Abstract
Xenorhabdus and Photorhabdus are gram-negative bacteria that produce a range of proteins that are toxic to insects. We recently identified a novel 42-kDa protein from Xenorhabdus nematophila that was lethal to the larvae of insects such as Galleria mellonella and Helicoverpa armigera when it was injected at doses of 30 to 40 ng/g larvae. In the present work, the toxin gene txp40 was identified in another 59 strains of Xenorhabdus and Photorhabdus, indicating that it is both highly conserved and widespread among these bacteria. Recombinant toxin protein was shown to be active against a variety of insect species by direct injection into the larvae of the lepidopteran species G. mellonella, H. armigera, and Plodia interpunctella and the dipteran species Lucilia cuprina. The protein exhibited significant cytotoxicity against two dipteran cell lines and two lepidopteran cell lines but not against a mammalian cell line. Histological data from H. armigera larvae into which the toxin was injected suggested that the primary site of action of the toxin is the midgut, although some damage to the fat body was also observed.
Xenorhabdus and Photorhabdus are gram-negative bacteria that are highly pathogenic to insects (10). These bacteria live in symbiosis with rhabditoid nematodes belonging to the genera Steinernema and Heterorhabditis. The nematodes and bacteria share a complex life cycle which involves both symbiotic and pathogenic stages. After nematode infection of an insect host, the nematodes release the bacteria into the insect hemocoel, where the bacteria grow to the stationary phase. The bacteria and nematodes work together to kill the insect host, although in most cases the bacteria alone are highly virulent once they are circulating in the insect hemocoel (16). The final stage of development is the reassociation of the bacteria and nematodes to form nonfeeding infective juveniles, which emerge from the insect carcass to find new hosts.
The bacterium-nematode associations have a wide insect host range (23) and are used for biological control of some lepidopteran, dipteran, and coleopteran pests of commercial crops (10). The susceptibility of a particular insect host to a nematode-bacterium association depends on a number of factors, including the species of bacteria or nematodes, the ease of finding and entering the insect host, the stage of the insect in the life cycle, the response of the insect immune system, and other biochemical and physiological responses to the various metabolites produced by the nematodes and bacteria (2). In particular, both the bacteria and the nematodes produce a range of toxins that are responsible for killing the insect host (2). Analysis of the genome of Photorhabdus luminescens resulted in identification of more putative toxin genes than have been found in any other bacterium sequenced to date (15). The only toxins that have been studied in detail from Xenorhabdus and Photorhabdus bacteria are the Tc toxins from P. luminescens strain W14 (7, 8, 17). The Tc toxins form a large protein complex consisting of about 10 polypeptides ranging in size from 30 to 200 kDa that is toxic to insects after either ingestion or injection (7, 8). Some work has also been performed with a 39-kDa toxin from Xenorhabdus nematophila (29), the large Xin toxin from X. nematophila strain BJ (27), and the 17-kDa pilin subunit from X. nematophila (20). We recently described a novel toxin, A24tox, from X. nematophila strain A24. This 42-kDa toxin is a secreted protein that is lethal to the larvae of insects such as Galleria mellonella and Helicoverpa armigera when it is injected at doses of 30 to 40 ng/g larvae (9).
The details of the modes of action of the various toxins of Xenorhabdus and Photorhabdus are still not known. Injection of the Tca complex of P. luminescens strain W14 or ingestion by Manduca sexta larvae damaged the midgut cells, resulting in shedding of the midgut epithelium into the gut lumen, followed by lysis of the epithelium (5). Injection of A24tox into lepidopteran larvae caused the larvae to cease feeding almost immediately, and preliminary histological studies indicated that the main site of action for the toxin was the insect midgut (9). Many other insecticidal toxins are known to act on the insect gut, including the well studied δ-endotoxins from Bacillus thuringiensis and cholesterol oxidase from Streptomyces (5). In this study we used histology and cytotoxicity assays to obtain further insight into the mode of action of the novel insecticidal toxin A24tox. We also investigated the occurrence of genes closely related to the A24tox gene in 59 strains of Xenorhabdus and Photorhabdus bacteria that encompass most of the genetic diversity currently known for these two genera.
MATERIALS AND METHODS
Isolation of the txp40 toxin gene from P. luminescens strain V16.
High-molecular-weight genomic DNA was isolated from P. luminescens strain V16 (31). The DNA was partially digested with Sau3AI and used to create a cosmid library, as described previously (9). This library was screened by hybridization using the previously discovered A24tox gene from X. nematophila strain A24 as a probe (9). Two hundred cosmid clones were grown overnight at 37°C on Luria-Bertani (LB) medium-ampicillin plates, transferred to nylon membrane disks (Colony/Plaque Screen; NEN DuPont), lysed in situ by treatment with 0.5 N NaOH, and neutralized with 1.0 M Tris-Cl (pH 7.5). After air drying, the filters were prehybridized in a solution consisting of 5× SSPE (1× SSPE is 180 mM NaCl, 10 mM sodium phosphate [pH 7], and 1 mM EDTA) containing 0.2% (wt/vol) skim milk powder, 0.5% (wt/vol) sodium dodecyl sulfate (SDS), and 0.2 mg/ml denatured salmon sperm DNA at 68°C for 3 h. A hybridization probe was prepared by radiolabeling approximately 100 ng of isolated A24tox gene with 50 μCi [α-32P]dATP by random primed synthesis using a Gigaprime DNA labeling kit (GPK-1; Bresatec). Filters were incubated with the A24tox probe in the same buffer that was used for the prehybridization step at 68°C overnight. The filters were rinsed briefly in 2× SSC (1× SSC is 150 mM NaCl plus 15 mM sodium citrate, pH 7) and washed once for 15 min at room temperature in 2× SSC containing 0.1% (wt/vol) SDS and once at 68°C for 30 min in 0.5× SSC containing 0.2% SDS. After a final rinse in 0.5× SSC the filters were autoradiographed for 24 h at −80°C. For each clone that hybridized with the A24tox probe, cultures were grown and the cell lysates were assayed for toxicity using the G. mellonella injection bioassay (9). Clones of interest were analyzed by further restriction enzyme mapping (NotI, EcoRI, HindIII, EcoRV, and SmaI), cloned into the plasmid vector pBluescipt II (KS)+ (Stratagene), transformed into Escherichia coli strain DH10B (Stratagene), and tested with the G. mellonella injection bioassay. As described previously for the A24tox gene (9), both strands of the smallest deletion clone that retained insecticidal activity, the DNA immediately surrounding the toxin gene, and the DNA at the 5′ and 3′ ends of the cosmid clones and the intermediate deletion clones were sequenced using a combination of vector- and gene-specific primers. Similarity searches of the nonredundant nucleotide and protein databases (April 2005) were performed using the various BLAST programs available via NCBI (http://www.ncbi.nlm.nih.gov/BLAST/). Analysis of the P. luminescens subsp. laumondii strain TT01 genome was performed using the utilities available at PhotoList (http://genolist.pasteur.fr/PhotoList/index.html; release date, 25 September 2003). Similarity searches were also performed against the partially completed genome sequences of Xenorhabdus bovienii and X. nematophila strain ATCC 19061 (http://xenorhabdus.danforthcenter.org/; release dates, 8 November 2004 and 21 March 2005, respectively), as well as Photorhabdus asymbiotica (http://www.sanger.ac.uk/Projects/P_asymbiotica/; release date, 14 March 2005).
Identification of the txp40 toxin gene in other Xenorhabdus and Photorhabdus strains.
Forty-seven Xenorhabdus strains and 17 Photorhabdus strains were tested for the occurrence of the toxin gene by PCR and Southern blot analysis. The strains were streaked from glycerol stocks onto LB agar plates and grown for 2 to 3 days at 28°C. Two-milliliter portions of LB medium were then inoculated with single colonies, and the cultures were grown overnight at 28°C and 200 rpm. The genomic DNA was isolated from all strains using the protocol for gram-negative bacteria of a DNeasy kit (QIAGEN). The concentration of DNA was determined using genomic DNA samples (200 μl) and the DNA-binding dye Hoechst 33258 (fluorescent DNA quantitation kit; Bio-Rad) in a 96-well plate (Maxisorp; Nunc). The fluorescence was measured by using a POLARstar fluorescence plate reader (BMG Labtechnologies) with an excitation wavelength of 355 nm and an emission wavelength of 460 nm. The genomic DNA was analyzed using a combination of PCR and low-stringency Southern blot analysis. For Xenorhabdus strains, the A24tox clone originally constructed from X. nematophila strain A24 (9) was used to design two pairs of primers. The first pair of primers was designed to anneal to the 5′ and 3′ untranslated regions of the A24tox gene (AC15F [5′-ATTAGCACCTCAATTTTCCGG], 141 bp upstream of the A24tox gene start codon; A24PE3 [5′-CTGAGCGGTCACCTCT], 72 bp downstream of the A24tox gene stop codon). If a PCR product was not observed with these primers, then the PCR was repeated using a second pair of primers that were internal to the A24tox gene (ToxF4 [5′-AGAGTAACGCCTGATGATAA], 54 bp downstream of the A24tox gene start codon; AC1 [5′-CCTGTGTTCTTGGCTTAGTC], 66 bp upstream of the A24tox gene stop codon). For Photorhabdus strains, the primers were designed to anneal to the start codon (V16AC9F [5′-ATGGTTATACAATTAACACCTG] or V16AC15F [5′-AAGCTTATGGTTATACAATTAACACCTG]) and to the 3′ untranslated region of the toxin gene from P. luminescens strain V16 (V16AC1 [5′-GGTAGTAAAGTGTATTGGC], 127 bp downstream of the stop codon). The PCR was performed using 100 ng of bacterial genomic DNA, 50 pmol of each of the appropriate primers, Taq DNA polymerase (Gibco), and the PCR mixture supplied with the polymerase. The thermal cycler was programmed as follows: 94°C for 3 min; 94°C for 30 s, 50°C for 30 s, and 72°C for 2 min for 33 cycles; 72°C for 10 min; and 25°C for 1 min. The PCR products were analyzed by agarose gel electrophoresis.
For the Xenorhabdus strains for which no PCR product was detected with either set of primers, the genomic DNA was analyzed by Southern blotting under low-hybridization-stringency conditions, using the A24tox gene as the probe. The genomic DNA was digested with EcoRI, fractionated by agarose gel electrophoresis, and transferred to Hybond N+ nylon membranes (Amersham) using the Southern blot method (25). Hybridization was carried out as described above, except that the prehybridization solution contained 5× SSPE, 5× Denhardt's solution (1× Denhardt's solution is 0.2 mg/ml Ficoll 400, 0.2 mg/ml polyvinylpyrrolidone, and 0.2 mg/ml bovine serum albumin), 0.5% (wt/vol) SDS, and 0.2 mg/ml denatured herring sperm DNA, overnight incubation was performed at 55°C, and washing was performed three times at 55°C in 2× SSC containing 0.1% (wt/vol) SDS.
Txp40 toxin sequencing.
PCR products from 17 Xenorhabdus strains (X. nematophila strains A24, All, AllD, AN6, BK, Cb, F1, and Mex; X. bovienii strains F5, F7, T319, and T363; X. japonica strain Kushidai; and Xenorhabdus sp. strains C8401, CB-G, CB-W, and SaV) and four Photorhabdus strains (P. luminescens strain HI, P. luminescens subsp. luminescens strain V16, P. luminescens subsp. akhurstii strain Tetuan, and Photorhabdus sp. strain Q621) that showed the presence of a toxin gene were cloned into the pGEM-T Easy vector (Promega) and sequenced using vector primers T7 and SP6 and primers specific to the A24tox gene. Note that the sequences for Xenorhabdus strains Kushidai, C8401, Cb, CB-G, CB-W, F5, F7, SaV, and T319 were not full length as the sequences were determined using primers internal to the toxin gene. DNA sequencing was performed using an ABI Prism dideoxy dye terminator sequencing mixture (Applied Biosystems) with an automated DNA sequencer (Applied Biosystems model 377) according to the manufacturer's instructions. The translated protein sequences were aligned using the program ClustalW (34). The gene was designated txp40 (toxin from Xenorhabdus and Photorhabdus, 40 kDa), and the gene product was designated Txp40. Thus, the new designations of the A24tox protein from X. nematophila strain A24 and the V16tox protein from P. luminescens strain V16 are Txp40A24 and Txp40V16, respectively. Nucleotide and amino acid sequence similarities were determined using the GCG program GAP (26).
Purification and characterization of recombinant Txp40 toxin.
Recombinant proteins corresponding to the toxins from X. nematophila strains A24 and Mex, P. luminescens strain V16, and Photorhabdus sp. strain HI were prepared using the IMPACT system (New England Biolabs), as described previously (9). The IMPACT system expresses the recombinant protein as a self-splicing intein fused to a chitin binding domain, where the fusion partner is cleaved during the purification process, resulting in a recombinant protein consisting of the toxin sequence only. The identities of the recombinant proteins were confirmed by whole-protein and tryptic digest mass spectroscopy as described previously (9). Detailed bioinformatic analyses of the Txp40V16 toxin, including similarity searches and motif and fold recognition, were also performed as described previously (9). Recombinant toxin was used in the insect bioassays, the cytotoxicity assays, and the histological experiments described below; E. coli maltose binding protein (MBP) prepared in the same manner with the IMPACT system was used as a control.
A six-histidine-tagged version of the toxin from P. luminescens strain V16 (His6V16tox) was also purified. The EcoRV/SmaI restriction fragment of the active cosmid clone from P. luminescens strain V16 was cloned into the N-terminal six-histidine-tag vector pQE-31 (QIAGEN) and transformed into E. coli XL1-Blue cells (Stratagene) by electroporation. The cells were grown, harvested, and lysed by sonication using standard methods, and the cell extract was purified on an Ni-nitrilotriacetic acid agarose resin (QIAGEN) column. The protein was further purified by visualization of fractions by SDS-polyacrylamide gel electrophoresis (PAGE) using zinc stain (Bio-Rad), excision of the toxin bands from the gel, and electroelution using an Electroeluter 422 (Bio-Rad). The purified protein was analyzed by SDS-PAGE and Western blotting (9), using an Ni-nitrilotriacetic acid horseradish peroxidase conjugate (QIAGEN) and the substrate 3-amino-9-ethyl-carbazole. The final concentration of toxin was estimated by comparison to a bovine serum albumin standard using SDS-PAGE. The identity of the toxin was confirmed by tryptic digest mass spectroscopy as described previously (9). The His6V16tox protein was used for preparation of toxin antiserum (9).
Insect bioassays.
Insect rearing and bioassays with G. mellonella, H. armigera, and Lucilia cuprina were performed as described previously (9). Plodia interpunctella was reared at 25°C on an artificial diet containing 10 parts wheat bran, 2 parts wheat germ, 1 part yeast, and 2 parts glycerol. The activities of a recombinant protein corresponding to the toxin from P. luminescens strain V16 (Txp40V16) were determined with G. mellonella, H. armigera, and L. cuprina; these analyses included determination of the 50% lethal doses (LD50) for G. mellonella and H. armigera. The activities of recombinant proteins corresponding to the toxins from X. nematophila strain Mex and P. luminescens strain HI were determined using the G. mellonella injection bioassay with doses of 10, 20, and 200 ng for Txp40Mex and 50 and 500 ng for Txp40HI. Recombinant Txp40V16 was also assayed with P. interpunctella by using a dose of 100 ng.
Cytotoxicity assay.
A CyQUANT cell proliferation assay kit (Molecular Probes) was used to test the cytotoxicity of recombinant Txp40A24 protein against in vitro-cultured cell lines Sf9 and Sf21 (Spodoptera frugiperda), Schneider line 2 (S2) (Drosophila melanogaster), Aedes aegypti, and SP2/0 (mouse myeloma). Sf9 and Sf21 cells were cultured in Grace's medium at 28°C, S2 and A. aegypti cells were cultured in Schneider's medium at 28°C, and SP2/0 cells were cultured in Dulbecco's modified Eagle's medium at 37°C. The cells were cultured using standard methods and were counted using a hemocytometer. A 96-well tissue culture plate was prepared with 1 × 104 cells per well (190 μl) and with each sample in quadruplicate. Stocks of Txp40A24 and MBP were filtered through a 0.4-μm membrane (Millipore), diluted in phosphate-buffered saline (pH 7.4), and added to the wells (10 μl) to obtain final protein concentrations ranging from 2,000 ng/ml to 0.002 ng/ml. The plate was incubated overnight at 28°C (Sf21, S2, and A. aegypti) or 37°C (SP2/0) without shaking or at 28°C and 200 rpm (Sf9). The plate was centrifuged (200 × g, 10 min), the pellets were washed once with phosphate-buffered saline, and the plate was frozen at −70°C for at least 1 h or overnight. After the plate was thawed, fluorescent CyQUANT dye (400-fold dilution, 200 μl) was added to each well to allow quantitation of the nucleic acid released by cell lysis in each well. A standard curve was constructed for each cell line by setting up a dilution series consisting of 0 to 6 × 104 or 8 × 104 cells. The fluorescence was measured by using a POLARstar fluorescence plate reader with an excitation wavelength of 485 nm and an emission wavelength of 520 nm. For all cell lines the standard curves (fluorescence intensity versus cell number) were linear in the range from 0 to 8 × 104 cells, indicating that there was a direct correlation between the measured fluorescence and the number of cells in a well. The Txp40A24 and MBP results were compared by the analysis of variance method using the program Statview SE, version 1.04 (Abacus Concepts).
Txp40 toxin histopathology in H. armigera.
Fourth-instar H. armigera larvae were injected with 10 μl of recombinant Txp40A24 toxin (10 ng or 100 ng) or MBP (100 ng). After 0, 6, 12, 18, 24, 30, 40, and 48 h, fixative (4% paraformaldehyde in 100 mM phosphate buffer [pH approximately 7.2]) was injected into three larvae, holes were punctured in the cuticle, and the larvae were transferred to fixative and left overnight at 4°C. The larvae were embedded in paraffin wax and cut into 6-μm sections, and the sections were stained with hematoxylin and eosin. Images were captured with a ProgRes 3012 digital camera on a Leica Diaplan microscope. For peritrophic matrix visualization, mid-third-instar H. armigera larvae were inoculated as described above, and the embedded larvae were cut into 6-μm longitudinal sections. For imaging the sections were cleared, hydrated, treated with citric acid (2.1 g/liter, pH 6.1) at 90°C for 20 min, and allowed to cool before a 5-min wash in distilled water and mounting in 70% glycerol. Autofluorescence images were captured with a Leica TCS SP2 confocal laser scanning microscope using an excitation wavelength of 488 nm.
Nucleotide sequence accession numbers.
A subset of the sequences determined has been deposited in the GenBank database under accession numbers DQ242618 to DQ242629.
RESULTS
Identification and sequencing of txp40 toxin gene.
We used the recently identified toxin gene txp40A24 (previously designated A24tox) (9) to identify a related toxin in a cosmid library of the bacterium P. luminescens strain V16. The Txp40V16 toxin was 83% and 75% identical to Txp40A24 at the nucleotide and protein levels, respectively. It also exhibited 98% identity to the hypothetical plu2326 protein encoded in the genome sequence of P. luminescens subsp. laumondii strain TT01. Further bioinformatic analyses of the Txp40V16 protein, including motif recognition or fold recognition, resulted in no significant matches to proteins with known structures or functions, as found for Txp40A24 (9).
PCR and Southern blot analysis were then used to detect the presence of the toxin gene in 64 Xenorhabdus and Photorhabdus isolates. A PCR product that was the same size as the txp40A24 or txp40V16 control was detected in 43 Xenorhabdus strains and 12 Photorhabdus strains (Table 1). Note that the primary and secondary phases of X. nematophila strain Mex both produced a PCR product with the external primers. Genomic DNA from the four Xenorhabdus strains which did not produce a PCR product (strains K77, Q58, T228, and W2/5.2) were shown to hybridize to the txp40A24 probe by low-stringency Southern blot analysis (data not shown). This indicated that 47 of the Xenorhabdus strains studied and 12 of the Photorhabdus strains studied contained a gene related to txp40A24.
TABLE 1.
Forty-seven Xenorhabdus strains and 12 Photorhabdus strains which were shown to contain the txp40 gene using PCR or hybridization experiments
| Species or subspecies | Strain(s)a | Gene product detected by:b
|
||
|---|---|---|---|---|
| External primers | Internal primers | Hybridization | ||
| X. nematophila | A24, All, AllD, AN6, BK, Cb, F1, Mex, Ohio, Pi, S. carp It | + | ND | ND |
| XnNach | − | + | ND | |
| X. poinarii | ATCC 49121, ATCC 49122, NC32, NC33, NC40, NC513 | − | + | ND |
| X. bovienii | F7, T363 | + | ND | ND |
| F3, F5, F9, Si, SK, T319 | − | + | ND | |
| T228 | − | − | + | |
| X. beddingii | Q58 | − | − | + |
| X. japonica | Kushidai | − | + | ND |
| Xenorhabdus sp. | ED3 | + | ND | ND |
| C8401, C8503, CB 19, CB/2A/W, CB2B, CB-G, CB-W, EC1, K78, NC270, NC276, Q1, SaR, SaV, W1 | − | + | ND | |
| K77, W2/5.2 | − | − | + | |
| P. luminescens | C8406, HI, K80 | ND | + | ND |
| P. luminescens subsp. luminescens | Hb, V16 | ND | + | ND |
| P. luminescens subsp. laumondii | HP88 | ND | + | ND |
| P. luminescens subsp. akhurstii | D1, Tetuanc | ND | + | ND |
| P. temperata | C1, HW79, NZH3 | ND | + | ND |
| Photorhabdus sp. | Q621 | ND | + | ND |
The toxin gene in the following strains was sequenced: A24, All, AllD, AN6, BK, Cb, F1, Mex, F7, T363, F5, T319, Kushidai, C8401, CB-G, CB-W, SaV, HI, V16, Tetuan, and Q621.
The primers for Xenorhabdus strains were designed from txp40A24, and the primers for Photorhabdus were designed from txp40V16. +, gene product detected; −, gene product not detected; ND, experiment not done.
The PCR product observed for P. luminescens subsp. akhurstii strain D1 was obtained using the internal primers designed from the txp40A24 gene.
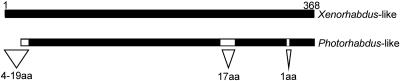
The toxin gene (txp40) was cloned from 17 Xenorhabdus strains and four Photorhabdus strains and sequenced. The predicted toxin proteins in all of these strains exhibited high levels of similarity to Txp40A24. Due to the high levels of similarity between the toxins from different strains, the diversity of the 21 sequences could be minimally represented by the eight sequences from Xenorhabdus strains A24, Kushidai, F1, Mex, AN6, and T363 and Photorhabdus strains HI and V16. Alignment of these eight sequences (see Fig. S1 in the supplemental material) revealed that there are three distinct forms of the toxin protein which differ by small insertions and deletions dispersed throughout the protein (Fig. 1).
FIG. 1.
Schematic diagram showing the regions where amino acid insertions and deletions occur in the Txp40 toxin. The diagram was constructed from the alignment of the amino acid sequences of toxins from 23 species of Xenorhabdus and Photorhabdus. aa, amino acids.
Nucleotide sequence surrounding the txp40 toxin gene in P. luminescens strain V16.
The genetic context of the txp40V16 gene was investigated by limited sequencing of the DNA flanking this gene (9). The region around the toxin gene that was sequenced was 4,180 bp long (average G+C content, about 37%), including 1,693 bp upstream and 2,210 bp downstream of txp40V16, as well as some single sequences (approximately 450 to 550 bp) from the original cosmid clones or the intermediate deletion clones that were produced when txp40V16 was isolated. All of the sequences obtained were also present in the genome of P. luminescens subsp. laumondii strain TT01, with ≥91% identity. The sequences matching the 5′ and 3′ ends of the cosmid clone were approximately 44 kb apart in the Photorhabdus genome, a distance consistent with the size of the cosmid clone, and the plu2326 toxin gene was situated between the two end sequences. This suggests that the general arrangements of genes near the toxin gene are similar in Photorhabdus strains V16 and TT01. However, many of the open reading frames identified in this 44 kb of sequence in the Photorhabdus genome are annotated as open reading frames that encode unknown proteins, particularly downstream of the toxin gene (plu2327 to plu2346). The only annotated open reading frames in this region are upstream of the toxin gene and are a locus that includes eight genes annotated as a cluster that is very similar to the Yersinia pestis yersiniabactin cluster (plu2316 and plu2318 to plu2324). Similarity searches using all experimentally obtained sequences produced only three other convincing matches in more than 6,000 bp of sequence. The first match was an open reading frame that ended approximately 500 bp upstream of the txp40V16 gene and that was very similar to the Bacillus subtilis yvrK gene (blastx; gi:7445182; 948 bp; expect value, 2 × 10−87). The second match was well upstream of txp40V16 and corresponded to DNA invertase transposon Tn5393 from Erwinia amylovora (blastx; gi:420963; 132 bp; expect value, 8 × 10−10). The third match was downstream of the toxin gene and corresponded to the gene encoding the hypothetical protein PpoE, which is part of a flagellum operon (gi:AY422684) in P. luminescens (blastx; gi:AAQ97873; 135 bp; expect value, 3 × 10−13).
Searches for proteins or DNA with similarity to the Txp40 toxin or gene were also conducted with the partially completed genomes of X. bovienii, X. nematophila strain ATCC 19061, and P. asymbiotica. The toxin gene was found in the genome of X. nematophila strain ATCC 19061 and was shown to be identical to the gene encoding the toxin protein from X. nematophila strain AN6. The sequence surrounding the toxin gene was also very similar to that obtained for X. nematophila strain A24 (9), indicating that the toxin gene is in the same context in these two X. nematophila strains. The toxin gene was not found in the partially completed genomes of X. bovienii or P. asymbiotica, although in P. asymbiotica some matches to parts of the DNA sequence flanking the toxin gene were found.
Txp40 toxin activity.
Recombinant proteins corresponding to the toxins from X. nematophila strain Mex (Txp40Mex), P. luminescens strain V16 (Txp40V16), and Photorhabdus sp. strain HI (Txp40HI) were purified using the IMPACT system. The identities and purities of the proteins were confirmed by SDS-PAGE and mass spectroscopy (data not shown). LD50s of 34 ng (95% confidence limits, 18 to 59 ng) and 64 ng (95% confidence limits, 36 to 114 ng) were determined for the interaction of recombinant Txp40V16 with G. mellonella and H. armigera, respectively (data not shown). Since the average larval mass at the time of injection was approximately 800 mg for G. mellonella and 140 mg for H. armigera, the LD50s were 43 and 460 ng/g for G. mellonella and H. armigera, respectively. Qualitative results were also obtained for the activity of recombinant toxins against other insect species (data not shown). Txp40V16 was active against L. cuprina and P. interpunctella, and Txp40Mex and Txp40HI were active against G. mellonella. The typical toxin dose required to cause ≥50% mortality was in the 5- to 100-ng range, although the dose required for Txp40HI was significantly higher (500 ng). These results show that the toxins are active against a variety of insect species, although the lethal dose varies with the toxin and the insect species.
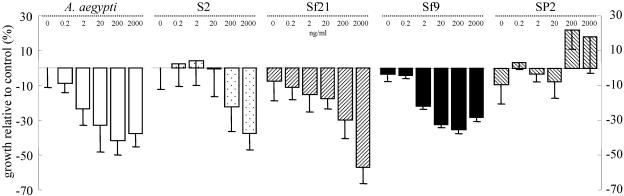
A CyQUANT cell proliferation assay kit was used to determine the effect of 0.002 to 2,000 ng/ml of recombinant Txp40A24 on insect and mammalian cell lines (Fig. 2). Txp40A24 caused statistically significant reductions (up to 60%) in the number of cells (compared to the MBP control) for the Spodoptera cell lines Sf9 and Sf21 and the mosquito cell line (A. aegypti) at concentrations greater than 2 ng/ml and for the D. melanogaster cell line (S2) at the highest concentration tested (2,000 ng/ml). In contrast, the mammalian myeloma (SP2) cell line was not significantly affected by Txp40A24 at the range of toxin concentrations tested.
FIG. 2.
Inhibition of cell growth observed for various doses of recombinant Txp40A24 protein in the in vitro cytotoxicity assay with A. aegypti, S2, Sf21, Sf9, and SP2 cells. Inhibition is expressed as a percentage relative to the growth of the control cells, which were treated with MBP. For each cell type, the inhibition with 0, 0.2, 2, 20, 200, and 2,000 ng/ml of Txp40A24 is shown. As there was no significant difference between the growth of the toxin-treated cells and the growth of the MBP-treated cells at doses of 0.002 and 0.02 ng/ml, the data for these concentrations were omitted for clarity.
Txp40 toxin histopathology.
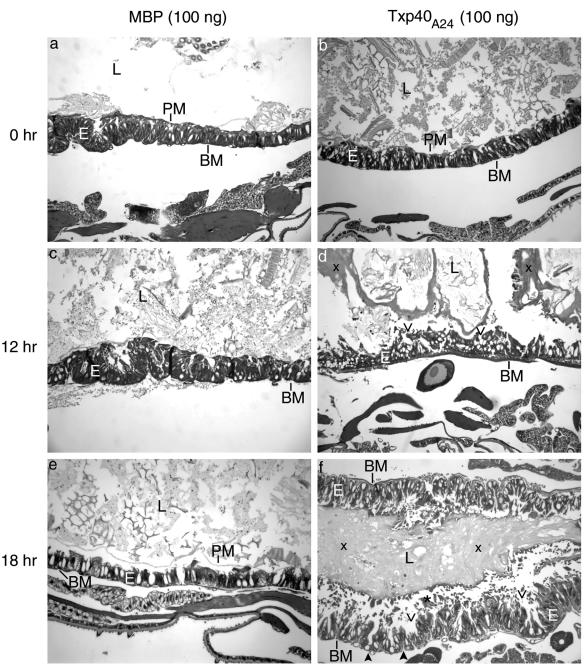
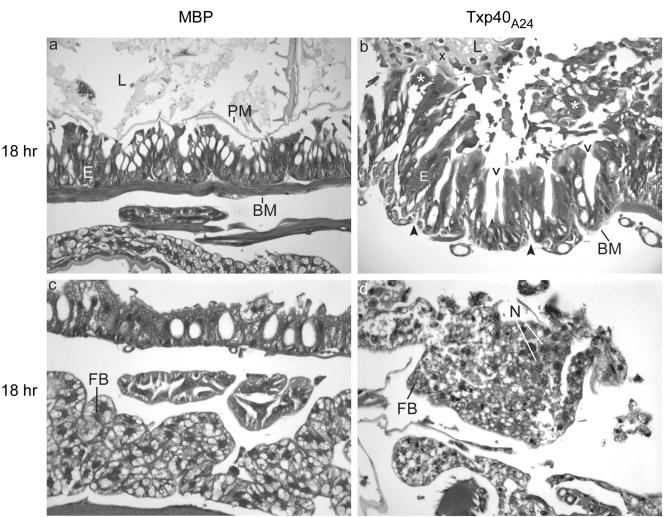
H. armigera larvae treated with 10 ng or 100 ng of Txp40A24 were examined by microscopy (Fig. 3 to 5). The control larvae inoculated with 100 ng of MBP showed the histology expected for healthy H. armigera larvae (Fig. 3 to 5). At 6 h after injection of the toxin the only observable difference between the toxin-treated and control larvae was that the peritrophic matrix had started to disintegrate in the toxin-treated larvae. Twelve hours after injection with 100 ng of toxin, there were large gaps between the midgut cells, which were starting to separate from the basement membrane and each other (Fig. 3). The peritrophic matrix was also no longer visible (Fig. 3), a result that was confirmed by analysis of the citric acid- and heat-treated sections (Fig. 4). This treatment resulted in strong autofluorescence of the peritrophic matrix and revealed the presence and absence of the peritrophic matrix in the control and toxin-treated larvae, respectively. By 18 h with either concentration of toxin, the damage to the midgut was obvious, with numerous cells sloughed into the lumen and a large amount of unidentified material appearing in the lumen (Fig. 5). At the same time, the fat body showed signs of nuclear degradation, as shown by chromatin condensation (Fig. 5). By 24 h postinjection the midgut was quite disrupted and the basement membrane had started to disintegrate. The damage was evenly distributed throughout the midgut, and there was damage to the foregut or hindgut and no evidence of general cell lysis. All of these effects of the toxin were observed in at least two larvae from independent experiments.
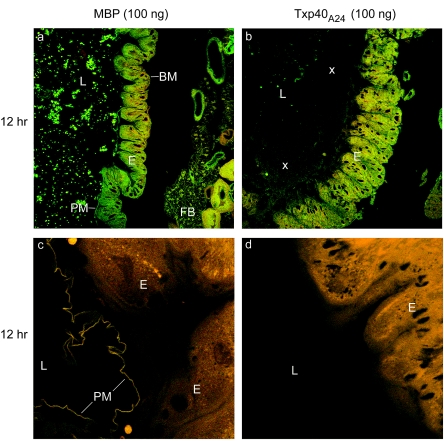
FIG. 3.
Histological images of toxin-treated H. armigera larvae, showing the effect of recombinant toxin on the midgut. All images (magnification, ×90) are images of longitudinal sections through the anterior region of the midguts of larvae inoculated with 100 ng of either MBP or Txp40A24 toxin. BM, basement membrane; E, midgut epithelium; L, midgut lumen; PM, peritrophic matrix. The open arrowheads indicate spaces between cells of the gut epithelium, the solid arrowheads indicate breakdown of the basement membrane, the asterisk indicates cells sloughed into the midgut lumen, and the multiplication signs indicate rafts of unidentified material in the midgut lumen. (a, c, and e) Larvae treated with MBP. (b, d, and f) Larvae treated with Txp40A24. Tissues were fixed at zero time (a and b) and at 12 h (c and d) and 18 h (e and f) after toxin injection.
FIG. 5.
Histological images of toxin-treated H. armigera larvae, showing the effect of recombinant toxin on the midgut and the fat body. All images are images of longitudinal sections through the anterior region of the midguts of larvae inoculated with either MBP or Txp40A24 toxin. BM, basement membrane; E, midgut epithelium; L, midgut lumen; FB, fat body; PM, peritrophic matrix. The open arrowheads indicate spaces between cells of the gut epithelium, the solid arrowheads indicate breakdown of the basement membrane, the asterisks indicate cells sloughed into the midgut lumen, the multiplication sign indicates rafts of unidentified material in the midgut lumen, and N indicates fat body nuclei that show signs of damage. (a and c) Larvae following treatment with 100 ng MBP. (b) Larvae following treatment with 100 ng Txp40A24. (d) Larvae after treatment with 10 ng Txp40A24. Tissues were fixed at 18 h after injection of toxin. (a and b) Magnification, ×230. (c and d) Magnification, ×370.
FIG. 4.
Histological images of toxin-treated H. armigera larvae, showing the effect of recombinant toxin on the peritrophic matrix. All images are images of longitudinal sections through the anterior region of the midguts of larvae inoculated with 100 ng of either MBP or Txp40A24 toxin. Tissues were fixed 12 h after injection of the toxin, and autofluorescence was monitored after sections were treated with citric acid and heat. BM, basement membrane; E, midgut epithelium; L, midgut lumen; PM, peritrophic matrix. The multiplication signs indicate rafts of unidentified material in the midgut lumen. (a and c) Larvae following MBP treatment. (b and d) Larvae following Txp40A24 treatment. (a and b) Magnification, ×370. (c and d) Magnification, ×1,100.
DISCUSSION
Forty-seven Xenorhabdus strains and 12 Photorhabdus strains were shown to contain a gene closely related to txp40A24, and the toxin gene was also found in the genomes of X. nematophila strain ATCC 19061 and P. luminescens subsp. laumondii strain TT01. These results suggest that the toxin is ubiquitous among Xenorhabdus and Photorhabdus bacteria. The toxin gene was found in both phase 1 and phase 2 cells of X. nematophila strain Mex, which is consistent with evidence that there are no differences at the DNA level (either chromosomal or plasmid) between the different phases of this bacterium (3, 6). For Xenorhabdus, the highest level of similarity to txp40A24 (as shown by the production of a PCR product with primers external to txp40A24) was observed with strains of X. nematophila (Table 1). There was no apparent correlation between the geographic location of the Xenorhabdus strain and the method that was needed to detect the txp40A24 homolog (Table 1; see Table S1 in the supplemental material).
The txp40 toxin gene was sequenced for 17 Xenorhabdus strains and four Photorhabdus strains. The toxins from five Photorhabdus strains were very similar to Txp40V16, differing by at most five amino acids. With the exception of toxin from X. bovienii strain T363, the toxin sequences from Xenorhabdus strains were all very similar to the sequence of Txp40A24, differing by a maximum of nine amino acids between residues 19 and 344 (Txp40A24 numbering). These results confirm that the toxin protein is highly conserved. The toxin from X. bovienii strain T363 was anomalous in that the sequence was more similar to the sequences of the Photorhabdus toxins, yet biochemical analysis (colony morphology, 16S RNA sequence, and negative catalase activity) clearly classified this organism as a Xenorhabdus strain. There appears to be heterogeneity within the species X. bovienii (1, 6), which may explain this result.
The schematic diagram of the toxin in Fig. 1 shows that there are three regions in the protein that can tolerate insertions or deletions, the N terminus, a 17-amino-acid region with low complexity (residues 254 to 270 for Txp40A24), and a single amino acid (residue 332 in Txp40A24). Based on these insertion-deletion regions, it is possible to classify the toxin proteins as either Xenorhabdus- or Photorhabdus-like, where a longer N terminus, the presence of the sequence with the low level of complexity, and the extra amino acid at position 332 classify the protein as Xenorhabdus-like. As mentioned above, X. bovienii strain T363 is anomalous according to this classification. Since toxins from both Xenorhabdus and Photorhabdus strains were shown to kill insects, the insertion-deletion regions cannot be essential for the lethality of the protein. This was confirmed by the observation that insertion of a myc epitope tag at the N terminus or at position 254 of Txp40A24 did not significantly affect the injection bioassay results (data not shown). Although the variable regions or the single amino acid substitutions are not essential for lethality, they may contribute to the differences in relative potency for particular insect species.
There were three interesting observations that could be made about the broader genetic context in which the txp40A24 and txp40V16 genes occur. First, for txp40V16, the sequence matches indicated that P. luminescens strain V16 had shared genetic material with other bacteria, that mobile genetic factors such as transposons were present, and that the G+C content of the DNA region containing the toxin (37%) was different from the G+C content estimated for the whole genome (43%) (15). This implied that the toxin gene was part of a genomic island involved in pathogenicity, as concluded previously for txp40A24 (9). Other toxins in Xenorhabdus and Photorhabdus, including the Tc toxins and the mcf gene product, are also part of putative genomic islands involved in pathogenicity (36). Second, the analysis of the genetic context of the two Photorhabdus toxin genes, txp40V16 and plu2326, resulted in identification of several proteins with functions associated with virulence. These proteins included the yersiniabactin-like cluster, which is responsible for iron acquisition using siderophores (11, 15, 37), the flagellum protein PpoE (28), and three pseudogene-encoded proteins related to the TccB toxin (proteins encoded by plu2333 to plu2335) (15). The similar locations of different genes implicated in virulence are consistent with the idea that the txp40 gene is part of a genomic island involved in pathogenicity. Third, despite the high degree of similarity between the txp40A24 and txp40V16 genes, the DNA surrounding the toxin genes exhibited no significant similarity, indicating that the context of the toxin genes in the two genera is quite different. This is consistent with the observation that the txp40 gene product appears to be active as a single protein, unlike the Tc proteins, which are encoded by multiple genes arranged in multiple loci (35, 36). This is also another example of the molecular and biochemical differences between Xenorhabdus and Photorhabdus (16).
Recombinant Txp40 toxin was active against a range of lepidopteran species (G. mellonella, H. armigera, and P. interpunctella) and cell lines (Spodoptera cell lines Sf9 and Sf21), as well as dipteran species (L. cuprina) and cell lines (A. aegypti and D. melangoaster cell line S2). Khandelwal and colleagues also observed that an outer membrane vesicle preparation from X. nematophila was toxic to Sf21 cells (19). The broad insecticidal activity of the Txp40 toxin is consistent with observations that the nematode-bacterium association is active against a range of different insect species (23). It suggests that the toxin has a target that is common to many different insects.
The primary site of action of the Txp40 toxin appeared to be the insect midgut, as revealed by the histology of H. armigera larvae inoculated with the toxin (Fig. 3 to 5). The results are consistent with reports which described the colonization of Spodoptera littoralis by green fluorescent protein-labeled X. nematophila (32) and the colonization of M. sexta by P. luminescens (33). Both reports suggested that the bacteria occupy a niche in the extracellular matrix surrounding the midgut, so that the bacteria are associated closely with the midgut epithelial cells. Bacterial colonization appears to start in the anterior midgut and hemolymph, progress to the posterior midgut, and finally spread to other tissues, such as the fat body (32, 33). This is consistent with our observations which showed that there was damage to the midgut at the early stages of infection and damage to other tissues, such as the fat body, at the later stages of infection.
The histopathological effects on lepidopteran larvae of three other toxins from Xenorhabdus or Photorhabdus have been examined previously (5, 13, 20). All three toxins (the Tca toxins, the mcf gene product, and a 17-kDa pilin subunit) also damage the midgut. The Tc toxins from Photorhabdus cause shedding of the midgut epithelium of M. sexta larvae into the gut lumen and lysis of the epithelial cells (5). The Tca toxin has been reported to cause no damage to other tissues of M. sexta larvae (5). The mcf gene from P. luminescens was cloned into E. coli, and the bacteria were injected into M. sexta larvae, resulting in shedding of the insect midgut epithelium and destructive blebbing of hemocytes (13). The peritrophic matrix was intact 24 h after ingestion of the bacteria (13). E. coli expressing the mcf gene product was also shown to trigger apoptosis in mammalian cells (14). The damage to the lepidopteran midgut and other tissues caused by the Tc toxins and the mcf gene product is different than the damage seen with the Txp40 toxin. The third toxin studied, the 17-kDa pilin subunit from X. nematophila, was administered orally to H. armigera larvae and was shown to break down the midgut epithelial lining and basement membrane and to cause sloughing of cell debris into the midgut lumen (20). The limited analysis of the previously published microscopy images that was possible suggested that the histopathology is somewhat similar to that seen with the Txp40 toxin. This indicates that there are now at least four toxin proteins from Xenorhabdus or Photorhabdus that are known to cause significant damage to the insect midgut.
The Txp40 toxin also caused damage to the fat body (Fig. 5). No damage has been reported to tissues other than the midgut for the other three toxins from Xenorhabdus or Photorhabdus that have been studied. The lack of reported damage to the fat body may be due to the fact that large numbers of Photorhabdus bacteria are not recovered from the M. sexta fat body until >48 h after bacterial infection (33), suggesting that damage to the fat body occurs late in the infection process. One study reported that Xenorhabdus bacteria disrupt the fat body, silk glands, hypoderma, and epithelia (18). Proteins or extracts from Xenorhabdus and Photorhabdus have also been shown to damage insect hemocytes, which is consistent with observations that the bacteria manage to evade the cellular immune response of the insect (4). However, in general, reports have focused on the damage that Xenorhabdus or Photorhabdus bacteria and their toxins cause to the insect midgut.
Due to the limited amount of work on the histopathology of Xenorhabdus and Photorhabdus toxins, it is informative to examine the larger body of work for B. thuringiensis and its toxins. The Txp40 toxin caused a significant decrease in midgut intercellular adhesion, degradation of the peritrophic matrix lining the midgut cells, and degradation of the fat body nuclei (Fig. 3 to 5). All of these tissues have also been reported to be damaged by B. thuringiensis or isolated B. thuringiensis toxins, although the data are somewhat sparse and sometimes contradictory. Several reports indicate that treatment of insects with B. thuringiensis or its toxins disrupts the peritrophic matrix (12, 30). The B. thuringiensis toxins have been reported to damage the intercellular junctions of midgut cells of A. aegypti (12) and M. sexta (22) but not Pieris brassicae (24). The fat body of H. armigera larvae was shown to be damaged by B. thuringiensis (30). Thus, although many known insect toxins appear to target the insect midgut, it is clear that bacteria such as Xenorhabdus and B. thuringiensis and their toxins can also cause significant damage to other insect tissues.
The results suggest that the Txp40 toxin has multiple target sites in H. armigera and that the primary site of action is the midgut, although there is also damage to other tissues, such as the fat body. It is intriguing to consider how the toxin can act at several different sites yet leave sites such as the foregut and hindgut unaffected. Such tissue specificity is accompanied by activity against a range of insects but not against a mammalian cell line. An attractive hypothesis to explain these results is that the toxin targets a protein that is found in smooth septate (continuous) intercellular junctions, which are found in the insect midgut but not in mammals or other insect tissues, such as the foregut and hindgut (21). Regardless of speculation about the possible mode of action of the toxin, it is clear that Txp40 is a widely occurring and highly conserved toxin in Xenorhabdus and Photorhabdus bacteria. Txp40 is important for the broad insecticidal activity of these bacteria and is a significant component of the extensive array of toxins that the bacteria and nematodes use to destroy their insect hosts.
Supplementary Material
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Akhurst, R. J., and N. E. Boemare. 1988. A numerical taxonomic study of the genus Xenorhabdus (Enterobacteriaceae) and proposed elevation of the subspecies of X. nematophilus to species. J. Gen. Microbiol. 134:1835-1845. [DOI] [PubMed] [Google Scholar]
- 2.Akhurst, R. J., and G. B. Dunphy. 1993. Tripartite interactions between symbiotically associated entomopathogenic bacteria, nematodes, and their insect hosts, p. 1-23. In N. E. Beckage, S. N. Thompson, and B. A. Federici (ed.), Parasites and pathogens of insects, vol. 2. Academic Press, Inc., San Diego, Calif. [Google Scholar]
- 3.Akhurst, R. J., A. J. Smigielski, J. Mari, N. Boemare, and R. G. Mourant. 1992. Restriction analysis of phase variation in Xenorhabdus spp. (Enterobacteriaceae), entomopathogenic bacteria associated with nematodes. Syst. Appl. Microbiol. 15:469-473. [Google Scholar]
- 4.Au, C., P. Dean, S. E. Reynolds, and R. H. ffrench-Constant. 2004. Effect of the insect pathogenic bacterium Photorhabdus on insect phagocytes. Cell. Microbiol. 6:89-95. [DOI] [PubMed] [Google Scholar]
- 5.Blackburn, M., E. Golubeva, D. Bowen, and R. H. ffrench-Constant. 1998. A novel insecticidal toxin from Photorhabdus luminescens, Toxin complex a (Tca), and its histopathological effects on the midgut of Manduca sexta. Appl. Environ. Microbiol. 64:3036-3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boemare, N. E., R. J. Akhurst, and R. G. Mourant. 1993. DNA relatedness between Xenorhabdus spp. (Enterobacteriaceae), symbiotic bacteria of entomopathogenic nematodes, and a proposal to transfer Xenorhabdus luminescens to a new genus, Photorhabdus gen. nov. Int. J. Syst. Bacteriol. 43:249-255. [Google Scholar]
- 7.Bowen, D., T. A. Rocheleau, M. Blackburn, O. Andreev, E. Golubeva, R. Bhartia, and R. H. ffrench-Constant. 1998. Insecticidal toxins from the bacterium Photorhabdus luminescens. Science 280:2129-2132. [DOI] [PubMed] [Google Scholar]
- 8.Bowen, D. J., and J. C. Ensign. 1998. Purification and characterization of a high-molecular-weight insecticidal protein complex produced by the entomopathogenic bacterium Photorhabdus luminescens. Appl. Environ. Microbiol. 64:3029-3035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Brown, S. E., A. T. Cao, E. R. Hines, R. J. Akhurst, and P. D. East. 2004. A novel secreted protein toxin from the insect pathogenic bacterium Xenorhabdus nematophila. J. Biol. Chem. 279:14595-14601. [DOI] [PubMed] [Google Scholar]
- 10.Burnell, A. M., and S. P. Stock. 2000. Heterorhabditis, Steinernema and their bacterial symbionts—lethal pathogens of insects. Nematology 2:31-42. [Google Scholar]
- 11.Carniel, E. 2001. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 3:561-569. [DOI] [PubMed] [Google Scholar]
- 12.Charles, J.-F., and H. de Barjac. 1983. Action des cristaux de Bacillus thuringiensis var. israelensis sur l'intestin moyen des larves de Aedes aegypti L., en microscopie électronique. Ann. Inst. Pasteur Microbiol. 134A:197-218. [PubMed] [Google Scholar]
- 13.Daborn, P. J., N. Waterfield, C. P. Silva, C. P. Y. Au, S. Sharma, and R. H. ffrench-Constant. 2002. A single Photorhabdus gene, makes caterpillars floppy (mcf), allows Escherichia coli to persist within and kill insects. Proc. Natl. Acad. Sci. USA 99:10742-10747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dowling, A. J., P. J. Daborn, N. R. Waterfield, P. Wang, C. H. Streuli, and R. H. ffrench-Constant. 2004. The insecticidal toxin makes caterpillars floppy (Mcf) promotes apoptosis in mammalian cells. Cell. Microbiol. 6:345-353. [DOI] [PubMed] [Google Scholar]
- 15.Duchaud, E., C. Rusniok, L. Frangeul, C. Buchrieser, A. Givaudan, S. Taourit, S. Bocs, C. Boursaux-Eude, M. Chandler, J.-F. Charles, E. Dassa, R. Derose, S. Derzelle, G. Freyssinet, S. Gaudriault, C. Médigue, A. Lanois, K. Powell, P. Siguier, R. Vincent, V. Wingate, M. Zouine, P. Glaser, N. Boemare, A. Danchin, and F. Kunst. 2003. The genome sequence of the entomopathogenic bacterium Photorhabdus luminescens. Nat. Biotechnol. 21:1307-1313. [DOI] [PubMed] [Google Scholar]
- 16.Forst, S., and K. Nealson. 1996. Molecular biology of the symbiotic-pathogenic bacteria Xenorhabdus spp. and Photorhabdus spp. Microbiol. Rev. 60:21-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo, L., R. O. Fatig III, G. L. Orr, B. W. Schafer, J. A. Strickland, K. Sukhapinda, A. T. Woodsworth, and J. K. Petell. 1999. Photorhabdus luminescens W-14 insecticidal activity consists of at least two similar but distinct proteins—purification and characterization of toxin A and toxin B. J. Biol. Chem. 274:9836-9842. [DOI] [PubMed] [Google Scholar]
- 18.Jarosz, J., M. Balcerzak, and H. Skrzypek. 1991. Involvement of larvicidal toxins in pathogenesis of insect parasitism with the rhabditoid nematodes, Steinernema feltiae and Heterorhabditis bacteriophora. Entomophaga 36:361-368. [Google Scholar]
- 19.Khandelwal, P., and N. Banerjee-Bhatnagar. 2003. Insecticidal activity associated with the outer membrane vesicles of Xenorhabdus nematophilus. Appl. Environ. Microbiol. 69:2032-2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Khandelwal, P., D. Choudhury, A. Birah, M. K. Reddy, G. P. Gupta, and N. Banerjee. 2004. Insecticidal pilin subunit from the insect pathogen Xenorhabdus nematophila. J. Bacteriol. 186:6465-6476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lane, N. J., R. Dallai, and D. E. Ashhurst. 1996. Structural macromolecules of the cell membranes and the extracellular matrices of the insect midgut, p.115-150. In M. J. Lehane and P. F. Billingsley (ed.), Biology of the insect midgut. Chapman & Hall, London, United Kingdom.
- 22.Lane, N. J., J. B. Harrison, and W. M. Lee. 1989. Changes in microvilli and Golgi-associated membranes of lepidopteran cells induced by an insecticidally active bacterial δ-endotoxin. J. Cell Sci. 93:337-347. [Google Scholar]
- 23.Laumond, C., H. Mauléon, and A. Kermarrec. 1979. Données nouvelles sur le spectre d'hôtes et le parasitisme du nématode entomophage Neoaplectana carpocapsae. Entomophaga 24:13-27. [Google Scholar]
- 24.Lüthy, P., and H. R. Ebersold. 1981. Bacillus thuringiensis delta-endotoxin: histopathology and molecular mode of action, p. 235-267. In E. W. Davidson (ed.), Pathogenesis of invertebrate microbial diseases. Allenheld, Osmun & Co. Publishers, Totowa, N.J.
- 25.Maniatis, T., E. F. Fritsch, and J. Sambrook. 1982. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 26.Needleman, S. B., and C. D. Wunsch. 1970. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J. Mol. Biol. 48:443-453. [DOI] [PubMed] [Google Scholar]
- 27.Pan, Y., H. Jian, J. Zhang, Z. Liu, Z. Chen, X. Yang, H. Yang, and D. Huang. 2002. An intracellular toxic protein (Xin) isolated from Xenorhabdus nematophilus strain BJ. Prog. Nat. Sci. 12:310-312. [Google Scholar]
- 28.Ramos, H. C., M. Rumbo, and J.-C. Sirard. 2004. Bacterial flagellins: mediators of pathogenicity and host immune responses in mucosa. Trends Microbiol. 12:509-517. [DOI] [PubMed] [Google Scholar]
- 29.Ryu, K. G., J. S. Bae, Y. S. Yu, and S. H. Park. 2000. Insecticidal toxin from Xenorhabdus nematophilus, symbiotic bacterium associated with entomopathogenic nematode Steinernema glaseri. Biotechnol. Bioprocess Eng. 5:141-145. [Google Scholar]
- 30.Salama, H. S., and A. Sharaby. 1985. Histopathological changes in Heliothis armigera infected with Bacillus thuringiensis as detected by electron microscopy. Insect Sci. Appl. 6:503-511. [Google Scholar]
- 31.Scott, K. F., B. G. Rolfe, and J. Shine. 1981. Biological nitrogen fixation: primary structure of the Klebsiella pneumoniae nifH and nifD genes. J. Mol. Appl. Genet. 1:71-81. [PubMed] [Google Scholar]
- 32.Sicard, M., K. Brugirard-Ricaud, S. Pages, A. Lanois, N. E. Boemare, M. Brehélin, and A. Givaudan. 2004. Stages of infection during the tripartite interaction between Xenorhabdus nematophila, its nematode vector, and insect hosts. Appl. Environ. Microbiol. 70:6473-6480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Silva, C. P., N. R. Waterfield, P. J. Daborn, P. Dean, T. Chilver, C. P. Y. Au, S. Sharma, U. Potter, S. E. Reynolds, and R. H. ffrench-Constant. 2002. Bacterial infection of a model insect: Photorhabdus luminescens and Manduca sexta. Cell. Microbiol. 4:329-339. [DOI] [PubMed] [Google Scholar]
- 34.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Waterfield, N. R., D. J. Bowen, J. D. Fetherston, R. D. Perry, and R. H. ffrench-Constant. 2001. The tc genes of Photorhabdus: a growing family. Trends Microbiol. 9:185-191. [DOI] [PubMed] [Google Scholar]
- 36.Waterfield, N. R., P. J. Daborn, and R. H. ffrench-Constant. 2002. Genomic islands in Photorhabdus. Trends Microbiol. 10:541-545. [DOI] [PubMed] [Google Scholar]
- 37.Watson, R. J., S. A. Joyce, G. V. Spencer, and D. J. Clarke. 2005. The exbD gene of Photorhabdus temperata is required for full virulence in insects and symbiosis with the nematode Heterorhabditis. Mol. Microbiol. 56:763-773. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.