Abstract
The growth of pure cultures of Bacteroides thetaiotaomicron LMG 11262 and Bacteroides fragilis LMG 10263 on fructose and oligofructose was examined and compared to that of Bifidobacterium longum BB536 through in vitro laboratory fermentations. Gas chromatography (GC) analysis was used to determine the different fractions of oligofructose and their degradation during the fermentation process. Both B. thetaiotaomicron LMG 11262 and B. fragilis LMG 10263 were able to grow on oligofructose as fast as on fructose, succinic acid being the major metabolite produced by both strains. B. longum BB536 grew slower on oligofructose than on fructose. Acetic acid and lactic acid were the main metabolites produced when fructose was used as the sole energy source. Increased amounts of formic acid and ethanol were produced when oligofructose was used as an energy source at the cost of lactic acid. Detailed kinetic analysis revealed a preferential metabolism of the short oligofructose fractions (e.g., F2 and F3) for B. longum BB536. After depletion of the short fractions, the larger oligofructose fractions (e.g., F4, GF4, F5, GF5, and F6) were metabolized, too. Both Bacteroides strains did not display such a preferential metabolism and degraded all oligofructose fractions simultaneously, transiently increasing the fructose concentration in the medium. This suggests a different mechanism for oligofructose breakdown between the strain of Bifidobacterium and both strains of Bacteroides, which helps to explain the bifidogenic nature of inulin-type fructans.
The human colon harbors a complex microbial ecosystem which is indispensable for the well-being of its host (40). Both culture-based methods and advanced molecular techniques revealed that some predominant genera can be distinguished, such as Eubacterium, Bacteroides, and Bifidobacterium, although the human microbiota is host specific (10, 20, 29, 42, 46, 49). Members of this microbial ecosystem are capable of metabolizing nondigested polysaccharides, thereby producing a variety of metabolites such as short-chain fatty acids (SCFA) (e.g., acetic acid, propionic acid, and butyric acid), other organic acids (e.g., lactic acid and succinic acid), and gases (e.g., H2, H2S, CO2, and CH4). Despite intensive research carried out during the past decade, the exact composition and metabolic interactions of the colon microbiota remain largely unknown.
Bifidobacteria account for approximately 3% of the human microbiota (10, 23, 28). They are not pathogenic and contribute to the degradation of starch and other plant polysaccharides, as they harbor specific enzymes and sugar uptake systems (2, 39, 43). Bacteroides species account for approximately 20% of the human microbiota (10, 29, 34, 42). Several species are thought to be pathogenic, and Bacteroides spp. in general are probably involved in the horizontal gene transfer of antibiotic resistance genes in the human colon (36, 37, 41). Bacteroides spp. are known to ferment numerous substrates (16, 38), a fact that was recently confirmed by analysis of both the Bacteroides thetaiotaomicron and Bacteroides fragilis genomes (22, 48). These genome analyses revealed the presence of highly evolved strategies to assure their survival in the human colon. Most noteworthy is the presence of an abundant machinery capable of acquiring and metabolizing a large variety of simple and complex carbohydrates (5).
An important class of complex, nondigestible polysaccharides is referred to as prebiotics. A prebiotic can be defined as a nondigestible food ingredient that beneficially affects the host by selectively stimulating the growth and/or activity of one or a limited number of bacteria in the colon, thus improving host health (12, 13). Based on existing scientific data, three types of carbohydrates can be classified as prebiotics, namely inulin-type fructans, trans-galacto-oligosaccharides, and lactulose (12). Inulin-type fructans are linear D-fructose polymers linked by β(2-1)-glycosidic bonds, often with a terminal glucose moiety that is linked by an α(1-2)-glycosidic bond, as in sucrose (35). Inulin can be obtained by extraction of specific plant parts such as chicory roots. Oligofructose can be produced either enzymatically through fructosyltransferase action on sucrose or by partial enzymatic hydrolysis of the inulin chains. Oligofructose derived from chicory roots contains both fructose chains (Fm type) and fructose chains with a terminal glucose unit (GFn type) (6). Proof has been given of the prebiotic potential of inulin-type fructans through both in vitro tests and well-controlled human intervention trials (11, 24, 30, 33, 44, 47). Bifidobacteria are the target organisms for prebiotic action of inulin and oligofructose because they ferment inulin-type fructans and are considered beneficial for host health. Besides the stimulation of bifidobacteria (the so-called bifidogenic effect), some in vitro and mice studies indicate that also other members of the human colon microbiota might consume and even be stimulated upon ingestion of inulin-type fructans (1, 8, 15). Certain lactobacilli, e.g., strains of Lactobacillus paracasei, are indeed able to grow on these prebiotics (19, 26).
The growth of pure cultures of Bacteroides on inulin-type fructans has hardly been investigated. Only one study reports on the growth of a Bacteroides strain on commercial oligofructose, which was slower than growth rates of pure bifidobacterial cultures (14). In vitro mixed culture fermentations, often using fecal slurries, and in vivo tests revealed that in most cases Bacteroides spp. are neither stimulated nor repressed through administration of inulin-type fructans (1, 24, 31). However, one in vivo test indicates a stimulatory effect of inulin-type fructans on Bacteroides spp., apart from its clear stimulation of bifidobacterial growth (33).
The aim of this study was to examine the growth of strains of B. thetaiotaomicron and B. fragilis, both predominant human commensals, on oligofructose through in vitro laboratory fermentations. A detailed kinetic analysis of their oligofructose breakdown was carried out. Furthermore, this study was performed to compare the degradation of oligofructose between Bacteroides spp. and Bifidobacterium spp. to give a more mechanistic insight into the bifidogenic effect of oligofructose. We have shown before that strains of bifidobacteria, in particular, Bifidobacterium animalis subsp. lactis DN-173 010, preferentially metabolize the short oligofructose fractions (45).
MATERIALS AND METHODS
Microorganisms and media.
One Bifidobacterium strain and two strains of Bacteroides (two species) were used throughout this study. Both the Bacteroides thetaiotaomicron LMG 11262 and the Bacteroides fragilis LMG 10263 strains were obtained from the BCCM/LMG Bacteria Collection (Ghent, Belgium). The Bifidobacterium longum BB536 strain was obtained from Morinaga Industry Co., Ltd. (Tokyo, Japan). All strains were stored at −80°C; the Bacteroides strains were stored in Wilkins-Chalgren broth (WCB; Oxoid, Ltd., Basingstoke, United Kingdom), and the Bifidobacterium strain was stored in de Man-Rogosa-Sharpe medium (MRS; Oxoid). Both media were supplemented with 25% (vol/vol) glycerol as a cryoprotectant. Solid media were prepared by the addition of 1.5% (wt/vol) agar (Oxoid) to WCB or MRS.
The fermentations were performed in a complex medium, allowing good growth of human colon bacteria when supplemented with an appropriate energy source (R. Van der Meulen and L. De Vuyst, unpublished results), containing (in grams per liter): bacteriological peptone (Oxoid), 6.5; soy peptone (Oxoid), 5.0; tryptone (Oxoid), 2.5; yeast extract (VWR International, Darmstadt, Germany), 3.0; KCl, 2.0; NaHCO3, 0.2; NaCl, 4.5; MgSO4 · 7H2O, 0.5; CaCl2 · 2H2O, 0.45; MnSO4 · H2O, 0.2; FeSO4 · 7H2O, 0.005; ZnSO4 · 7H2O, 0.005; cysteine-HCl, 0.4; hemin, 0.005; menadion, 0.005. The medium also contained H3PO4, 0.5 ml liter−1 and Tween 80, 2 ml liter−1. The pH of the medium was adjusted to 5.80 before sterilization (210 kPa, 121°C, 20 min). Fructose or oligofructose was used as the sole energy source, at a concentration of 15 g liter−1, sterilized separately and aseptically added to the fermentation medium. Fructose was sterilized in an autoclave (210 kPa, 121°C, 20 min), while the oligofructose was filter sterilized with Sartolab P-20 filters (0.2 μm; Sartorius AG, Goettingen, Germany). The oligofructose (RaftiloseP95) was kindly provided by ORAFTI N.V. (Tienen, Belgium). RaftiloseP95 is a commercial powder produced through the enzymatic hydrolysis of chicory inulin. The powder contains mainly oligofructose (>93.2% [wt/wt]) and small amounts of glucose, fructose, and sucrose. The degree of polymerization of the oligofructose chains varies between 2 and 8, with an average of 4.
Fermentation experiments.
Kinetic analyses of growth of B. thetaiotaomicron LMG 11262, B. fragilis LMG 10263, and B. longum BB536 in a complex medium with fructose or oligofructose as the sole energy source were carried out with a 1.5-l Biostat B-DCU fermentor (Sartorius AG). For inoculum buildup, the strains were transferred from −80°C to WCB (Bacteroides strains) or MRS (Bifidobacterium strain) and incubated anaerobically at 37°C for 24 h in a Modular Atmosphere Controlled system (MG Anaerobic Work Station; Don Withley Scientific, West Yorkshire, United Kingdom) that was continuously sparged with a mixture of 80% N2, 10% CO2, and 10% H2 (Air Liquide, Paris, France). Afterwards, the strains were propagated twice in the complex medium used later either with fructose or oligofructose as the sole energy source and finally added to the fermentor. The transfer volume was always 5% (vol/vol). All fermentations were carried out anaerobically by sparging the medium with a mixture of 90% N2 and 10% CO2 (Air Liquide) at a temperature of 37°C for 48 h. During the fermentation, a linear pH profile was applied, starting from pH 5.80 at time zero and ending at pH 6.80 after 48 h to simulate the pH change in the colon. The pH was controlled through automatic addition of 1.5 M NaOH. A gentle stirring (100 rpm) was applied to keep the medium homogenous. Temperature, pH, and agitation speed were on-line controlled (MFCS/win 2.1; Sartorius AG). Samples were withdrawn at regular time intervals for further analysis.
All fermentations were carried out in duplicate. The results and figures presented are representative of both fermentations.
Analysis of growth.
Cellular growth was followed by plate counting throughout the fermentation. The numbers of CFU per milliliter were obtained through enumeration on WCB (Bacteroides strains) or MRS (Bifidobacterium strain) agar after anaerobic incubation (Modular Atmosphere Controlled system) at 37°C for 48 h.
Analysis of fructose and oligofructose breakdown.
Residual fructose and oligofructose were quantified throughout the fermentations. The amounts of fructose were determined by high-performance liquid chromatography (HPLC) analysis with a chromatograph (Waters Corp., Milford, MA) equipped with a 2414 differential refractometer, a 600S controller, a column oven, and a 717plus autosampler. An ICSep ICE ORH-801 column (Interchim, Montluçon, France) was used with 10 mN H2SO4 as mobile phase at a flow rate of 0.4 ml min−1. The column temperature was kept at 35°C. Samples were centrifuged (16,060 × g for 15 min), and an equal volume of 20% (vol/vol) of trichloroacetic acid was added to remove proteins. After centrifugation (16,060 × g for 15 min), the supernatant was filtered (0.2-μm filters, Minisart RC4; Sartorius AG) before injection.
Detailed GC analysis of the different oligofructose fractions was performed on an HRGC 5300-HT Mega (Carlo Erba, Rodina, Italy) as described previously (18, 26, 45). The GC was equipped with a SGE Aluminum Clad-5 capillary column (Achrom NV/SA, Zulte, Belgium), and the oven temperature was programmed from 105 to 440°C at 10°C min−1. The samples were derivatized using a procedure involving oxymation and silylation of the sugars (18). The oxime-trimethylsilyl sugar derivatives were extracted using iso-octane and the extract was injected. RaftiloseP95 (ORAFTI N.V.), glucose, fructose, and sucrose were used as external standards.
Analysis of metabolites.
Volatiles (ethanol and SCFA) and nonvolatile organic acids (lactic acid and succinic acid) were analyzed throughout the fermentations.
The amounts of lactic acid and formic acid were determined by HPLC as described above.
Succinic acid was determined with a Waters 2695 HPLC coupled to a Quattro Micro mass spectrometer (Waters). The column (Atlantis; Waters) was kept at 35°C. The mobile phase, at a flow rate of 0.2 ml min−1, was composed of ultrapure water (eluent A), acetonitrile (eluent B), and 10 mM ammonium acetate (pH 6.5; eluent C). The gradient (vol/vol) used was as follows: 0.0 min, 85% A, 5% B, and 10% C; 15.0 min, 40% A, 50% B, and 10% C; 15.1 min, 10% A, 80% B, and 10% C; 23.0 min, 10% A, 80% B, and 10% C; 23.1 min, 85% A, 5% B, and 10% C; 30.0 min, 85% A, 5% B, and 10% C. Samples were centrifuged (16,060 × g for 15 min), and 100 μl of internal standard (3,4-dihydroxybenzoic acid) was added to 500 μl of supernatant. Afterwards, 600 μl of acetonitrile was added, and the samples were again centrifuged (16,060 × g for 15 min). The supernatant was filtered (0.2-μm filters) and injected.
The SCFA (acetic acid, propionic acid, butyric acid, valeric acid, and capronic acid) were measured by gas chromatography on an Agilent 6890 gas chromatograph (Agilent Technologies, Palo Alto, CA) coupled to an Agilent 5973N mass spectrometer (Agilent Technologies). A capillary column (DB-WAXetr; Agilent Technologies) was used together with the following oven temperature program: 0.0 min, 90°C; 5.0 min, 90°C; 7.3 min, 125°C; 12.3 min, 125°C; 16.0 min, 180°C; 31.3 min, 180°C; 34.3 min, 230°C; and 47.3 min, 230°C. Helium (Air Liquide) was used as carrier gas at a flow rate of 1.1 ml min−1. The samples were centrifuged (16,060 × g for 15 min), and 100 μl of an internal standard (2,6-dimethylphenol) and 50 μl of H2SO4 were added to 500 μl of supernatant. After being mixed for 15 s, 750 μl of diethyl ether was added to the sample and mixed thoroughly (30 min). Afterwards, the organic phase was transferred to a vial. The extraction procedure with diethyl ether was performed twice, after which the samples were injected.
Ethanol was determined using the GC-mass spectrometry apparatus described above. The same column was used with the following temperature program: 0.0 min, 40°C; 5.0 min, 40°C; 9.29 min, 100°C; 10.37 min, 230°C; and 15 min, 230°C. The sample preparation was similar to the analysis of SCFA, except that the addition of H2SO4 was skipped, chloroform was used as the organic phase instead of diethyl ether, and methanol (0.5% [wt/vol] in ultrapure water) was used as an internal standard.
RESULTS
Growth of Bacteroides thetaiotaomicron LMG 11262 on fructose or oligofructose.
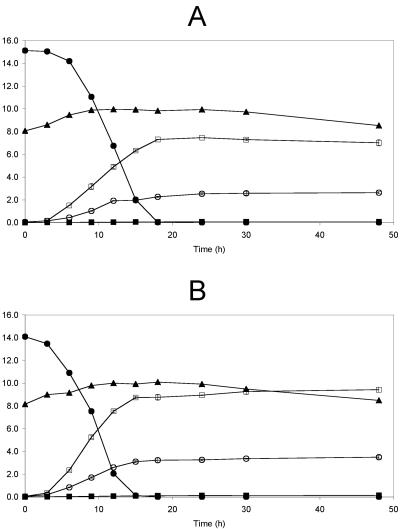
B. thetaiotaomicron LMG 11262 metabolized all fructose (15 g liter−1) within 15 h of fermentation with an accompanying cell growth of 2.4 log CFU ml−1 (Fig. 1A). The main metabolite of the fructose metabolism was succinic acid with a concentration of 8.4 ± 0.4 g liter−1. Acetic acid was produced at a concentration of 3.4 ± 0.1 g liter−1. Propionic acid was not produced.
FIG. 1.
Fermentation of B. thetaiotaomicron LMG 11262 with 15 g liter−1 fructose (A) or 15 g liter−1 oligofructose (RaftiloseP95) (B). ▴, log CFU per milliliter; •, fructose (grams per liter) (A) or oligofructose (grams per liter) (B); □, succinic acid (grams per liter); ○, acetic acid (grams per liter); ▪, propionic acid (grams per liter).
B. thetaiotaomicron LMG 11262 metabolized oligofructose as fast as fructose (Fig. 1B). Within 15 h of fermentation, all oligofructose (15 g liter−1) was metabolized, corresponding to cell growth of 1.9 log CFU ml−1. Succinic acid was again the main metabolite produced throughout fermentation, at a concentration of 9.3 ± 0.1 g liter−1 after 15 h of fermentation. Besides succinic acid, acetic acid and propionic acid were produced at concentrations of 3.1 ± 0.1 g liter−1 and 77 ± 5 mg liter−1, respectively.
Growth of Bacteroides fragilis LMG 10263 on fructose or oligofructose.
B. fragilis LMG 10263 metabolized all fructose (15 g liter−1) within 24 h of fermentation (Fig. 2A), with an accompanying cell growth of 1.9 log CFU ml−1. Succinic acid was the main metabolite produced, at a concentration of 7.5 ± 0.1 g liter−1 after 24 h of fermentation. Acetic acid and propionic acid were produced at concentrations of 2.5± 0.1 g liter−1 and 75 ± 3 mg liter−1, respectively.
FIG. 2.
Fermentation of B. fragilis LMG 10263 with 15 g liter−1 fructose (A) or 15 g liter−1 oligofructose (RaftiloseP95) (B). ▴, log CFU per milliliter; •, fructose (grams per liter) (A) or oligofructose (grams per liter) (B); □, succinic acid (grams per liter); ○, acetic acid (grams per liter); ▪, propionic acid (grams per liter).
Fermentation with oligofructose as the sole energy source showed a pattern similar to that with fructose (Fig. 2b). Within 18 h of fermentation, all oligofructose (15 g liter−1) was metabolized, corresponding to cell growth of 2.0 log CFU ml−1. Succinic acid was again the main metabolite produced throughout fermentation with a concentration of 8.7 ± 0.2 g liter−1 after 18 h of fermentation. Acetic acid and propionic acid were produced at concentrations of 3.3 ± 0.1 g liter−1 and 121 ± 5 mg liter−1, respectively.
Growth of Bifidobacterium longum BB536 on fructose or oligofructose.
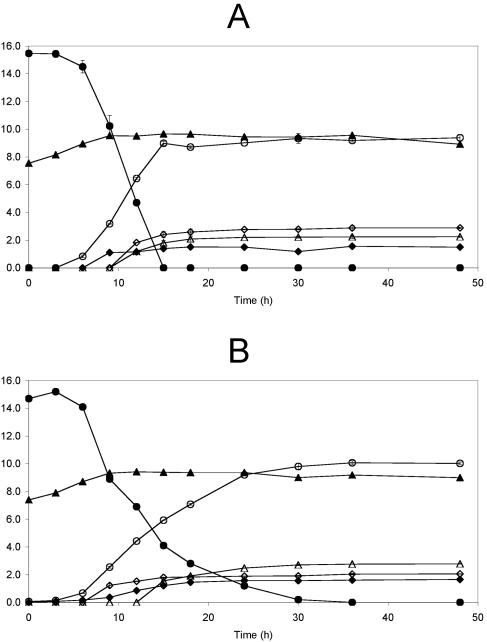
All fructose (15 g liter−1) was fermented by B. longum BB536 within 15 h of fermentation, corresponding to cell growth of 2.1 log CFU ml−1 (Fig. 3A). The most important sugar metabolite produced was acetic acid, at a concentration of 9.0 ± 0.1 g liter−1. Besides acetic acid, other metabolites were also produced, such as lactic acid, formic acid, and ethanol, at concentrations of 2.4 ± 0.2 g liter−1, 1.8 ± 0.2 g liter−1, and 1.4 ± 0.1 g liter−1, respectively.
FIG. 3.
Fermentation of B. longum BB536 with 15 g liter−1 fructose (A) or 15 g liter−1 oligofructose (RaftiloseP95) (B). ▴, log CFU per milliliter; •, fructose (g liter−1) (A) or oligofructose (g liter−1) (B); ○, acetic acid (grams per liter); ⋄, lactic acid (grams per liter); ⧫, ethanol (grams per liter); ▵, formic acid (grams per liter).
The growth of B. longum BB536 on oligofructose was slower than on fructose (Fig. 3B). After 36 h of fermentation, all oligofructose (15 g liter−1) was metabolized, with an accompanying cell growth of 1.8 log CFU ml−1. Acetic acid was the main metabolite produced throughout the fermentation, at a concentration of 10.1 ± 0.1 g liter−1 after 36 h of fermentation. Lactic acid was produced mainly at the beginning of the fermentation, reaching a concentration of 1.8 ± 0.1 g liter−1 after 15 h of fermentation. Formic acid production started when lactic acid production stagnated. After 36 h of fermentation, a concentration of 2.8 ± 0.1 g liter−1 of formic acid was produced. Although the concentration of ethanol increased throughout the whole fermentation course, most of it was produced simultaneously with formic acid, the former reaching a concentration of 1.7 ± 0.1 g liter−1.
Fructan analysis.
The analysis of the oligofructose samples for the fermentation with B. thetaiotaomicron LMG 11262 showed that during the first 9 h of fermentation, oligofructose was almost completely degraded (Table 1). At the same time, an increase in the concentrations of fructose and F2 was observed. This was the result of the breakdown of the longer oligofructose fractions, e.g., F4, GF4, and F5. The concentration of free fructose even increased to 5.9 g liter−1. From 9 to 15 h of fermentation, fructose was the main energy source, and degradation of the remaining oligofructose into fructose continued further. B. thetaiotaomicron LMG 11262 did not show any preference for degradation of one or more oligofructose fractions, since all of them were degraded from the start of the fermentation.
TABLE 1.
Fructan degradation during the growth of Bacteroides thetaiotaomicron LMG 11262 on oligofructose as 1.5% (wt/vol) RaftiloseP95
| Componenta | Concn (g liter−1) at time (h)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 3 | 6 | 9 | 12 | 15 | 24 | 48 | |
| F | 0.90 | 0.80 | 5.93 | 3.39 | 0.12 | 0.00 | 0.00 | 0.00 |
| G | 0.30 | 0.20 | 0.10 | 0.33 | 0.00 | 0.00 | 0.00 | 0.00 |
| GF | 0.10 | 0.10 | 0.30 | 0.11 | 0.00 | 0.00 | 0.00 | 0.00 |
| F2 | 0.70 | 0.70 | 2.11 | 0.11 | 0.12 | 0.00 | 0.00 | 0.00 |
| GF2 | 0.10 | 0.10 | 0.70 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| F3 | 3.60 | 3.52 | 0.90 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| GF3 | 0.30 | 0.30 | 0.10 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| F4 | 3.80 | 3.62 | 0.80 | 0.11 | 0.00 | 0.00 | 0.00 | 0.00 |
| GF4 | 0.90 | 0.80 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| F5 | 1.50 | 1.51 | 0.30 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| GF5 | 0.70 | 0.60 | 0.10 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| F6 | 1.00 | 1.00 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| GF6 | 0.40 | 0.40 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| F7 | 0.40 | 0.40 | 0.10 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
F, fructose; G, glucose.
The fermentation with B. fragilis LMG 10263 showed similar results concerning oligofructose degradation (Table 2). During the first 9 h of fermentation, a large part of oligofructose (>85%) was degraded, resulting in a large concentration of free fructose (5.4 g liter−1) in the fermentation medium. From 9 to 15 h of fermentation, the remaining oligofructose was further degraded, and the free fructose was metabolized. During the degradation of oligofructose, no specific preferences for degrading certain oligofructose fractions could be detected.
TABLE 2.
Fructan degradation during the growth of Bacteroides fragilis LMG 10263 on oligofructose as 1.5% (wt/vol) RaftiloseP95
| Componenta | Concn (g liter−1) at time (h)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 3 | 6 | 9 | 12 | 15 | 24 | 48 | |
| F | 0.90 | 1.11 | 3.12 | 5.38 | 1.81 | 0.12 | 0.00 | 0.00 |
| G | 0.30 | 0.10 | 0.00 | 0.11 | 0.00 | 0.00 | 0.00 | 0.00 |
| GF | 0.10 | 0.10 | 0.10 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| F2 | 0.70 | 1.01 | 1.96 | 0.43 | 0.12 | 0.00 | 0.00 | 0.00 |
| GF2 | 0.10 | 0.10 | 0.30 | 0.11 | 0.00 | 0.00 | 0.00 | 0.00 |
| F3 | 3.40 | 3.12 | 0.91 | 0.11 | 0.00 | 0.00 | 0.00 | 0.00 |
| GF3 | 0.30 | 0.30 | 0.10 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| F4 | 3.60 | 3.22 | 1.21 | 0.11 | 0.00 | 0.00 | 0.00 | 0.00 |
| GF4 | 0.80 | 0.70 | 0.50 | 0.11 | 0.00 | 0.00 | 0.00 | 0.00 |
| F5 | 1.50 | 1.41 | 0.80 | 0.22 | 0.00 | 0.00 | 0.00 | 0.00 |
| GF5 | 0.60 | 0.60 | 0.50 | 0.22 | 0.00 | 0.00 | 0.00 | 0.00 |
| F6 | 1.00 | 1.01 | 0.80 | 0.32 | 0.12 | 0.00 | 0.00 | 0.00 |
| GF6 | 0.40 | 0.40 | 0.30 | 0.22 | 0.00 | 0.00 | 0.00 | 0.00 |
| F7 | 0.40 | 0.30 | 0.30 | 0.22 | 0.00 | 0.00 | 0.00 | 0.00 |
F, fructose; G, glucose.
The growth of B. longum BB536 on oligofructose was slower than both Bacteroides strains, as the time needed for complete degradation of oligofructose was twice as long for the Bifidobacterium strain as for the Bacteroides strains. Detailed analysis of oligofructose degradation revealed that the growth of B. longum BB536 during the first 9 h of fermentation was the result of the metabolism of mainly fructose, F2, and F3 (Table 3). When these components were almost depleted, the strain started to metabolize the longer oligofructose fractions, such as F4, GF4, F5, F6, and others. During this degradation, hardly any increase in fructose concentration was observed, indicating the immediate consumption of the oligofructose degradation products. After 36 h of fermentation, all oligofructose (15 g liter−1) was metabolized. This indicates that the Bifidobacterium strain degraded the oligofructose much more slowly than either Bacteroides strain, probably due to its sequential consumption of the different fractions.
TABLE 3.
Fructan degradation during the growth of Bifidobacterium longum BB536 on oligofructose as 1.5% (wt/vol) RaftiloseP95
| Componenta | Concn (g liter−1) at time (h)
|
|||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 3 | 6 | 9 | 12 | 15 | 24 | 48 | |
| F | 0.90 | 0.90 | 0.71 | 0.10 | 0.23 | 0.24 | 0.25 | 0.00 |
| G | 0.30 | 0.20 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 |
| GF | 0.10 | 0.10 | 0.10 | 0.10 | 0.00 | 0.00 | 0.12 | 0.00 |
| F2 | 0.60 | 0.60 | 0.51 | 0.10 | 0.12 | 0.12 | 0.12 | 0.00 |
| GF2 | 0.10 | 0.10 | 0.10 | 0.00 | 0.00 | 0.12 | 0.00 | 0.00 |
| F3 | 3.70 | 3.81 | 3.34 | 0.52 | 0.23 | 0.24 | 0.00 | 0.00 |
| GF3 | 0.30 | 0.40 | 0.40 | 0.31 | 0.46 | 0.36 | 0.12 | 0.00 |
| F4 | 3.80 | 4.01 | 3.94 | 3.46 | 2.55 | 1.30 | 0.37 | 0.00 |
| GF4 | 0.80 | 0.90 | 0.91 | 0.84 | 0.81 | 0.12 | 0.12 | 0.00 |
| F5 | 1.50 | 1.60 | 1.62 | 1.47 | 1.39 | 0.83 | 0.12 | 0.00 |
| GF5 | 0.70 | 0.70 | 0.71 | 0.63 | 0.58 | 0.47 | 0.12 | 0.00 |
| F6 | 1.10 | 1.10 | 1.11 | 1.05 | 0.93 | 0.59 | 0.12 | 0.00 |
| GF6 | 0.40 | 0.40 | 0.40 | 0.31 | 0.35 | 0.24 | 0.00 | 0.00 |
| F7 | 0.40 | 0.40 | 0.40 | 0.42 | 0.35 | 0.24 | 0.00 | 0.00 |
F, fructose; G, glucose.
DISCUSSION
Bacteroides species are considered to be predominant in the human colon (34). Growth experiments using fecal slurries or in vivo clinical trials showed that inulin-type fructans do not display a stimulatory effect on members of the Bacteroides group, although Bacteroides species possibly degrade these prebiotics (1, 24, 31). One in vitro study mentioned the capability of a B. thetaiotaomicron strain to grow on inulin-type fructans, but this strain grew better on glucose than on oligofructose (14). As commercial prebiotics were used in the latter study, it is not clear if the presence of contaminating sugars, due to the production process of the manufacturer, was taken into account. Indeed, detailed kinetic analysis of the growth of different important human commensals on the commercial prebiotic RaftiloseP95 showed that the presence of contaminating sugars (e.g., glucose, fructose, and sucrose) cannot be neglected during in vitro studies (reference 45 and this study). These sugars may support limited growth of the strain(s) under study, an observation that is often misinterpreted as the presence of growth on the oligosaccharide used. Therefore, care has to be taken not to draw wrong conclusions from such pure culture growth experiments, and efforts should be made to quantify the oligosaccharides (45).
Our in vitro study shows that pure cultures of B. thetaiotaomicron LMG 11262 and B. fragilis LMG 10263 are capable of degrading and growing on oligofructose and that this growth appeared to be at least as good as growth on fructose (this study) and glucose (R. Van der Meulen and L. De Vuyst, unpublished results). Moreover, these strains grew much better on oligofructose than did B. longum BB536. One explanation could be that B. thetaiotaomicron and B. fragilis have more potential to grow on a large variety of substrates, oligosaccharides in particular, than B. longum has, as revealed by information from their genomes (22, 39, 48). Putting together the facts and uncertainties about the colon microbiota and its metabolism, in vitro experiments with pure bacterial cultures indicate that bacteria other than bifidobacteria might also grow on oligofructose, either directly or indirectly through cross-feeding on metabolites of prebiotic degraders (7, 8). However, caution is needed when extrapolating findings from in vitro tests to the in vivo situation, since the selective and stimulatory effect of oligofructose on bifidobacteria has been demonstrated in numerous human clinical trials (11, 24, 30, 33, 44, 47).
Hydrolysis of the longer oligofructose fractions by B. thetaiotaomicron LMG 11262 and B. fragilis LMG 10263 resulted in the release of free fructose in the fermentation medium. Although the genome information does not reveal the presence of an inulinase or β-fructofuranosidase for both Bacteroides strains (22, 48), a cell wall-attached fructanase capable of hydrolyzing both inulin-type and levan-type fructans has already been isolated from a B. fragilis strain (4). Fructanase action will result in the simultaneous consumption of fructose and oligofructose fractions by a nonspecific microbiota, as there are different species, e.g., Bacteroides spp., capable of metabolizing these simple and complex sugars. In contrast with the Bacteroides strains, B. longum BB536 showed a preferential metabolism of the short oligofructose fractions (both the F2 and F3 fractions of oligofructose). After depletion of the contaminating monosaccharides (glucose and fructose) present in the commercial prebiotic (RaftiloseP95) used and of the short F2 and F3 fractions of oligofructose, the longer oligofructose fractions (e.g., F4, GF4, F5, and F6) were metabolized as well. A similar preferential metabolism of the short oligofructose fractions has already been described for Bifidobacterium animalis subsp. lactis DN-173 010 (45). Hence, this preferential metabolism of shorter oligofructose fractions appears to be a specific characteristic of bifidobacteria.
There are several possible explanations why a preferential metabolism of the short oligofructose fractions is observed for the B. longum strain and other bifidobacteria (45), whereas the Bacteroides strains tested did not display such a preferential metabolism in this in vitro setup. A first explanation is the expression and localization of the enzyme responsible for oligofructose degradation. This enzyme is probably expressed constitutively and located extracellularly or attached to the cell wall in the case of the Bacteroides strains (4), whereas for the Bifidobacterium strains this enzyme is inducible and located intracellularly (9, 14, 17, 32). Consequently, both Bacteroides strains degraded oligofructose extracellularly, thereby releasing fructose in the culture medium. This is supported by our kinetic analysis of oligofructose consumption, which clearly showed a large increase in free fructose during oligofructose breakdown for both Bacteroides strains, in contrast with the Bifidobacterium fermentations (reference 45 and this study). This suggests that the bifidobacteria degrade oligofructose intracellularly and metabolize the released fructose moieties simultaneously. The necessary uptake of oligofructose prior to hydrolysis may also explain the slower growth of B. longum strain BB536 on oligofructose than on fructose, while the Bacteroides strains metabolized oligofructose as fast as fructose in our in vitro test. A second explanation for the differences in oligofructose metabolism may be ascribed to differences between the oligofructose transport system(s) in Bacteroides spp. and Bifidobacterium spp. It has been shown that a strain of B. longum possesses a specific permease type of transporter for oligofructose, although an ABC type or phosphotransferase-type transporter is more common for other bacterial genera (3, 39). However, the oligofructose uptake mechanism has not yet been studied for Bacteroides strains. It is likely that Bacteroides strains will not take full profit from an extracellular degradation because of the loss of oligofructose digestion products to other commensals, e.g., bifidobacteria. This may explain why in vivo studies do not reveal a clear stimulatory effect of oligofructose on Bacteroides spp. (1, 24). The presence of specific transport systems for short oligofructose fractions may give bifidobacteria the surplus needed to compete with other commensals for this substrate, besides their acidification through the production of lactic acid, acetic acid, and formic acid. The acidic pH in the proximal colon, where most of the undigested carbohydrates are fermented, may inhibit the growth of certain commensals (e.g., Bacteroides), while bifidobacteria are more acid tolerant and hence less affected.
Succinic acid was the main metabolite produced throughout all fermentations with B. thetaiotaomicron LMG 11262 and B. fragilis LMG 10263. These results are in agreement with previous in vitro studies that show that succinic acid is the main metabolite for Bacteroides spp. if generation times are short (21, 25), which was the case in our study. However, generation times will be much longer in the human colon, due to limited substrate availability; propionic acid will probably be the main metabolite for Bacteroides strains in the human colon (21, 36). Due to the low generation times and the sparging of the medium with CO2 during the in vitro experiments, propionic acid was produced only in very small amounts by both Bacteroides strains. The increased production of acetic acid, formic acid, and ethanol at the cost of lactic acid confirms the switch of metabolism of bifidobacteria at lower growth rates to obtain a higher amount of ATP per mole of sugar consumed (27, 45).
To conclude, this study has shown for the first time that pure cultures of two Bacteroides strains were able to degrade oligofructose. Both strains grew as fast on fructose as they did on oligofructose. The latter was not the case for a Bifidobacterium strain that grew much slower on oligofructose than the Bacteroides strains. The Bifidobacterium strain completely metabolized oligofructose with a clear preference for the shorter oligofructose fractions (e.g., F2 and F3), probably due to the presence of appropriate uptake systems. Both Bacteroides strains did not show such a preferential metabolism and simultaneously degraded the different oligofructose fractions extracellularly, transiently increasing the fructose concentration of the fermentation medium. These results indicate important differences in oligofructose degradation kinetics between dominant members of the human colon. This in vitro study further contributes to the understanding of the bifidogenic properties of inulin-type fructans.
Acknowledgments
This work was supported by the Research Council of the Vrije Universiteit Brussel, the Fund for Scientific Research—Flanders, the European Community (Quality of Life grant QLK1-CT-2001-01179), and the Flemish Institute for the Encouragement of Scientific and Technological Research in the Industry (GBOU project 10054).
Further, we thank the people of the analytical laboratory of ORAFTI N.V. (Tiense Suikerraffinaderij, Tienen, Belgium).
REFERENCES
- 1.Apajalahti, J. H. A., H. Kettunen, A. Kettunen, W. E. Holben, P. H. Nurminen, N. Rautonen, and M. Mutanen. 2002. Culture-independent microbial community analysis reveals that inulin in the diet primarily affects previously unknown bacteria in the mouse cecum. Appl. Environ. Microbiol. 68:4986-4995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ballongue, J. 1998. Bifidobacteria and probiotic action, p. 519-587. In S. Salminen and A. von Wright (ed.), Lactic acid bacteria: microbiology and functional aspects. Marcel Dekker, Inc., New York, N.Y.
- 3.Barrangou, R., E. Altermann, R. Hutkins, R. Cano, and T. R. Klaenhammer. 2003. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc. Natl. Acad. Sci. USA 100:8957-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blatch, G. L., and D. R. Woods. 1993. Molecular characterization of a fructanase produced by Bacteroides fragilis Bf-1. J. Bacteriol. 175:3058-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Comstock, L. E., and M. J. Coyne. 2003. Bacteroides thetaiotaomicron: a dynamic, niche-adapted human symbiont. BioEssays 25:926-929. [DOI] [PubMed] [Google Scholar]
- 6.De Leenheer, L. 1996. Production and use of inulin: industrial reality with a promising future, p. 67-92. In H. Van Bekkum, H. Röper, and F. Voragen (ed.), Carbohydrates as organic raw materials, vol. 3. VCH Publishing Inc., New York, N.Y. [Google Scholar]
- 7.Duncan, S. H., P. Louis, and H. J. Flint. 2004. Lactate-utilizing bacteria, isolated from human feces, that produce butyrate as a major fermentation product. Appl. Environ. Microbiol. 70:5810-5817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan, S. H., K. P. Scott, A. G. Ramsay, H. J. M. Harmsen, G. W. Welling, C. S. Stewart, and H. J. Flint. 2003. Effects of alternative dietary substrates on competition between human colonic bacteria in an anaerobic fermentor system. Appl. Environ. Microbiol. 69:1136-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrmann, M. A., M. Korakli, and R. F. Vogel. 2003. Identification of the gene for beta-fructofuranosidase of Bifidobacterium lactis DSM 10140T and characterization of the enzyme expressed in Escherichia coli. Curr. Microbiol. 46:391-397. [DOI] [PubMed] [Google Scholar]
- 10.Franks, A. H., H. J. M. Harmsen, G. C. Raangs, G. J. Jansen, F. Schut, and G. W. Welling. 1998. Variations of bacterial populations in human feces measured by fluorescent in situ hybridization with group-specific 16S rRNA-targeted oligonucleotide probes. Appl. Environ. Microbiol. 64:3336-3345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gibson, G. R., E. R. Beatty, X. Wang, and J. H. Cummings. 1995. Selective stimulation of bifidobacteria in the human colon by oligofructose and inulin. Gastroenterology 108:975-982. [DOI] [PubMed] [Google Scholar]
- 12.Gibson, G. R., H. M. Probert, J. Van Loo, R. A. Rastall, and M. B. Roberfroid. 2004. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr. Res. Rev. 17:259-275. [DOI] [PubMed] [Google Scholar]
- 13.Gibson, G. R., and M. B. Roberfroid. 1995. Dietary modulation of the human colonic microbiota—introducing the concept of prebiotics. J. Nutr. 125:1401-1412. [DOI] [PubMed] [Google Scholar]
- 14.Gibson, G. R., and X. Wang. 1994. Bifidogenic properties of different types of fructo-oligosaccharides. Food Microbiol. 11:491-498. [Google Scholar]
- 15.Hartemink, R., K. M. J. Van Laere, and F. M. Rombouts. 1997. Growth of enterobacteria on fructo-oligosaccharides. J. Appl. Microbiol. 83:367-374. [DOI] [PubMed] [Google Scholar]
- 16.Hooper, L. V., T. Midtvedt, and J. I. Gordon. 2002. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu. Rev. Nutr. 22:283-307. [DOI] [PubMed] [Google Scholar]
- 17.Imamura, L., K. Hisamitsu, and K. Kobashi. 1994. Purification and characterization of beta-fructofuranosidase from Bifidobacterium infantis. Biol. Pharmacol. Bull. 17:596-602. [DOI] [PubMed] [Google Scholar]
- 18.Joye, D., and H. Hoebregs. 2000. Determination of oligofructose, a soluble dietary fiber, by high-temperature capillary gas chromatography. J. AOAC Int. 83:1020-1025. [PubMed] [Google Scholar]
- 19.Kaplan, H., and R. W. Hutkins. 2000. Fermentation of fructooligosaccharides by lactic acid bacteria and bifidobacteria. Appl. Environ. Microbiol. 66:2682-2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kleessen, B., E. Bezirtzoglou, and J. Mättö. 2000. Culture-based knowledge on biodiversity, development and stability of human gastrointestinal microflora. Microb. Ecol. Health Dis. Suppl. 2:53-63. [Google Scholar]
- 21.Kotarski, S. F., and A. A. Salyers. 1981. Effect of long generation times on growth of Bacteroides thetaiotaomicron in carbohydrate-limited continuous culture. J. Bacteriol. 146:853-860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kuwahara, T., A. Yamashita, H. Hirakawa, H. Nakayama, H. Toh, N. Okada, S. Kuhara, M. Hattori, T. Hayashi, and Y. Ohnishi. 2004. Genomic analysis of Bacteroides fragilis reveals extensive DNA inversions regulating cell surface adaptation. Proc. Natl. Acad. Sci. USA 101:14919-14924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Langendijk, P. S., F. Schut, G. J. Jansen, G. C. Raangs, G. R. Kamphuis, M. H. F. Wilkinson, and G. W. Welling. 1995. Quantitative fluorescence in-situ hybridization of Bifidobacterium spp. with genus-specific 16S ribosomal-RNA-targeted probes and its application in fecal samples. Appl. Environ. Microbiol. 61:3069-3075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langlands, S. J., M. J. Hopkins, N. Coleman, and J. H. Cummings. 2004. Prebiotic carbohydrates modify the mucosa associated microflora of the human large bowel. Gut 53:1610-1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macy, J. M., L. G. Ljungdahl, and G. Gottschalk. 1978. Pathway of succinate and propionate formation in Bacteroides fragilis. J. Bacteriol. 134:84-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makras, L., G. Van Acker, and L. De Vuyst. 2005. Lactobacillus paracasei subsp. paracasei 8700:2 degrades inulin-type fructans of different degrees of polymerization. Appl. Environ. Microbiol. 71:6531-6537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marx, S. P., S. Winkler, and W. Hartmeier. 2000. Metabolization of beta-(2,6)-linked fructose-oligosaccharides by different bifidobacteria. FEMS Microbiol. Lett. 182:163-169. [DOI] [PubMed] [Google Scholar]
- 28.Matsuki, T., K. Watanabe, J. Fujimoto, Y. Miyamoto, T. Takada, K. Matsumoto, H. Oyaizu, and R. Tanaka. 2002. Development of 16S rRNA-gene-targeted group-specific primers for the detection and identification of predominant bacteria in human feces. Appl. Environ. Microbiol. 68:5445-5451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matsuki, T., K. Watanabe, J. Fujimoto, T. Takada, and R. Tanaka. 2004. Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl. Environ. Microbiol. 70:7220-7228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Menne, E., N. Guggenbuhl, and M. Roberfroid. 2000. Fn-type chicory inulin hydrolysate has a prebiotic effect in humans. J. Nutr. 130:1197-1199. [DOI] [PubMed] [Google Scholar]
- 31.Palframan, R. J., G. R. Gibson, and R. A. Rastall. 2002. Effect of pH and dose on the growth of gut bacteria on prebiotic carbohydrates in vitro. Anaerobe 8:287-292. [DOI] [PubMed] [Google Scholar]
- 32.Perrin, S., M. Warchol, J. P. Grill, and F. Schneider. 2001. Fermentations of fructo-oligosaccharides and their components by Bifidobacterium infantis ATCC 15697 on batch culture in semi-synthetic medium. J. Appl. Microbiol. 90:859-865. [DOI] [PubMed] [Google Scholar]
- 33.Rao, V. A. 2001. The prebiotic properties of oligofructose at low intake levels. Nutr. Res. 21:843-848. [Google Scholar]
- 34.Rigottier-Gois, L., V. Rochet, N. Garrec, A. Suau, and J. Doré. 2003. Enumeration of Bacteroides species in human faeces by fluorescent in situ hybridisation combined with flow cytometry using 16S rRNA probes. Syst. Appl. Microbiol. 26:110-118. [DOI] [PubMed] [Google Scholar]
- 35.Roberfroid, M. 1993. Dietary fiber, inulin, and oligofructose—a review comparing their physiological effects. Crit. Rev. Food Sci. Nutr. 33:103-148. [DOI] [PubMed] [Google Scholar]
- 36.Salyers, A. A. 1984. Bacteroides of the human lower intestinal tract. Annu. Rev. Microbiol. 38:293-313. [DOI] [PubMed] [Google Scholar]
- 37.Salyers, A. A., A. Gupta, and Y. P. Wang. 2004. Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12:412-416. [DOI] [PubMed] [Google Scholar]
- 38.Salyers, A. A., J. R. Vercellotti, S. E. H. West, and T. D. Wilkins. 1977. Fermentation of mucin and plant polysaccharides by strains of Bacteroides from the human colon. Appl. Environ. Microbiol. 33:319-322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schell, M. A., M. Karmirantzou, B. Snel, D. Vilanova, B. Berger, G. Pessi, M. C. Zwahlen, F. Desiere, P. Bork, M. Delley, R. D. Pridmore, and F. Arigoni. 2002. The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract. Proc. Natl. Acad. Sci. USA 99:14422-14427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shanahan, F. 2002. The host-microbe interface within the gut. Best Pract. Res. Clin. Gastroenterol. 16:915-931. [DOI] [PubMed] [Google Scholar]
- 41.Shoemaker, N. B., H. Vlamakis, K. Hayes, and A. A. Salyers. 2001. Evidence for extensive resistance gene transfer among Bacteroides spp. and among Bacteroides and other genera in the human colon. Appl. Environ. Microbiol. 67:561-568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Suau, A., R. Bonnet, M. Sutren, J. J. Godon, G. R. Gibson, M. D. Collins, and J. Doré. 1999. Direct analysis of genes encoding 16S rRNA from complex communities reveals many novel molecular species within the human gut. Appl. Environ. Microbiol. 65:4799-4807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tamura, Z. 1983. Nutriology of bifidobacteria. Bifidobacteria Microflora 2:3-16. [Google Scholar]
- 44.Tuohy, K. M., R. K. Finlay, A. G. Wynne, and G. R. Gibson. 2001. A human volunteer study on the prebiotic effects of HP-inulin-faecal bacteria enumerated using fluorescent in situ hybridization (FISH). Anaerobe 7:113-118. [Google Scholar]
- 45.Van der Meulen, R., L. Avonts, and L. De Vuyst. 2004. Short fractions of oligofructose are preferentially metabolized by Bifidobacterium animalis DN-173 010. Appl. Environ. Microbiol. 70:1923-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vanhoutte, T., G. Huys, E. De Brandt, and J. Swings. 2004. Temporal stability analysis of the microbiota in human feces by denaturing gradient gel electrophoresis using universal and group-specific 16S rRNA gene primers. FEMS Microbiol. Ecol. 48:437-446. [DOI] [PubMed] [Google Scholar]
- 47.Van Loo, J., J. Cummings, N. Delzenne, H. Englyst, A. Franck, M. Hopkins, N. Kok, G. Macfarlane, D. Newton, M. Quigley, M. Roberfroid, T. van Vliet, and E. van den Heuvel. 1999. Functional food properties of non-digestible oligosaccharides: a consensus report from the ENDO project (DGXII AIRII-CT94-1095). Br. J. Nutr. 81:121-132. [DOI] [PubMed] [Google Scholar]
- 48.Xu, J., M. K. Bjursell, J. Himrod, S. Deng, L. K. Carmichael, H. C. Chiang, L. V. Hooper, and J. I. Gordon. 2003. A genomic view of the human-Bacteroides thetaiotaomicron symbiosis. Science 299:2074-2076. [DOI] [PubMed] [Google Scholar]
- 49.Zoetendal, E. G., A. D. L. Akkermans, and W. M. De Vos. 1998. Temperature gradient gel electrophoresis analysis of 16S rRNA from human fecal samples reveals stable and host-specific communities of active bacteria. Appl. Environ. Microbiol. 64:3854-3859. [DOI] [PMC free article] [PubMed] [Google Scholar]