)− ions of m/z 339.4, 337.4, 371.5, and 367.4, respectively; the maximum UV wavelengths were 290, 289, 293, and 293 nm, respectively.
Sequence alignments.
The BLASTP2.2.1 algorithm (1) (National Center for Biotechnology Information) was used to compare the predicted protein sequence of AflX to sequences in the GenBank database. In some cases, individual short conserved motifs were searched against the database using the “search for short, nearly exact matches” method for protein-protein BLAST analyses. Sequences with expectation value E < e−0.09 were aligned using the full alignment method in DNAMAN.
RESULTS
Generation and molecular analysis of an A flavus aflX knockout strain.
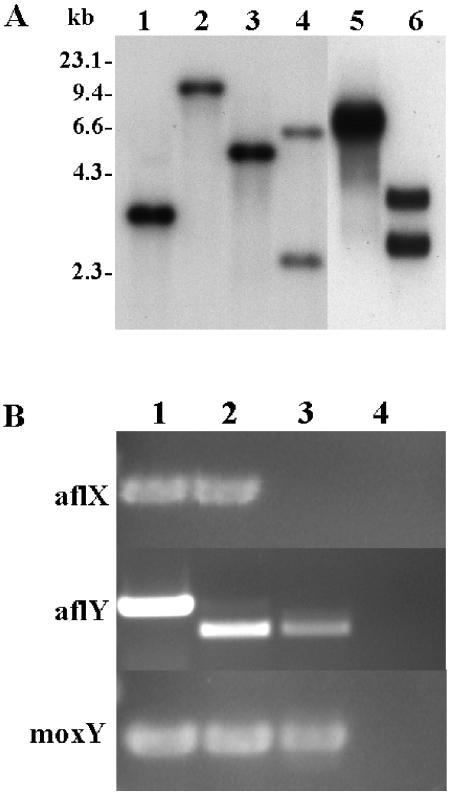
Of 30 randomly selected transformants of the A. flavus AF13 niaD mutant with XhoI-SphI-linearized pAFLX-niaD plasmid DNA (Fig. 2A), 6 demonstrated a significant reduction in aflatoxin production (Fig. 2B). Initial Southern hybridization results using the A. flavus aflX gene coding region as probe indicated that the aflX gene region of four of the six putative aflX knockout transformants had undergone homologous recombination with the pAFLX-niaD plasmid. Southern hybridization results for one of the transformants (O1) are shown in Fig. 3A. DNA from A. flavus AF13 niaD gave hybridization bands at 3.5, 5.3, and 6.7 kb after XhoI, EcoRI, and HindIII digestion, respectively, whereas O1 gave bands at 9.8 kb for XhoI, 2.6 and 6.4 kb for EcoRI, and 2.9 and 3.9 kb after HindIII digestion.
FIG. 3.
Results of Southern blot and RT-PCR analyses. (A) Southern blot results. Lanes: 1, AF13, XhoI; 2, Transformant O1, XhoI; 3, AF13, EcoRI; 4, O1, EcoRI; 5, AF13, HindIII; 6, O1, HindIII. (B) RT-PCR results. RT was carried out on total RNA digested with RNase-free DNase I with oligonucleotide primers specific for the coding regions of aflX and the flanking genes aflY and moxY. Lanes: 1, AF13 genomic DNA control; 2, AF13 cDNA; 3, aflX knockout transformant O1 cDNA; 4, no template negative control.
Loss of aflX gene expression in the aflX knockout O1 was confirmed by the lack of a PCR product after reverse transcription-PCR (RT-PCR) of total RNA with primers for aflX sequence (Fig. 3B), while PCR products of the correct sizes were obtained when RT-PCR was performed with primers to aflY and moxY, the genes that flank aflX in the aflatoxin gene cluster. Therefore, aflX was selectively inactivated. As controls, PCR of genomic DNA and cDNA from A. flavus AF13 niaD− gave products of the expected sizes for aflX (505 bp), moxY (453 bp), and aflY (731 bp for genomic and 643 bp for cDNA due to 51- and 37-bp introns). Sequencing of the product obtained from RT-PCR of aflY confirmed that the intron sequence was not present (data not shown). No PCR products were detected in the no-template DNA negative control lane (lane 4).
LC-MS analysis of transformants and feeding studies.
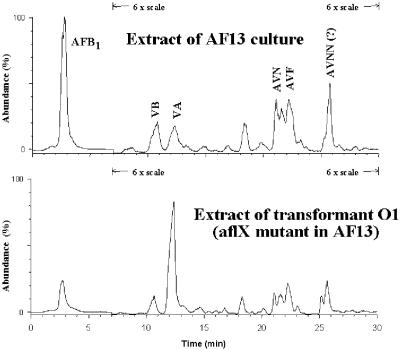
LC-MS results confirmed that production of AFB1 was reduced in O1 by ∼4-fold compared to production by the wild-type A. flavus AF13. A concomitant increase occurred in accumulation of VA in the aflX knockout transformant (Fig. 4). When transformant O1 was incubated with the AF precursor metabolites, the only metabolites that increased the level of AF produced by the disruptant culture were ST and OMST, confirming the TLC and LC-MS results that the metabolic block is after formation of VA (Table 1).
FIG. 4.
LC-MS analysis of metabolite extracts from AF13 niaD mutant and transformant O1 cultures. Identification of metabolites was by elution times and comparison of ion peaks from MS to peaks from authentic samples. The scale was 1× for detection of AFB1 and 6× for detection of the other metabolites, as shown on the plot.
TABLE 1.
AF production by A. parasiticus O1 (aflX disruptant) by precursor feeding
| Expt (precursor feeding)a | AF produced (ng/5-ml culture)b |
|---|---|
| None | 220 |
| AVN | 210 |
| AVF | 280 |
| HAVN | 270 |
| VA | 240 |
| ST | 620 |
| OMST | 750 |
Abbreviations: AVN, averantin; AVF, averufin; HAVN, hydroxyaverantin; ST, sterigmatocystin; OMST, O-methylsterigmatocystin.
Quantification of AF was by comparison of fluorescence on silica gel TLC plates to that of known amounts of authentic standards. The limit of detection was 5 ng. Experiments were repeated twice with similar results.
BLAST searches and alignment of AflX with AvfA.
The sequence that most resembles AflX in the GenBank protein database, other than the A. nidulans homolog StcQ, is AvfA (AflO, 27% amino acid identity) and its A. nidulans homolog (StcO), an enzyme that was shown to catalyze one of the steps in the conversion of averufin to versicolorin A (23). AflX possesses a catalytic domain characteristic of an NADH-flavin reductase (COG2910.1). Most of the proteins in COG2910.1 with closest sequence match had no ascribed function. Alignment of AflX with AvfA and two other COG2910.1-like proteins is shown in Fig. 5. Several regions show sequence conservation: the N-terminal sequences, AxxGATGxTG and RxxxKL, the sequences in the middle of the protein, LxSA, and YxD, while the C terminus demonstrated little conservation. The function of A. nidulans AN0152 is not known. Colletotrichum lagenarium Cad2 is a protein encoded by 1 of 11 genes that are presumed to be involved in appressorium differentiation (12).
FIG. 5.
Alignments of the predicted sequence for AflX with closely related sequences. Abbreviations: Af, A. flavus; Anid, A. nidulans; Clag, Colletotrichum lagenarium. The color scheme for amino acid similarity is: black, 100%; dark gray, >75%; light gray, >50%. Accession numbers for the AflX and AvfA sequences were as follows: A. flavus AF13, AAS90109 and AY510451.1; A. nidulans AN0152.2, XM404289; and Colletotrichum lagenarium Cad2, AF250382. The sequence of AN0152.2 is truncated at aa 298 (predicted length, 674 aa).
DISCUSSION
Our results show that aflX encodes an enzyme involved in the conversion of VA to DMST. This conclusion is based on the observation that disruption of aflX in an A. flavus isolate capable of AFB1 production gave transformants in which AFB1 accumulation was markedly reduced, whereas the only metabolite that accumulated significantly was VA (Fig. 4). The feeding studies also support this conclusion, since only ST or OMST, metabolites that are formed after VA, are capable of substantially increasing AF production when fed to an aflX knockout transformant (Table 1). No known VA precursor metabolites were able to be converted to AFB1 in the feeding studies, and such precursors did not accumulate at higher levels in extracts of the culture medium from the knockout cultures compared to their levels in the wild-type cultures. Therefore, the metabolic block in the aflX knockout transformants is in the conversion of VA to DMST. Support for this function of aflX could be confirmed by retransformation of the A. flavus aflX knockout strain with a plasmid-encoded aflX, which we predict would restore wild-type levels of AF production. (This experiment was not included here because the authors' lab was physically destroyed while this study was in review [Editor's note].)
Conversion of VA to DMST is predicted to involve multiple steps as shown in Fig. 1. Enzymes involved in some of these steps have been characterized, including Ver-1 (StcU), VerA (StcS), and AflY (StcR), and plausible enzymatic functions consistent with the chemistry required for the conversion of VA to DMST have been ascribed for these proteins (9, 10). However, the conversion process as shown in Fig. 1 requires cleavage of the epoxide ring formed by VerA oxidation and the decarboxylation/dehydration shown as the step prior to formation of DMST (Fig. 1). It is plausible that these steps are also mediated by enzymes. To date, most of the genes in the AF cluster have been found to encode enzymes associated with expected chemical steps required for the formation of intermediates produced during AF biosynthesis (22). AflX is among the few oxidoreductases encoded by a gene in the AF cluster whose function has not yet been assigned. Although not proven by the present study, our data and the structure of AflX are consistent with its being the enzyme catalyzing the epoxide ring-opening depicted in Fig. 1.
The enzyme encoded by aflX is predicted to contain an NADH-binding domain based on protein database searches and shows highest similarity to members of COG2910 which includes NADH-flavin reductase-like proteins from bacteria. The conserved Tyr and Asp residues in the amino acid motif YxD could catalyze the reductive epoxide ring cleavage. Such residues have been shown to be required for catalysis by known epoxide hydrolases (2, 17). Another Asp residue closer to the C terminus of the protein could also be involved in the catalysis. Catalysis of epoxide ring-opening by known epoxide hydrolases requires a His residue to release the covalently bound Asp formed during ring cleavage. The only possible conserved His residues in AflX are 11 amino acids (aa) on the N-terminal side of the YxD motif and 13 and 27 aa on the C-terminal side of this motif. The product of typical fungal and bacterial epoxide hydrolases is a diol, whereas, with AflX, we predict that the product is a hydroxydienone. Therefore, it is not surprising that AflX does not show significant overall sequence similarity to typical epoxide hydrolases.
Additional support for AflX catalyzing the presumed epoxide ring-opening step comes from the observation that AflX has a structure and expected catalytic motifs similar to those of AvfA, an enzyme shown to be required for an earlier conversion step in AF biosynthesis, namely, conversion of averufin to hydroxyversicolorone, a process that may involve an epoxide intermediate (19, 21). The similarity of these two proteins suggests that they may have overlapping functions. If this is true, less efficient catalysis of the AflX step by AvfA could partly bypass the gene disruption and explains why mutation of aflX gives rise to a “leaky” phenotype in the transformants. Generation of an A. flavus aflX-avfA double mutant would likely provide evidence for AvfA to also have the ability to catalyze conversion of VA to DMST. However, at this time a strain of A. flavus with two mutations allowing for the use of two different selectable markers required to generate an aflX-avfA double mutant is not available. Search of the A. nidulans protein database revealed one other high-scoring protein other than the putative homolog, StcQ. This protein is encoded by a hypothetical gene AN0152.2 outside of the ST biosynthetic cluster. The N-terminal sequence of this hypothetical protein aligns with the A. nidulans AflX and AvfA homologs, StcQ and StcO, respectively, but the sequence of the protein encoded by AN0152.2 is predicted to be much longer. Therefore, proteins with possible similar functionality to AflX may be in the genomes of fungal species and have unrelated function to AF/ST biosynthesis. In Colletotrichum lagenarium the gene CAD2 encodes a 278-aa protein with relatively high homology to AflX and AvfA. This gene is involved in development of appressorium from conidia, but its catalytic function has not been identified (12). Another gene in the conversion of VA to DMST, ver-1, encodes an enzyme that shows a high degree of similarity to the protein, tetrahydronapthalene reductase, an enzyme required for melanin biosynthesis (18). Melanization is necessary for appressoria to penetrate host plant cell walls during infection. Our results suggest that both appressorium development and aflatoxin formation may share similar biosynthesis genes.
In conclusion, A. flavus AflX is most likely involved in a ring-opening rearrangement of the epoxide produced by the VerA cytochrome P450 monooxygenase-catalyzed oxidation. We concluded in a previous study that ST and AF type clusters diverged over 100 Mya (8). In the ST cluster of A. nidulans, the genes involved in the VA to DMST conversion form a mini-cluster within the ST cluster. It is tempting to speculate that the primordial cluster in species ancestral to both Nidulantes and Flavi allowed biosynthesis of anthraquinones. As the AF/ST clusters further evolved, the machinery for oxidation of the anthraquinone moiety was introduced, thereby possibly providing a survival advantage.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zheng, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barth, S., M. Fischer, R. D. Schmid, and J. Pleiss. 2004. Sequence and structure of epoxide hydrolases: a systematic analysis. Proteins 55:846-855. [DOI] [PubMed] [Google Scholar]
- 3.Bhatnagar, D., R. Brown, K. Ehrlich, and T. E. Cleveland. 2002. Mycotoxins contaminating cereal grain crops: their occurrence and toxicity, p. 171-196. In G. G. Khachatourians and D. K. Arora (ed.), Applied mycology and biotechnology, vol. 2. Elsevier B.V., New York, N.Y. [Google Scholar]
- 4.Chang, P.-K., K. C. Ehrlich, J. E. Linz, D. Bhatnagar, T. E. Cleveland, and J. W. Bennett. 1996. Characterization of the Aspergillus parasiticus niaD and niiA gene cluster. Curr. Genet. 30:68-75. [DOI] [PubMed] [Google Scholar]
- 5.Reference deleted.
- 6.Ehrlich, K. C., B. G. Montalbano, S. M. Boue, and D. Bhatnagar. 2005. An aflatoxin biosynthesis cluster gene encodes a novel oxidase required for conversion of versicolorin A to sterigmatocystin. Appl. Environ. Microbiol. 71:8963-8965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ehrlich, K. C., B. G. Montalbano, and P. J. Cotty. 2003. Sequence comparison of aflR from different Aspergillus species provides evidence for variability in regulation of aflatoxin production. Fungal Genet. Biol. 38:63-74. [DOI] [PubMed] [Google Scholar]
- 8.Ehrlich, K. C., J. Yu, and P. J. Cotty. 2005. Aflatoxin biosynthesis gene clusters and flanking regions. J. Appl. Microbiol. 99:518-527. [DOI] [PubMed] [Google Scholar]
- 9.Henry, K. M., and C. A. Townsend. 2005. Ordering the reductive and cytochrome P450 oxidative steps in demethylsterigmatocystin formation yields general insights into the biosynthesis of aflatoxin and related fungal metabolites. J. Am. Chem. Soc. 127:3724-3733. [DOI] [PubMed] [Google Scholar]
- 10.Henry, K. M., and C. A. Townsend. 2005. Synthesis and fate of o-carboxybenzophenones in the biosynthesis of aflatoxin. J. Am. Chem. Soc. 127:3300-3309. [DOI] [PubMed] [Google Scholar]
- 11.Horng, J. S., P.-K. Chang, J. J. Pestka, and J. E. Linz. 1990. Development of a homologous transformation system for Aspergillus parasiticus with the gene encoding nitrate reductase. Mol. Gen. Genet. 224:294-296. [DOI] [PubMed] [Google Scholar]
- 12.Inagaki, A., Y. Takano, Y. Kubo, K. Mise, and I. Furusawa. 2000. Construction of an equalized cDNA library from Colletotrichum lagenarium and its application to the isolation of differentially expressed genes. Can. J. Microbiol. 46:150-158. [DOI] [PubMed] [Google Scholar]
- 13.Keller, N. P., N. J. Kantz, and T. H. Adams. 1994. Aspergillus nidulans verA is required for production of the mycotoxin sterigmatocystin. Appl. Environ. Microbiol. 60:1444-1450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keller, N. P., S. Segnar, D. Bhatnagar, and T. H. Adams. 1995. stcS, a putative P-450 monooxygenase, is required for the conversion of versicolorin A to sterigmatocystin in Aspergillus nidulans. Appl. Environ. Microbiol. 61:3628-3632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Liang, S.-H., C. D. Skory, and J. E. Linz. 1996. Characterization of the function of the ver-1A and ver-1B genes involved in aflatoxin biosynthesis in Aspergillus parasiticus. Appl. Environ. Microbiol. 62:4568-4575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Skory, C. D., P. K. Chang, J. Cary, and J. E. Linz. 1992. Isolation and characterization of a gene from Aspergillus parasiticus associated with the conversion of versicolorin A to sterigmatocystin in aflatoxin biosynthesis. Appl. Environ. Microbiol. 58:3527-3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smit, M. S. 2004. Fungal epoxide hydrolases: new landmarks in sequence-activity space. Trends Biotechnol. 22:123-129. [DOI] [PubMed] [Google Scholar]
- 18.Vidal-Cros, A., F. Viviani, G. Labesse, M. Boccara, and M. Gaudry. 1994. Polyhydroxynaphthalene reductase involved in melanin biosynthesis in Magnaporthe grisea: purification, cDNA cloning, and sequencing. Eur. J. Biochem. 219:985-992. [DOI] [PubMed] [Google Scholar]
- 19.Yabe, K., and H. Nakajima. 2004. Enzyme reactions and genes in aflatoxin biosynthesis. Appl. Microbiol. Biotechnol. 64:745-755. Epub 2004 Mar 12. [DOI] [PubMed] [Google Scholar]
- 20.Yu, J., D. Bhatnagar, and T. E. Cleveland. 2004. Completed sequence of the aflatoxin pathway gene cluster in Aspergillus parasiticus. FEBS Lett. 564:126-130. [DOI] [PubMed] [Google Scholar]
- 21.Yu, J., D. Bhatnagar, and K. C. Ehrlich. 2002. Aflatoxin biosynthesis. Rev. Iberoam. Micol. 19:191-200. [PubMed] [Google Scholar]
- 22.Yu, J., P.-K. Chang, K. C. Ehrlich, J. W. Cary, D. Bhatnagar, T. E. Cleveland, G. A. Payne, J. E. Linz, C. P. Woloshuk, and J. W. Bennett. 2004. Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 70:1253-1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yu, J., C. P. Woloshuk, D. Bhatnagar, and T. E. Cleveland. 2000. Cloning and characterization of avfA and omtB genes involved in aflatoxin biosynthesis in three Aspergillus species. Gene 248:157-167. [DOI] [PubMed] [Google Scholar]