Abstract
Examination of variation in ecological communities can lead to an understanding of the forces that structure communities, the consequences of change at the ecosystem level, and the relevant scales involved. This study details spatial and seasonal variability in the composition of nitrogen-fixing and cyanobacterial (i.e., oxygenic photosynthetic) functional groups of a benthic, hypersaline microbial mat from Salt Pond, San Salvador Island, Bahamas. This system shows extreme annual variability in the salinity of the overlying water and the extent of water coverage. Analysis of molecular variance and FST tests of genetic differentiation of nifH and cyanobacterial 16S rRNA gene clone libraries allowed for changes at multiple taxonomic levels (i.e., above, below, and at the species level) to inform the conclusions regarding these functional groups. Composition of the nitrogen-fixing community showed significant seasonal changes related to salinity, while cyanobacterial composition showed no consistent seasonal pattern. Both functional groups exhibited significant spatial variation, changing with depth in the mat and horizontally with distance from the shoreline. The patterns of change suggest that cyanobacterial composition was more insensitive to water stress, and consequently, cyanobacteria dominated the nitrogen-fixing community during dry months but gave way to a more diverse community of diazotrophs in wet months. This seasonal pattern may allow the mat community to respond quickly to water-freshening events after prolonged dry conditions (system recovery) and maintain ecosystem function in the face of disturbance during the wet season (system resilience).
Communities from hypersaline microbial mats have been the focus of much research (1, 4, 11, 14, 17, 33, 44). Several unique features of these systems make them attractive models for studies involving community dynamics, ecosystem function, and the links between the two. Because the hypersaline environmental conditions are detrimental to many multicellular organisms, the interspecific interactions in these mats are almost entirely microbial (43). The result is confinement of biogeochemically important processes to microscale habitats where they can proceed under the influence of environmental gradients controlled by microbial consortial processes (31, 32). For example, laminated microbial mats typically exhibit very steep vertical gradients in pH, Eh, oxygen, light regime, and chemical forms of carbon, nitrogen, sulfur, etc. (31). These gradients largely result from microbial activity (7). Along these gradients, a great deal of phylogenetic and metabolic diversity is packed into a small space, facilitating the interspecific exchange of organic and inorganic materials upon which ecosystem function depends. Organisms that inhabit these mats are known to participate in specific carbon and nitrogen transformations (32, 44), so microbial mats are ideal systems to study the link between species composition and the cycling of these fundamental biological elements.
The islands of the Bahamas contain numerous shallow, hypersaline lagoons that support laminated microbial mats (21). These laminated communities often reside in nutrient-poor ecosystems (30), and so microbes must supply and recycle carbon and nitrogen. On the supply side of this equation, two taxonomically diverse functional groups are important. Cyanobacteria are often the dominant structural organisms comprising the matrix of these mats (44), and they provide a bulk of the organic matter for mat growth and accretion by oxygenic photosynthesis. Thus, cyanobacteria play important functional roles by supporting mat microstructure, metabolism, and the resultant development of microenvironments, e.g., oxygen gradients (32). A second important functional group in these nitrogen-poor habitats consists of the nitrogen-fixing prokaryotes (“diazotrophs”) that reduce atmospheric nitrogen to biologically available ammonia (45). They include diverse members of the α-, β-, and δ-Proteobacteria as well some species of cyanobacteria and comprise both aerobic and anaerobic organisms (46-48, 50). The existence of molecular probes specific for each of these functional groups (25, 50) allows for the investigation of their roles in community composition and activity.
The climate of the Bahamas is influenced by large, periodic storm events. Previous work on a microbial mat on the island of San Salvador has shown that storm-induced changes in water salinity, including two major hurricanes in the past 7 years (Floyd in 1999 and Frances in 2004), have had large effects on microbial community metabolism, growth, and mat accretion (29). The sensitivity of hypersaline lake communities to such environmental perturbations has proven them to be useful indicators of short-term, and possibly longer-term, climate change (29). Clarification of the range of microbial community structural variation under typical conditions is a precursor to understanding the physiological and ecological responses of these systems to extreme climatic forcing and other environmental perturbations.
Here, we report observations of spatial and temporal changes in the community structure of cyanobacterial and diazotrophic communities from the microbial mats located in Salt Pond, San Salvador, Bahamas. Statistical analyses developed for population genetics allowed for the integration of phylogenetic information from clone libraries into the assessment of community structure. Specific questions addressed include the following: (i) what is the phylogenetic composition of cyanobacteria and nitrogen fixers in this system, (ii) how do these functional groups change temporally between wet and dry seasons and spatially with regard to depth and water coverage, and (iii) what do these patterns reveal about the ecological responses of these functional groups to environmental change?
MATERIALS AND METHODS
Study site and sample collection.
The island of San Salvador, Bahamas (24°05′N, 74°30′W), has a subtropical, semiarid climate, with a mean annual rainfall of 100.7 cm. In a typical year, just under half of this rainfall is the result of episodic tropical storm activity during a relatively short “wet season” from mid-September through late November (40). Salt Pond (24°01′22"N, 74°27′02"W) is a shallow hypersaline pond located on the central eastern side of San Salvador. The surface area and salinity of Salt Pond vary dramatically with the wet and dry seasons, with salinity ranging in extremis from 60 to 350 practical salinity units (PSU). The lake sediments, including portions of the littoral zone that dry out over parts of the year, are covered by a microbial mat, which is dominated by benthic diatoms and nonheterocystous cyanobacteria of the orders Oscillatoriales, Chroococcales, and Pleurocapsales as described elsewhere previously (33, 36).
Three sampling transects were established on the eastern side of Salt Pond and maintained over the course of this study. The transects, separated by 15 m, ran perpendicular to the shoreline, extending into Salt Pond 26 m from the edge of the wet-season shoreline (i.e., when the water level is highest due to rainfall). Samples for molecular characterization of mat communities were collected in the dry season, 15 March 2002 and 6 March 2003, and in the wet season, 15 October 2003 (Table 1). Samples were collected along a desiccation gradient from three locations on each transect: “inshore” (0- or 1-meter marker), “midshore” (7- or 8-meter marker), and “offshore” (26-meter marker). On some occasions, the inshore samples were collected from above the water line (Table 1). The topmost 1.0 cm of mat material was collected by inserting a cylindrical coring device (interior diameter, 5.5 mm) into the mat. For samples collected in 2003, cores were further subdivided vertically with a razor blade for investigation of vertical community structure. For these samples, the “upper” layer refers to the uppermost 2 mm, a layer dominated by extracellular polymeric substances (5, 6, 15); the “middle” layer refers to the next 4 mm, a green, cyanobacterium-dominated layer; and the “lower” layer refers to the lowest 4 mm, a purple/pink layer dominated by purple sulfur bacteria. In all cases, samples were frozen (−20°C) in Tris-EDTA (TE) buffer (pH 8.0) for transport back to the laboratory, at which time they were stored at −80°C until they were processed as described below.
TABLE 1.
Salt Pond environmental conditions at the time of sample collection
| Date | Season | Salinity (PSU) | Water temp (°C) | Shoreline position along transect |
|---|---|---|---|---|
| March 2002 | Dry | 114 | 28.0 | 6-m mark |
| March 2003 | Dry | 104 | 30.5 | 4-m mark |
| October 2003 | Wet | 87 | 40.0 | 0-m mark |
Molecular methods.
Cell lysis for all mat samples was accomplished by incubation at 70°C in phenol followed by bead beating at high speed for 40 s. Pellets were then washed in high-salt (5 M NaCl) TE buffer (pH 8.0) to remove extracellular polymers. DNA purification began with two rounds of extraction with phenol and chloroform (19). Extracts were further purified by incubation (15 min) at 4°C in 0.5% hexadecyltrimethyl ammonium bromide and 0.3 M sodium acetate, followed by two subsequent rounds of phenol-chloroform-isoamyl alcohol (25:24:1) purification and precipitation with 70% ethanol (19). Pellets were resuspended in TE buffer and purified a final time using DNeasy spin columns, as described by the manufacturer of the DNeasy Plant Mini kit (QIAGEN Inc., Chatsworth, CA). The resulting extract was stored at −20°C in TE buffer (pH 8.0) prior to analysis.
PCR for nifH was performed using the Z1-Z2 primer set described previously by Zehr et al. (51, 53), and the cyanobacterial 16S sequence was amplified using the primer CYA359F and an equimolar mixture of primers CYA781R(a) and CYA781R(b) (25). PCR products were gel purified and cloned into the pCR2.1-TOPO vector (Invitrogen, San Diego, CA). Twenty-four nifH and 24 cyanobacterial 16S clones were sequenced from each sample for March 2002 and for each vertical layer of each sample for March and October 2003. Sequencing was done by the University of North Carolina at Chapel Hill Automated DNA Sequencing Facility with a model 373A DNA sequencer (Applied Biosystems, Foster City, CA) using the Taq DyeDeoxy Terminator Cycle Sequencing kit (Applied Biosystems, Foster City, CA).
Phylogeny.
Sequences were aligned using the Fast Aligner function in ARB (version 20030822-12; Lehrstuhl fuer Mikrobiologie, TU Muenchen [http://www.arb-home.de]) and then manually aligned with the ARB Editor. ARB alignment was facilitated by comparison with the nifH database described previously by Zehr and others (50) and, for 16S sequences, with the database described previously by Hugenholtz (12), with an additional 1,600 cyanobacterial 16S sequences from the Ribosomal Database Project, including the closest BLAST matches from GenBank.
For the purpose of phylogenetic tree construction, highly similar sequences were treated as part of a single operational taxonomic unit (OTU) determined using the farthest-neighbor criterion of DOTUR (version 1.51; Department of Plant Pathology, University of Wisconsin—Madison [http://www.plantpath.wisc.edu/fac/joh/DOTUR.html]), with a matrix of Kimura's two-parameter molecular distances (13, 16) used as input. To determine the farthest-neighbor distance cutoff approximating the species level, DOTUR runs were carried out with at least 50 sequences from cultured organisms, and the distance that grouped these sequences properly by species was applied to the sequences of the Salt Pond mat organisms. Based on these tests, it was determined that OTUs corresponded most closely to the species level at farthest-neighbor distances of 0.07 for nifH and 0.03 for cyanobacterial 16S sequences. DOTUR was also used to calculate the Chao1 estimator of minimal taxonomic diversity (2, 3) for each functional group in each of the sampling months.
Phylogenetic trees for mat OTUs were constructed with the program PAUP* (version 4.0b10 [Altivec]; Sinauer Associates, Inc. [http://paup.csit.fsu.edu/index.html]) using the maximum likelihood criterion. Prior to tree construction, evolutionary models and model parameters were evaluated using Posada and Crandall's Modeltest (version 3.7; University of Vigo, Vigo, Spain [http://darwin.uvigo.es/software/modeltest.html]), and the model judged to be the best by likelihood ratio tests was implemented in PAUP*. PAUP* searches were started by using trees constructed by ARB's neighbor-joining function. Clade credibility values were assessed with PAUP* by 1,000 bootstrap replicates using neighbor joining with the appropriate maximum likelihood model distance criterion.
Statistical approaches.
Gross differences in mat functional group composition between months were assessed using ∫-LIBSHUFF (version 1.21; Department of Plant Pathology, University of Wisconsin—Madison [http://www.plantpath.wisc.edu/fac/joh/S-LIBSHUFF.html]), with a matrix of Kimura's two-parameter molecular distances (13, 16) used as input. ∫-LIBSHUFF uses the integral form of the Cramer-von Mises statistic to determine whether two clone libraries are likely to represent samples drawn from statistically distinguishable mixes of species, as first described by Singleton and colleagues (41). The significance is tested by permutation of sequences between two libraries, libraries A and B. This test is asymmetrical, making it possible for library A versus library B to be significant while library B versus library A is not; this would indicate that the species mix from which library B was constructed is likely to be a subset of the species mix from which library A was constructed (39, 41).
∫-LIBSHUFF does not explicitly consider clone frequency. To further examine variation in the genetic structure of mat functional groups, analysis of molecular variance (AMOVA) and FST tests (20) were conducted with the program Arlequin (version 2.001; Genetics and Biometry Laboratory, University of Geneva [http://lgb.unige.ch/arlequin]). AMOVA, as formalized previously by Excoffier et al. (8), uses a hierarchically partitioned matrix of (squared) genetic distances to assess, by permutation, the significance of variance component associated with each level of the partitioning. It is therefore analogous to a nested analysis of variance for genetic data. In the present context, the input matrices (for nifH and cyanobacterial 16S) consisted of Kimura's two-parameter distances (13, 16) partitioned into each of four hierarchical schemes: (i) inshore, midshore, and offshore samples, nested within the sampling month; (ii) upper, middle, and lower layers, nested within the sampling month (2003 only); (iii) March 2003 and October 2003, nested within the vertical layer of the mat; and (iv) inshore, midshore, and offshore, nested within vertical layers for the same month.
FST tests, as described previously by Martin (20), were performed as tests of genetic differentiation between all pairs of samples and all monthly clone libraries. The FST test determines whether samples contain close phylogenetic relatives or more deeply divergent sequences. In the present context, a significant (by permutation) FST between any pair of samples indicates that the average genetic diversity within each sample is substantially less than what would be expected if the same underlying mix of species was being sampled. FST tests conducted using pairs of samples are analogous to post hoc pairwise comparisons in analysis of variance. FST can also be considered as a measure of “distance” between pairs of samples that takes into account both the frequency of identical or closely related sequences and the amount of evolved diversity within the sample pairs. With this in mind, the matrices of pairwise FST statistics were used as the basis of nonmetric multidimensional scaling plots in the program PRIMER 5 for Windows (version 5.2.7; PRIMER-E, Ltd. [http://www.primer-e.com]) to help visualize patterns in genetic composition between samples.
Nucleotide sequence accession numbers.
All sequences obtained during the course of this work have been submitted to GenBank under the accession numbers DQ140418 to DQ140728 (nifH) and DQ140729 to DQ141101 (cyanobacterial 16S).
RESULTS
General description of recovered phylotypes.
Clone libraries from March 2002, March 2003, and October 2003 yielded 490 nifH and 513 cyanobacterial 16S sequences representing 97 nifH OTUs and 93 cyanobacterial OTUs.
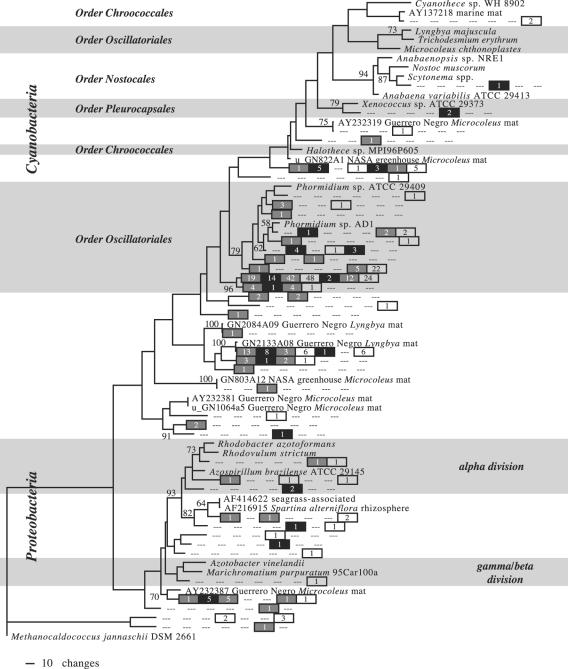
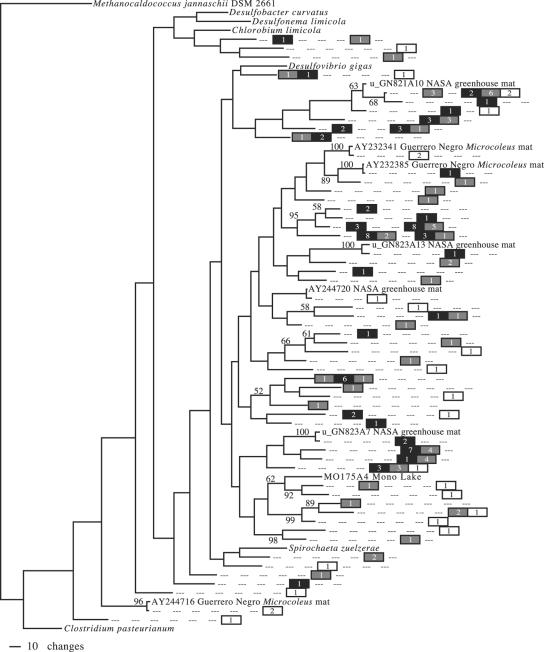
All nifH sequences recovered from the mat were related to type I or type III nitrogenases (Fig. 1 and 2). Type I nitrogenase representatives included members of the α-, β-, and γ-Proteobacteria as well as the cyanobacterial orders Chroococcales, Oscillatoriales, Pleurocapsales (seen only in October 2003), and Nostocales (one representative, in October 2003). No type III nitrogenases from the mat were closely related to cultured strains, although a few were broadly related to δ-Proteobacteria. Most mat sequences with very close BLAST matches in GenBank (i.e., high bit score similarity and an E value of 0.0) were related to environmental sequences obtained from other microbial mat systems (Fig. 1).
FIG. 1.
Maximum likelihood tree of type I nifH sequences found in the Salt Pond mat, San Salvador, Bahamas. Methanocaldococcus jannaschii was used as an outgroup to root the tree. Numbers at the nodes reflect clade confidence (percentage) based on 1,000 bootstrap replicates; only numbers greater than 50% are shown. Salt Pond mat sequences recovered for this study are represented by a seven-part symbol (boxes and/or dashes [“—”]) reflecting the temporal and vertical distribution of each OTU. From left to right, positions in this symbol represent March 2002 (gray box); March 2003, lower mat (black box); March 2003, middle mat (gray box); March 2003, upper mat (white box); October 2003, lower mat (black box); October 2003, middle mat (gray box); and October 2003, upper mat (white box). If a sequence was recovered from a particular spot on the mat, e.g., March 2003, lower mat third box from the left), the number in the box indicates the number of clones in the OTU found at that spot. Three dashes (“—”) indicate that no clones in that OTU were recovered at that spot.
FIG. 2.
Maximum likelihood tree of type III nifH sequences found in the Salt Pond mat. Methanocaldococcus jannaschii was used as an outgroup to root the tree. Clade confidence, symbols, and numbers are the same as those described in the legend of Fig. 1.
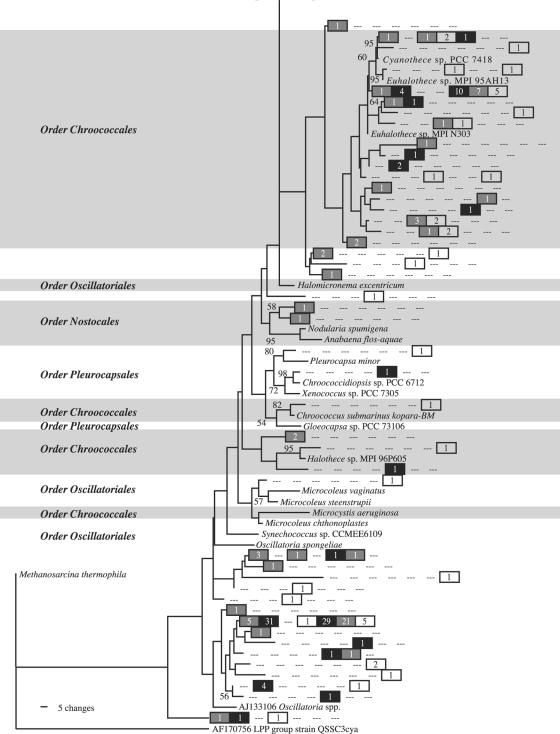
As with the nifH sequences, most of the cyanobacterial 16S sequences recovered belonged to the orders Chroococcales and Oscillatoriales or else clustered with unidentified organisms from environmental studies (Fig. 3 and 4). There were no Nostocales sequences, and Pleurocapsales sequences were infrequently seen. The most commonly seen 16S sequences had no very close BLAST matches in GenBank, although a sequence highly similar to those of Euhalothece spp. was moderately common in the October library (Fig. 4).
FIG. 3.
Maximum likelihood tree of cyanobacterial 16S sequences found in the Salt Pond mat. This is the top half of the tree, which is continued in Fig. 4. Methanosarcina thermophila, which joins the tree in Fig. 4, was used as an outgroup to root the tree. Clade confidence, symbols, and numbers are the same as those described in the legend of Fig. 1.
FIG. 4.
Maximum likelihood tree of cyanobacterial 16S sequences found in the Salt Pond mat. This is the bottom half of the tree, which is continued in Fig. 3. Methanosarcina thermophila was used as an outgroup to root the tree. Clade confidence, symbols, and numbers are the same as those described in the legend of Fig. 1.
Library composition by month.
Diversity of nifH libraries was 24, 36, and 70 distinct OTUs for March 2002, March 2003, and October 2003, respectively, and Chao1 nonparametric estimators of minimal mat nifH diversity for these times were 68.3 (95% confidence interval [CI], 36.1 to 180.0), 52.0 (95% CI, 40.3 to 89.2), and 127.6 (95% CI, 95.2 to 199.9). There were 22, 38, and 47 distinct cyanobacterial 16S OTUs found in the libraries from March 2002, March 2003, and October 2003, respectively, and Chao1 estimators for these times were 71.5 (95% CI, 46.1 to 146.5), 59.5 (95% CI, 42.0 to 114.8), and 71.1 (95% CI, 54.3 to 115.0). Thus, confidence intervals for Chao1 estimates suggested that the true species diversity may have been at least 1.1 to 7.7 times higher than that indicated by clone library OTU richness.
For nifH, ∫-LIBSHUFF could not reject the null hypothesis that March 2002 and March 2003 clone libraries contained the same mix of species (P = 0.381 for March 2002 versus March 2003 and P = 0.048 for March 2003 versus March 2002; this last test is not significant at an α (type I error rate) of ≤0.05 after Bonferroni correction for multiple comparisons). Both March 2002 and March 2003 libraries appeared to comprise subsamples from October 2003, which contained significantly more unique sequences than either March library (P = 0.959 for March 2002 versus October 2003, P = 0.068 for March 2003 versus October 2003, and P ≪ 0.001 for October versus either March library). In particular, the October library encompassed a greater diversity of noncyanobacterial nitrogenase sequences than either of the March libraries. FST tests between months mirrored these results (Table 2).
TABLE 2.
FST statistics for clone library comparisons by month
| Library | Date | FSTa
|
|
|---|---|---|---|
| March 2002 | March 2003 | ||
| nifH | March 2003 | 0.016 (NS) | |
| October 2003 | 0.103*** | 0.087*** | |
| Cyanobacterial 16S | March 2003 | 0.041** | |
| October 2003 | 0.021* | 0.084*** | |
Bonferroni-corrected significance levels. NS, not significant; *, α = 0.05; **, α = 0.01; ***, α ≤ 0.001.
For cyanobacterial 16S clone libraries, March 2003 and October 2003 libraries were determined to be significantly different from each other (P ≪ 0.001 for March 2003 versus October 2003 and vice versa). March 2002 and October 2003 libraries were not distinguishable at an α of ≤0.05 (P = 0.296 for March 2002 versus October, and P = 0.662 for October versus March 2002). The March 2003 library was not significantly different from the March 2002 library (P = 0.525), but the converse was not true (P = 0.001). Thus, the March 2002 and October 2003 libraries tended to overlap with each other, and each contained enough unique sequences to be deemed different than the March 2003 library. However, the March 2003 library only contained unique sequences compared with the October library. For FST tests carried out for cyanobacterial 16S, each month was found to be significantly different from each other month at the Bonferroni-adjusted α level of 0.05 (Table 2), although the type I error rate was substantially higher for the test of March 2002 versus October 2003 than for either of the others.
AMOVA and pairwise comparison of samples.
For both nifH and cyanobacterial 16S, nested AMOVA detected significant variation in genetic structure between different spots on the horizontal transects (Table 3). This was true whether permutations were confined to single vertical layers of the mat (in March and October 2003) or whether all vertical layers were considered together (data not shown). For cyanobacterial 16S, when comparisons were restricted to the same vertical layer, this “down-the-transect” variability constituted a substantial fraction (20 to 48%) of the total genetic variance of samples. In March and October 2003, both nifH and cyanobacterial 16S sequences also showed significant variation between different vertical layers of the mat (Table 3), with different species found in different mat layers (Fig. 1 to 3). For nested AMOVA in which permutations were restricted to samples within the same layer, there was significant variation in all layers of the mat between March and October 2003 (Table 3). A striking example of this can be seen by comparing upper mat communities from March and October (Fig. 3).
TABLE 3.
Nested AMOVA statistics showing variance between groups, percentage of total variance, and significance level
| Library | Category | Comparison | σ2ba | % of σ2Ta |
|---|---|---|---|---|
| nifH | Inshore/midshore/offshore | Month | 1.673*** | 4.06 |
| Upper/middle/lower | Month (2003 only) | 4.904*** | 11.25 | |
| March/October (2003) | Vertical layer | 5.803*** | 13.55 | |
| Cyanobacterial 16S | Inshore/midshore/offshore | Month | 1.533*** | 8.73 |
| Upper/middle/lower | Month (2003 only) | 3.184*** | 17.78 | |
| March/October (2003) | Vertical layer | 2.274*** | 12.62 |
Bonferroni-corrected significance levels. σb2, variance between groups; σT2, total variance; ***, α ≤ 0.001.
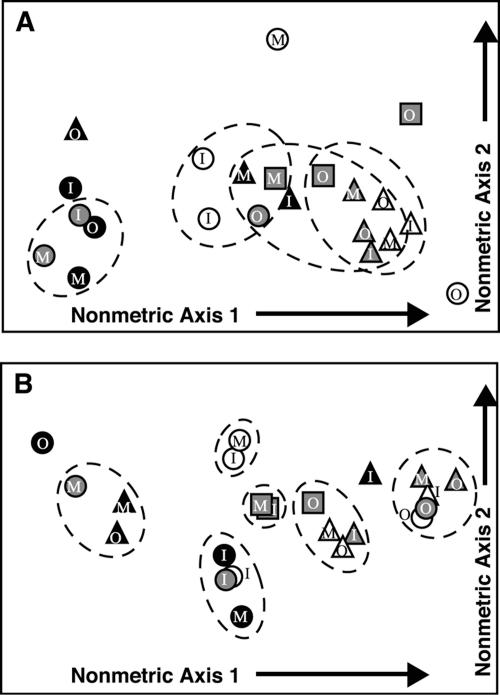
In nonmetric multidimensional scaling diagrams, March and October 2003 samples tended to plot on different sides of the graph, as did samples from the upper and lower mat (Fig. 5). For nifH (Fig. 5A), samples collected in March (both years) were statistically indistinguishable except for those samples collected offshore. In October, nifH communities from the middle and lower mat tended to be similar except for those samples collected offshore. Upper mat nifH communities in October tended to be different from each other (i.e., horizontal variation down transects) and from communities in different layers (Fig. 5A). Cyanobacterial communities in March 2003 (Fig. 5B) were significantly different, in each mat layer, between the inshore site and all other sites. In contrast, cyanobacterial communities in October 2003 showed significant differences between the offshore site and all other sites (Fig. 5B).
FIG. 5.
Nonmetric multidimensional scaling plot of genetic and community structure for (A) nifH and (B) cyanobacterial 16S. Distances between points are based upon the FST statistic for all sample pairs. Dashed circles group together all pairs of samples that are mutually indistinguishable by the FST test. Symbols: square, March 2002; triangle, March 2003; circle, October 2003; white, upper mat; gray, middle mat or whole mat core (for March 2002); black, lower mat; I, inshore; M, midshore; O, offshore.
DISCUSSION
Although species diversity in hypersaline microbial mats has been generally considered to be low (43), Chao1 estimates of minimal diversity in the Salt Pond mat indicated that this system may harbor as many as 200 and 115 distinct species of nitrogen fixers and cyanobacteria, respectively. Thus, the potential pool of participants in carbon and nitrogen sequestration may be quite large. While the sequencing effort in the present study was sufficient to characterize only a portion of this diversity, it is still possible to state some tentative conclusions regarding the composition of these two functional groups.
Most nifH sequences from the Salt Pond mat were related to cyanobacteria, most likely branching within the filamentous order Oscillatoriales, although a fair number of sequences related to single-celled cyanobacteria were also recovered (Fig. 1). Many of the closest relatives of these Salt Pond sequences were found in other hypersaline mat systems, particularly mats from Guerrero Negro in Baja California, Mexico (1, 27, 28). Thus, these systems harbor some of the same organisms. However, the relative proportions of sequences from these studies were not the same as those in the present study. For example, nifH libraries (DNA) from the Guerrero Negro mats were dominated by type III nifH genes instead of type I (28), and while reverse transcriptase PCR libraries from Guerrero Negro were dominated by type I nifH, most of these sequences were obtained from unicells instead of filamentous cyanobacteria (27). These discrepancies may result from differential biases introduced during clone library construction for those studies and the present one. However, they may also indicate that while these geographically separated mats share some of the same organisms, differences unique to each system allow different taxa to dominate. A more thorough investigation using quantitative methods will be necessary to distinguish these alternative hypotheses.
Cyanobacterial 16S sequences from the Salt Pond mat appeared to belong to orders previously identified in other mats by microscopy and by the analysis of diagnostic pigments (9, 23). Comparison of microscopically identified cyanobacteria and those found in the nifH and 16S libraries is hampered by the lack of reference strains closely related to the most common Salt Pond sequences (Fig. 1 to 4). However, it is noteworthy that cyanobacterial nifH sequences were dominated by a single lineage (Fig. 1), while multiple cyanobacterial 16S lineages were well represented (Fig. 3 and 4). This discrepancy suggests that several clades of nondiazotrophic cyanobacteria were prevalent in the Salt Pond mat.
Seasonal changes in community structure.
Consistent differences in community structure between March and October are likely to reflect the influences of environmental changes between the dry and wet seasons, the most prominent of these being salinity (Table 1). Analysis of clone libraries for nifH showed strong evidence that the composition of this functional group in the Salt Pond mat is different in March than in October (Tables 2 and 3 and Fig. 5A). October samples appeared to harbor more genetic diversity, and in particular, there was a greater diversity of noncyanobacterial diazotroph lineages in October (Fig. 1 and 2). Thus, freshening of the water appears to provide a favorable environment for certain transient microbial populations. Previous work in this system has shown that hypersalinity reduces the overall rate of nitrogen fixation in this mat (33, 34), and the lower rates detected in the dry season may be related to the absence of some of these lineages. Steppe and colleagues (47) demonstrated through reverse transcription studies of RNA that noncyanobacterial microbes can contribute more nitrogenase transcripts than cyanobacteria even when the latter dominate in biomass and numbers. Noncyanobacterial nitrogen fixation may be especially important in systems like Salt Pond, where nonheterocystous cyanobacterial predominate (32) and must reconcile oxygen-generating photosynthesis and oxygen-inhibited nitrogen fixation (31). Thus, a diverse community of diazotrophs, as seen in October, may show a higher overall rate of nitrogen fixation than one consisting almost entirely of cyanobacteria, as seen in March.
In contrast to nifH, cyanobacterial 16S libraries revealed no consistent seasonal differences. While March 2003 and October 2003 were clearly different (Tables 2 and 3), differences between March 2002 and October 2003 were marginally significant (FST test) (Table 2) or not significant (∫-LIBSHUFF). These results reflect the large number of sequences recovered from both March 2002 and October 2003 libraries but absent from the March 2003 library, with the most conspicuous example of this seen in a branch of the order Oscillatoriales related to the genera Phormidium and Leptolyngbya (Fig. 3). Given that March 2002 was the highest-salinity sampling period and October 2003 was the freshest, these results suggest that cyanobacterial taxonomic composition is not strongly influenced by salinity fluctuations in this system. This may indicate that all cyanobacterial clades present in the Salt Pond mat are well adapted to hypersaline conditions and are capable of maintaining viable populations during both wet and dry seasons. Studies of intertidal microbial mats have also shown that cyanobacteria appear to be more resilient to hypersaline and water-stressed conditions than other prokaryotes (38) and that factors other than salinity may be responsible for changes in dominant cyanobacterial taxa (35). However, hypersalinity is inhibitory to rates of primary productivity (33, 34). If hypersaline conditions adversely affect cyanobacterial activity but not taxonomic composition, this may position the mat microbes for a very rapid positive response to freshening events such as large storms, contributing to the overall resilience of the ecosystem.
Spatial changes in community structure.
There was significant (AMOVA) vertical and horizontal variation in the genetic structure of cyanobacterial and nitrogen fixer functional groups (Table 3). This heterogeneity may be produced by environmental gradients present in the mat at any given time. From top to bottom, laminated mats encompass extremely steep gradients in oxygen, pH, Eh, available solar radiation, and a variety of S, N, and C compounds (31, 32), and it is not surprising that microbial community composition changes to accommodate this variation at the microscale level (24). From inshore to offshore, the Salt Pond environment presents a variety of different environmental challenges that may affect community composition. For example, the annual expansion and contraction of the shoreline can be thought of as defining a desiccation gradient that would expose mat communities for increasingly longer periods of time as one moved further inshore. In addition to potential water stress, this exposure may also subject communities to more intense UV radiation and more extreme short-term temperature fluctuations. Finally, for submersed organisms, there may be a reduction in the level of UV radiation exposure as one moves deeper into the lake, allowing for microorganisms to position themselves accordingly (18, 22). Any of these factors may serve to structure communities from inshore to offshore.
A comparison of spatial and temporal variation (Fig. 5) offers some interesting suggestions about the ecology of this mat system. Of note is the fact that nonmetric multidimensional scaling plots place October and lower mat samples together at one end of the graph and March and upper mat samples together at the other end (Fig. 5). This configuration implies that the same organisms that distinguish October communities from March communities are those that distinguish lower mat from upper mat communities. For nifH, part of this pattern may be explained by the presence of more type III nifH in the October library and the fact that these sequences tended to be more common in lower mat samples (Fig. 2). However, as mat structure and components such as extracellular polymeric substances may serve to buffer microbes from exposure to hypersalinity (6, 37), there is another potential explanation for this pattern. Organisms that are poorly adapted to survive the osmotic stresses associated with hypersalinity may seek refuge in the lower mat layers during the dry season but may reposition themselves after sufficient freshening of the lake, causing October communities to look similar to lower mat communities. This pattern is exemplified by the type III nifH sequences, which tend to be seen in the bottom and middle layers of the mat when they occur in March 2003 but can be found all throughout the mat, including the top, in October 2003 (Fig. 2). Microbial mat organisms, particularly cyanobacteria and Beggiatoa, have been shown to make diel vertical migrations in response to changing mat conditions (10), and so it is not unreasonable to suspect that such migrations can occur over longer time scales. This may be an important strategy that allows some mat microbes to survive under otherwise harmful conditions.
Horizontal variation was not consistent for both genes, nor was the pattern of variation consistent in time. For nifH, there was considerably less down-the-transect variation in March compared with October (Fig. 5A). Thus, despite a spatially expanded desiccation gradient, the dry season appeared to homogenize the nitrogen-fixing community. The presence of October-like sequences in the lower offshore mat (Fig. 2 and 5A) suggests that this spot may serve as a refuge for nitrogen fixers susceptible to the environmental conditions prevalent elsewhere in the mat. In October, the nitrogen fixer community structure was more heterogeneous, especially in the upper mat (Fig. 5A). This heterogeneity suggests the presence of different microenvironments in the wet-season mat that allowed for the development of diverse mixtures of nitrogen-fixing organisms at different spots in the mat and maintained a high overall diversity of nitrogen fixers throughout the whole mat. Given that the whole mat was submersed at this time (Table 1), it is somewhat counterintuitive that October would demonstrate such high heterogeneity. A likely explanation for this is that freshened conditions relieve environmental stress, provide for higher overall microbial activity levels (33, 34, 42), and allow numerous microenvironments to develop (7).
For cyanobacteria, spatial variation was always apparent (Fig. 5B). The high percentage of genetic variance, 20% to 48% for some comparisons, attributable to transect variability (AMOVA) implies that different cyanobacterial lineages were found in different locations on the mat. From a seasonal perspective, however, the major patterns of variation were reversed in the dry and wet seasons. In March 2003, the inshore samples were different from the other samples (Fig. 5B). Because the inshore site at this time was located above the waterline (Table 1), this suggests that cyanobacterial community structure may be sensitive to aerial exposure. In October, the offshore samples are different from other samples in each layer. This observation is consistent with data showing that the proportions of diagnostic photosynthetic pigments in the Salt Pond mat change as the water depth increases (J. L. Pinckney, personal communication). Under increasing water cover, cyanobacterial communities may be responding to the changes in the quantity and spectral composition of light able to penetrate. If cyanobacterial community structure and activity are principal foundations of microenvironmental heterogeneity in the mat (7, 32), this higher-scale heterogeneity may have important consequences for other mat organisms.
Concluding remarks: ecological implications of mat community dynamics.
Previous work in this system has shown that rates of both primary productivity and nitrogen fixation are greatly reduced under hypersaline conditions (33, 34), which is consistent with observations for other microbial processes such as methanogenesis, sulfate reduction, and anoxygenic photosynthesis (42). Differential seasonal responses of nitrogen fixers and cyanobacteria suggest that the ecological mechanisms underlying this loss of function may be different. Nitrogen fixer community structure and diversity were significantly altered between dry-and wet-season months, so the noted reduction of nitrogen fixation may be related to changes in community structure and cell-specific activity levels. In contrast, since cyanobacterial community structure showed no consistent response to salinity changes, the reduction in primary productivity might be largely due to reduced activity at the cellular level. If so, cyanobacteria should be able to respond rapidly to freshening events. Since cyanobacterial nifH sequences tend to dominate dry-season libraries, these stimulated cyanobacterial communities would be capable of supplying new nitrogen to the mat system. As microbial activity creates a variety of microhabitats (7, 32), and as other organisms begin to respond to the freshened environment, a diverse community of potential nitrogen fixers appears to develop. This increased diversity may help to buffer the system against periodic disturbances such as the tropical storms that are more common in San Salvador at this time of year. The shift from low-diversity to high-diversity functional groups should afford an excellent opportunity to explore the relationship of microbial community composition and microbial function in this system. In addition, the changing microbial composition of the mat community may provide a useful model for exploring ecological successions over a variety of time scales (1, 35, 49).
It is noteworthy that many of the closest database relatives to the organisms of the present study were from other hypersaline systems, especially other mats, or from systems, like Antarctica, where water stress may be an overriding environmental constraint (Fig. 1 to 4). This may be an artifact of the current databases, particularly in the case of nifH, since an inordinately large proportion of currently available nifH sequences come from hypersaline, laminated mats (50). However, this also suggests that there may be a diversity of clades that are widely distributed and well adapted to life in hypersaline or water-stressed environments (26, 27, 52). If so, this diversity presents an enormous opportunity to study the physiological and ecological responses of organisms to water stress. These systems also afford the opportunity to test whether the patterns observed in the Salt Pond mat are of a general nature.
Acknowledgments
This work was funded by NSF Microbial Observatories Program project MCB-0132528.
We gratefully acknowledge V. Vogeli and the staff at the Gerace Research Center, San Salvador, Bahamas, for invaluable support during this project. Assistance in the field and in the laboratory was provided by B. Chang, N. Hall, R. Weaver, T. Gallo, P. Moisander, M. Leonard, and L. Cheshire. We are grateful to J. Zehr for graciously providing access to his database of nifH sequences and to A. Kent and K. McMahon for helpful suggestions during the preparation of the manuscript.
REFERENCES
- 1.Bebout, B. M., S. P. Carpenter, D. J. Des Marais, M. Discipulo, T. Embaye, F. Garcia-Pichel, T. M. Hoehler, M. Hogan, L. L. Jahnke, R. M. Keller, S. R. Miller, L. E. Prufert-Bebout, C. Raleigh, M. Rothrock, and K. Turk. 2002. Long-term manipulations of intact microbial mat communities in a greenhouse collaboratory: simulating Earth's present and past field environments. Astrobiology 2:383-402. [DOI] [PubMed] [Google Scholar]
- 2.Chao, A. 1984. Nonparametric-estimation of the number of classes in a population. Scand. J. Statistics 11:265-270. [Google Scholar]
- 3.Chao, A., M. C. Ma, and M. C. K. Yang. 1993. Stopping rules and estimation for recapture debugging with unequal failure rates. Biometrika 80:193-201. [Google Scholar]
- 4.Cohen, Y., and E. Rosenberg (ed.). 1989. Microbial mats: physiological ecology of benthic microbial mats. American Society for Microbiology, Washington, D.C.
- 5.Decho, A. W., and T. Kawaguchi. 1999. Confocal imaging of in situ natural microbial communities and their extracellular polymeric secretions using Nanoplast resin. BioTechniques 27:1246-1252. [PubMed] [Google Scholar]
- 6.Decho, A. W., P. T. Visscher, and R. P. Reid. 2005. Production and cycling of natural microbial exopolymers (EPS) within a marine stromatolite. Palaeogeogr. Palaeoclimatol. Palaeoecol. 219:71-86. [Google Scholar]
- 7.de Lomas, J. G., A. Corzo, C. M. Garcia, and S. A. van Bergeijk. 2005. Microbenthos in a hypersaline tidal lagoon: factors affecting microhabitat, community structure and mass exchange at the sediment-water interface. Aquat. Microb. Ecol. 38:53-69. [Google Scholar]
- 8.Excoffier, L., P. E. Smouse, and J. M. Quattro. 1992. Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial-DNA restriction data. Genetics 131:479-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fourans, A., T. G. de Oteyza, A. Wieland, A. Sole, E. Diestra, J. van Bleijswijk, J. O. Grimalt, M. Kühl, I. Esteve, G. Muyzer, P. Caumette, and R. Duran. 2004. Characterization of functional bacterial groups in a hypersaline microbial mat community (Salins-de-Giraud, Camargue, France). FEMS Microbiol. Ecol. 51:55-70. [DOI] [PubMed] [Google Scholar]
- 10.Garcia-Pichel, F., M. Mechling, and R. W. Castenholz. 1994. Diel migrations of microorganisms within a benthic, hypersaline mat community. Appl. Environ. Microbiol. 60:1500-1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoehler, T. M., B. M. Bebout, and D. J. Des Marais. 2001. The role of microbial mats in the production of reduced gases on the early Earth. Nature 412:324-327. [DOI] [PubMed] [Google Scholar]
- 12.Hugenholtz, P. 2002. Exploring prokaryotic diversity in the genomic era. Genome Biol. 3:1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jin, L., and M. Nei. 1990. Limitations of the evolutionary parsimony method of phylogenetic analysis. Mol. Biol. Evol. 7:82-102. [DOI] [PubMed] [Google Scholar]
- 14.Jonkers, H. M., R. Ludwig, R. De Wit, O. Pringault, G. Muyzer, H. Niemann, N. Finke, and D. De Beer. 2003. Structural and functional analysis of a microbial mat ecosystem from a unique permanent hypersaline inland lake: ‘La Salada de Chiprana’ (NE Spain). FEMS Microbiol. Ecol. 44:175-189. [DOI] [PubMed] [Google Scholar]
- 15.Kawaguchi, T., and A. W. Decho. 2000. Biochemical characterization of cyanobacterial extracellular polymers (EPS) from modern marine stromatolites (Bahamas). Prep. Biochem. Biotechnol. 30:321-330. [DOI] [PubMed] [Google Scholar]
- 16.Kimura, M. 1980. A simple method for estimating evolutionary rate of base substitution through comparative studies of nucleotide sequences. J. Mol. Evol. 16:111-120. [DOI] [PubMed] [Google Scholar]
- 17.Krumbein, W. E., A. A. Gorbushina, and E. Holtkamp-Tacken. 2004. Hypersaline microbial systems of Sabkhas: examples of life's survival in “extreme”' conditions. Astrobiology 4:450-459. [DOI] [PubMed] [Google Scholar]
- 18.Kruschel, C., and R. W. Castenholz. 1998. The effect of solar UV and visible irradiance on the vertical movements of cyanobacteria in microbial mats of hypersaline waters. FEMS Microbiol. Ecol. 27:53-72. [Google Scholar]
- 19.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 20.Martin, A. P. 2002. Phylogenetic approaches for describing and comparing the diversity of microbial communities. Appl. Environ. Microbiol. 68:3673-3682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Monty, C. 1976. The origin and development of cryptoalgal fabrics, p. 193-249. In M. Walter (ed.), Stromatolites. Elsevier, New York, N.Y.
- 22.Nadeau, T. L., C. Howard-Williams, and R. W. Castenholz. 1999. Effects of solar UV and visible irradiance on photosynthesis and vertical migration of Oscillatoria sp. (cyanobacteria) in an Antarctic microbial mat. Aquat. Microb. Ecol. 20:231-243. [Google Scholar]
- 23.Nübel, U., F. Garcia-Pichel, M. Kühl, and G. Muyzer. 1999. Quantifying microbial diversity: morphotypes, 16S rRNA genes, and carotenoids of oxygenic phototrophs in microbial mats. Appl. Environ. Microbiol. 65:422-430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nübel, U., F. Garcia-Pichel, M. Kühl, and G. Muyzer. 1999. Spatial scale and the diversity of benthic cyanobacteria and diatoms in a salina. Hydrobiologia 401:199-206. [Google Scholar]
- 25.Nübel, U., F. Garcia-Pichel, and G. Muyzer. 1997. PCR primers to amplify 16S rRNA genes from cyanobacteria. Appl. Environ. Microbiol. 63:3327-3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Olson, J. B., T. F. Steppe, R. W. Litaker, and H. W. Paerl. 1998. N-2-fixing microbial consortia associated with the ice cover of Lake Bonney, Antarctica. Microb. Ecol. 36:231-238. [DOI] [PubMed] [Google Scholar]
- 27.Omoregie, E. O., L. L. Crumbliss, B. M. Bebout, and J. P. Zehr. 2004. Comparison of diazotroph community structure in Lyngbya sp. and Microcoleus chthonoplastes dominated microbial mats from Guerrero Negro, Baja, Mexico. FEMS Microbiol. Ecol. 47:305-318. [DOI] [PubMed] [Google Scholar]
- 28.Omoregie, E. O., L. L. Crumbliss, B. M. Bebout, and J. P. Zehr. 2004. Determination of nitrogen-fixing phylotypes in Lyngbya sp. and Microcoleus chthonoplastes cyanobacterial mats from Guerrero Negro, Baja California, Mexico. Appl. Environ. Microbiol. 70:2119-2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Paerl, H. W., J. Dyble, P. H. Moisander, R. T. Noble, M. F. Piehler, J. L. Pinckney, T. F. Steppe, L. Twomey, and L. M. Valdes. 2003. Microbial indicators of aquatic ecosystem change: current applications to eutrophication studies. FEMS Microbiol. Ecol. 46:233-246. [DOI] [PubMed] [Google Scholar]
- 30.Paerl, H. W., S. B. Joye, and M. Fitzpatrick. 1993. Evaluation of nutrient limitation of CO2 and N2 fixation in marine microbial mats. Mar. Ecol. Prog. Ser. 101:297-306. [Google Scholar]
- 31.Paerl, H. W., and J. L. Pinckney. 1996. A mini-review of microbial consortia: their roles in aquatic production and biogeochemical cycling. Microb. Ecol. 31:225-247. [DOI] [PubMed] [Google Scholar]
- 32.Paerl, H. W., J. L. Pinckney, and T. F. Steppe. 2000. Cyanobacterial-bacterial mat consortia: examining the functional unit of microbial survival and growth in extreme environments. Environ. Microbiol. 2:11-26. [DOI] [PubMed] [Google Scholar]
- 33.Paerl, H. W., T. F. Steppe, K. C. Buchan, and M. Potts. 2003. Hypersaline cyanobacterial mats as indicators of elevated tropical hurricane activity and associated climate change. Ambio 32:87-90. [DOI] [PubMed] [Google Scholar]
- 34.Pinckney, J., H. W. Paerl, and B. M. Bebout. 1995. Salinity control of benthic microbial mat community production in a Bahamian hypersaline lagoon. J. Exp. Mar. Biol. Ecol. 187:223-237. [Google Scholar]
- 35.Pinckney, J., H. W. Paerl, and M. Fitzpatrick. 1995. Impacts of seasonality and nutrients on microbial mat community structure and function. Mar. Ecol. Prog. Ser. 123:207-216. [Google Scholar]
- 36.Pinckney, J. L., and H. W. Paerl. 1997. Anoxygenic photosynthesis and nitrogen fixation by a microbial mat community in a Bahamian hypersaline lagoon. Appl. Environ. Microbiol. 63:420-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Potts, M. 1999. Mechanisms of desiccation tolerance in cyanobacteria. Eur. J. Phycol. 34:319-328. [Google Scholar]
- 38.Rothrock, M. J., and F. Garcia-Pichel. 2005. Microbial diversity of benthic mats along a tidal desiccation gradient. Environ. Microbiol. 7:593-601. [DOI] [PubMed] [Google Scholar]
- 39.Schloss, P. D., B. R. Larget, and J. Handelsman. 2004. Integration of microbial ecology and statistics: a test to compare gene libraries. Appl. Environ. Microbiol. 70:5485-5492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shaklee, R. V. 1996. Weather and climate of San Salvador Island, Bahamas. Bahamian Field Station Publications, San Salvador, Bahamas.
- 41.Singleton, D. R., M. A. Furlong, S. L. Rathbun, and W. B. Whitman. 2001. Quantitative comparisons of 16S rRNA gene sequence libraries from environmental samples. Appl. Environ. Microbiol. 67:4374-4376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sorensen, K. B., D. E. Canfield, and A. Oren. 2004. Salinity responses of benthic microbial communities in a solar saltern (Eilat, Israel). Appl. Environ. Microbiol. 70:1608-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stal, L. J. 1994. Microbial mats in coastal environments, p. 21-23. In L. J. Stal and P. Caumette (ed.), Microbial mats: structure, development, and environmental significance, vol. G. Springer-Verlag, Berlin, Germany. [Google Scholar]
- 44.Stal, L. J., and P. Caumette (ed.). 1994. Microbial mats: structure, development, and environmental significance, vol. G. Springer-Verlag, Berlin, Germany.
- 45.Stal, L. J., S. Grossberger, and W. E. Krumbein. 1984. Nitrogen-fixation associated with the cyanobacterial mat of a marine laminated microbial ecosystem. Mar. Biol. 82:217-224. [Google Scholar]
- 46.Steppe, T. F., J. B. Olson, H. W. Paerl, R. W. Litaker, and J. Belnap. 1996. Consortial N2 fixation: a strategy for meeting nitrogen requirements of marine and terrestrial cyanobacterial mats. FEMS Microbiol. Ecol. 21:149-156. [Google Scholar]
- 47.Steppe, T. F., and H. W. Paerl. 2002. Potential N-2 fixation by sulfate-reducing bacteria in a marine intertidal microbial mat. Aquat. Microb. Ecol. 28:1-12. [Google Scholar]
- 48.Steppe, T. F., J. L. Pinckney, J. Dyble, and H. W. Paerl. 2001. Diazotrophy in modern marine Bahamian stromatolites. Microb. Ecol. 41:36-44. [DOI] [PubMed] [Google Scholar]
- 49.Yannarell, A. C., A. D. Kent, G. L. Lauster, T. K. Kratz, and E. W. Triplett. 2004. Temporal patterns in bacterial communities in three temperate lakes of different trophic status. Microb. Ecol. 46:391-405. [DOI] [PubMed] [Google Scholar]
- 50.Zehr, J. P., B. D. Jenkins, S. M. Short, and G. F. Steward. 2003. Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5:539-554. [DOI] [PubMed] [Google Scholar]
- 51.Zehr, J. P., and L. A. McReynolds. 1989. Use of degenerate oligonucleotides for amplification of the nifH gene from the marine cyanobacterium Trichodesmium thiebautii. Appl. Environ. Microbiol. 55:2522-2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zehr, J. P., M. Mellon, S. Braun, W. Litaker, T. Steppe, and H. W. Paerl. 1995. Diversity of heterotrophic nitrogen fixation genes in a marine cyanobacterial mat. Appl. Environ. Microbiol. 61:2527-2532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zehr, J. P., M. T. Mellon, and W. D. Hiorns. 1997. Phylogeny of cyanobacterial nifH genes: evolutionary implications and potential applications to natural assemblages. Microbiology 143:1443-1450. [DOI] [PubMed] [Google Scholar]