Abstract
Plants are naturally colonized by many fungal species that produce effects ranging from beneficial to pathogenic. However, how many of these fungi are linked with a single host plant has not been determined. Furthermore, the composition of plant-associated fungal communities has not been rigorously determined. We investigated these essential issues by employing the perennial wetland reed Phragmites australis as a model. DNA extracted from roots, rhizomes, stems, and leaves was used for amplification and cloning of internal transcribed spacer rRNA gene fragments originating from reed-associated fungi. A total of 1,991 clones from 15 clone libraries were differentiated by restriction fragment length polymorphism analyses into 345 operational taxonomical units (OTUs). Nonparametric estimators for total richness (Chao1 and ACE) and also a parametric log normal model predicted a total of about 750 OTUs if the libraries were infinite. Sixty-two percent of the OTUs sequenced were novel at a threshold of 3%. Several of these OTUs represented undocumented fungal species, which also included higher taxonomic levels. In spite of the high diversity of the OTUs, the mycofloras of vegetative organs were dominated by just a few typical fungi, which suggested that competition and niche differentiation influence the composition of plant-associated fungal communities. This suggestion was independently supported by the results of nested PCR assays specifically monitoring two OTUs over 3 years, which revealed significant preferences for host habitat and host organ.
The introduction of cultivation-independent, molecular approaches for assessing microbial diversity has had a tremendous impact on bacterial taxonomy and ecology. Many studies have documented that only a small fraction of bacteria present in a given habitat is retrievable by current cultivation techniques. For example, the magnitude of the bacterial diversity in just 1 g of soil has been estimated to be at least 6,400 different operational taxonomical units (OTUs) (7). Most cultivation-independent assessments of bacterial diversity have relied on phylogenetic analysis of rRNA genes that were directly cloned from environmental samples originating from various habitats. Such efforts have increased the number of known bacterial phyla from 12 in 1987 to 52 now (23).
The estimates of the total number of fungal species on Earth are about 1.5 million species, whereas the number of species that have been described is just about 7% of this number (13, 14). This calculation does not include species known only on the basis of the rRNA gene sequence. In contrast to the situation in bacteria, cultivation-independent, molecular methods have just recently been used to determine broad fungal diversity. Recent investigations of fungal communities in soil and plant roots indicated that the diversity is high and resulted in the discovery of novel fungal lineages at higher taxonomic levels (28, 36).
An important subject in ecology is the structure of communities. Recent evidence suggests that for bacterial communities a few species are not necessarily dominant over many other species that are also present in a habitat. Dominance indicates that there is competition for resources, so that only species that are well adapted to a given habitat can become more abundant. Molecular studies have suggested that in bacterial communities there can also be uniform species distributions and that a vast number of species can coexist, each at a low level (31, 44). For fungi, recent evidence has indicated that the mycofloras associated with plants may be highly diverse (10, 28, 36). However, in contrast to the situation in bacteria, practically no quantitative molecular data for assessing the structures of entire fungal communities are available, and which forces shape the structures is not known.
We use the common reed [Phragmites australis (Cav.) Trin. ex Steudel] as a model to study the diversity of plant-associated fungi and the structure of the fungal community. This perennial reed colonizes shallow shores of freshwater and brackish water habitats, often forming homogeneous reed belts. Propagation is mostly by rhizomes that send up new shoots each spring. Seeds are important only for colonizing new sites. Previously, we used a cultivation approach to assess the diversity of fungi living in close association with P. australis growing at Lake Constance in Germany (41). A total of 322 isolates were grouped according to morphology. Sequence analysis of the internal transcribed spacer (ITS) region, which is part of the rRNA gene cluster, indicated that these isolates belonged to at least 17 genera. This previous work and follow-up studies of two distinct genera, Cladosporium and Stagonospora, revealed that at least 30 fungal species colonized healthy P. australis (9, 41, 42). Our investigations were complemented by a PCR-based study targeting most of the currently known taxa in the Glomeromycota (40), which are the arbuscular mycorrhiza fungi found in the roots of many land plants. Twenty-one putative arbuscular mycorrhiza fungus species were observed within the range theoretically detectable by the approach used. This provided the first clue about how much of the total fungal diversity present in the habitats investigated might have been missed by previous cultivation-based studies and encouraged us to conduct a comprehensive molecular investigation.
In the work described in this paper, we examined the community structure and the diversity of fungi associated with P. australis by using an rRNA gene cloning approach. We targeted the fungal ITS region within the rRNA gene cluster since it is more variable than the 18S or 28S rRNA genes. For fungi, ITS sequences can distinguish species so that diversity and community structure can be analyzed at this level. By using cloning, restriction fragment length polymorphism (RFLP) typing, and sequence analysis of fungal ITS fragments originating from vegetative organs of the reed, we addressed the following questions. How many fungi can be associated with a single host species? How many of these fungi may belong to undocumented taxa and at what level? What is the general structure of reed-associated fungal communities?
MATERIALS AND METHODS
Sample collection and DNA extraction.
The ITS clone libraries originated from DNA extracts of the common reed (P. australis) that was harvested at the seasonal biomass peak on 9 August 2001 at Mainau Bay of Lake Constance (Germany). Details concerning the location have been described previously (41). Two contrasting sites were sampled; one site was on the lakeward front of the reed belt and was flooded (“flooded site”), and the other was on the landward side and was not flooded (“dry site”). At each site, three individual plants that were not more than 5 m apart were dug out. Within the next 3 h, the plants were divided by organ, intensively cleaned under running tap water, washed in sterile distilled water, and stored at −20°C until DNA isolation. After thawing, the samples were again vigorously washed, and several small tissue samples were taken from each organ of an individual plant and pooled. We used parts from the middle of the rhizome, 5- to 10-cm-long pieces from three to five roots, 2- to 3-cm-long pieces from the top, middle, and bottom of the stems, and leaves that were connected to the stem pieces. The pools were homogenized with a mixer mill (MM 300; QIAGEN GmbH, Hilden, Germany), and 100-mg aliquots were used for DNA extraction using a DNeasy plant mini kit (QIAGEN). DNA quality was controlled as described previously (42).
Template DNAs used in nested PCRs to monitor the occurrence of fungi at the two sites described above and two additional sites that were about 7 km from the sites described above (one dry and the other flooded) were isolated previously (9). The additional sites have been described previously (41).
Amplification and cloning of fungal ITS rRNA gene.
The PCR mixtures (total volume, 50 μl) contained primers ITS1F and ITS4 targeting fungi (0.3 μM each) (11, 39), 2 mM MgCl2, 0.5 μg/μl bovine serum albumin, each deoxynucleoside triphosphate at a concentration of 0.2 mM, 1× reaction buffer, 0.4 U/μl Expand High Fidelity PCR polymerase (Roche Diagnostics GmbH, Mannheim, Germany), and about 50 ng of template DNA. The cycling conditions were as follows: 94°C for 150 s, followed by 29 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 45 s plus one additional second per cycle and then a final extension at 72°C for 15 min. PCR fragments were purified with an EZNA Cycle-Pure kit (Peqlab Biotechnology GmbH, Erlangen, Germany) and cloned with a PCR cloning kit (QIAGEN) used according to the instructions in the supplied manuals. For each template DNA, duplicate libraries from independent PCRs were created and pooled.
Inserts were amplified from recombinant clones and digested separately with restriction enzymes MboI, MspI, and (in some instances) PalI (MBI Fermentas GmbH, St. Leon-Roth, Germany). The resulting RFLP patterns delineated OTUs. Two nonparametric approaches, ACE and Chao1, which weight the abundance of rare taxa differently and were implemented in the software EstimateS, version 6.0b1 (http://viceroy.eeb.uconn.edu/estimates), were used to estimate the “true” richness of the libraries (16). Analysis of diversity indices and prediction of the “true” richness with a truncated lognormal model were carried out with the software SDR, version 3.03 (Pisces Conservation Ltd., Pennington, United Kingdom). This software was also used to fit the experimentally obtained abundance distributions to the distributions from theoretical models. For additional statistical analyses we used tests implemented in the software JMP, version 4.04 (SAS Institute, Cary, NC).
Sequence and phylogenetic analysis.
DNA sequences were generated, assembled, aligned, and edited as described previously (42). We used two approaches to identify putative chimeras. First, we visually checked pairwise alignments with the closest matches in current sequence depositories that were found by BLASTN searches at http://www.ncbi.nlm.nih.gov/BLAST/. These alignments were created by the Martinez-Needleman-Wunsch algorithm implemented in the software package DNAStar (GATC GmbH, Konstanz, Germany). Abrupt changes in similarity within conserved regions indicated that there might be chimeric sequences. Second, we independently submitted the 18S, ITS1, 5.8S, ITS2, and 28S parts of each sequence as queries for BLASTN searches. Ambiguous results were considered a symptom of chimeric sequences. RFLP types were exempted from exclusion if several clones originating from independent libraries were available and were found to have the same sequence.
An alignment of 129 5.8S rRNA gene sequences was created by ClustalX (33) and then manually improved using the EDIT option of the MUST package (22). Potentially unambiguously aligned portions or low-complexity regions of the data were eliminated using the program g-blocks (5). Phylogenetic analyses, including two probabilistic model-based methods, maximum likelihood and Bayesian inference, were performed using the program Treefinder (17) and Bayesian analysis as implemented in MrBayes 3.0b4 (26), respectively. For both approaches the same complex model, GTR+G8+I, was used, where GTR indicates a general time-reversible substitution matrix, G8 indicates gamma-distributed rates with eight discrete categories, and I is the proportion of invariant positions. The Bayesian analysis was performed with default settings. The four-couple chains were run for a total of 3,000,000 generations, sampling one tree every 100 generations and discarding the first 7,500 trees as “burn in,” to ensure that the analysis converged to stable likelihood values. In order to test for consistent results, the MrBayes analyses were repeated three times; however, the results of the runs were too different (for certain bipartitions, >0.5). Therefore, we decided to exclude the MrBayes analyses since they did not converge. For the Treefinder analysis, confidence intervals were obtained by using the LRSH_RELL method, a modified version of the RELL bootstrap approach (18), in which for each node associated with local rearrangements, a Shimodaira-Hasegawa likelihood test (18) is performed for the three alternative topologies at the node. The final support value is 1 minus the worst (highest) probability value (P value) obtained in the Shimodaira-Hasegawa test, expressed as a percentage. A total of 10,000 RELL replicates were performed.
Nested PCR assays.
Two-step nested PCR assays were designed to monitor specifically the presence of two OTUs in field samples. For the first PCR step we used the conditions described above; for the second step we used specific primers and 5 μl of a 400-fold dilution of the first reaction mixture in a 25-μl (total volume) mixture. For OTU Ms7Mb4 the primers were 1365-for (GTGTAAAATAAAAACCTCTGTA) and 1365-rev (CACAGGAGTGAGAAGAATAC), and for OTU Ms43Mb21 the primers were 2526-for (AAACAAACACTGCCTTCTGGAG) and 2526-rev (CTGAGGGTTTTTGAGGACGC). The reaction parameters were 94°C for 150 s, followed by 35 cycles of 94°C for 30 s, 62°C (OTU Ms7Mb4) or 70°C (OTU Ms43Mb21) for 30 s, and 72°C for 45 s plus one additional second per cycle and then a final extension at 72°C for 10 min. The specificity of the assays was assessed by using 1:50 dilutions of PCR products amplified from plasmids carrying fungal ITS fragments. The results of nested PCR assays with field samples were scored as 0 or 1. For statistical analysis we used the JMP software to create a contingency table that compared pairwise scores for all habitat-organ combinations. Significance was determined by a binomial test (P = 0.05).
Nucleotide sequence accession numbers.
Sequences obtained during this study have been deposited in the EMBL database under accessions numbers AJ875338 to AJ875398.
RESULTS
Cultivation-independent characterization of the reed-associated mycoflora.
Fifteen DNA preparations from vegetative organs of the common reed were used to generate libraries of amplified ITS rRNA gene fragments originating from host-associated fungi. The libraries from triplicate plants growing in close proximity at the nonflooded site originated from rhizomes, roots, stems, and leaves. For comparison, triplicate plants growing at the flooded site nearby were used to create libraries from only roots. A total of 1,991 clones were typed by two separate restriction digestions using the enzymes MboI and MspI, which yielded a total of 348 distinct RFLP types.
In our initial sequencing efforts we focused on the five most abundant RFLP types in each library since rank-abundance plots indicated that the majority of types were rare (see below). If available, for each of these RFLP types at least two clones originating from independent libraries were sequenced. Additional clones were chosen at random from rare RFLP types to examine the potential for additional taxonomic novelty. A total of 159 clones representing 64 different RFLP types, as defined by their MboI and MspI restriction patterns, were sequenced. When several identical sequences were obtained, only one was used for further analysis. Sequencing revealed that in four cases digestion with only two restriction enzymes did not sufficiently distinguish them. All members of the affected RFLP types (Ms4Mb4, Ms9Mb43, Ms28Mb4, and Ms7Mb28) (Table 1)were therefore digested with a third enzyme, PalI, which differentiated each group into two or three separate subgroups. The results of segmental BLASTN searches and pairwise alignments with the closest database matches hinted that there were seven sequences that might have originated from PCR artifacts. These sequences, as well as the RFLP types that they corresponded to, were removed prior to all subsequent analyses. The remaining 345 sequences were defined as OTUs.
TABLE 1.
Database typing of ITS sequencesa
| Sum (%)b | OTU | Accession no. | Best BLAST hit
|
% Similaritye | ||||
|---|---|---|---|---|---|---|---|---|
| Taxon | Phylum | Class | Scorec | Accession no.d | ||||
| 10.58 | Ms9Mb4f | AJ875353 | Cladosporium sp. strain 4/97-17 | Ascomycota | Dothideomycetes | 1,158 | AJ279487 | 99.8 |
| 5.27 | Ms4Mb4af | AJ875341 | Unidentified isolate 4/97-9 | Ascomycota | Sordariomycetes | 1,088 | AJ279484 | 100 |
| 5.11 | Ms6Mb4 | AJ875345 | Chaetomium globosum | Ascomycota | Sordariomycetes | 476 | AY429056 | 80.5 |
| 3.70 | Ms28Mb4a | AJ875376 | Cercophora appalachianensis | Ascomycota | Sordariomycetes | 667 | AF177155 | 79.6 |
| 2.55 | Ms9Mb43a | AJ875357 | Stenella araguata | Ascomycota | Dothideomycetes | 359 | AF362066 | 81.0 |
| 2.45 | Ms39Mb28g | AJ875381 | Aureobasidium pullulans | Ascomycota | Dothideomycetes | 1,197 | AF455533 | 99.7 |
| 2.45 | Ms51Mb4 | AJ875391 | Thielavia intermedia | Ascomycota | Sordariomycetes | 603 | AJ271588 | 89.9 |
| 2.40 | Ms9Mb43b | AJ875358 | Cladosporium sp. | Ascomycota | Dothideomycetes | 581 | AJ222807 | 83.3 |
| 2.35 | Ms22Mb7 | AJ875369 | Ericoid mycorrhiza | Ascomycota | Leotiomycetes | 928 | AY046400 | 98.0 |
| 2.35 | Ms25Mb17 | AJ875374 | Terfezia leptoderma | Ascomycota | Pezizomycetes | 509 | AF396863 | 86.4 |
| 2.19 | Ms7Mb28bf | AJ875351 | Stagonospora sp. strain 4/99-18 | Ascomycota | Dothideomycetes | 1,185 | AJ496627 | 100 |
| 1.82 | Ms5Mb4 | AJ875343 | Podospora ellisiana | Ascomycota | Sordariomycetes | 458 | AY515360 | 79.6 |
| 1.77 | Ms7Mb4 | AJ875347 | Podospora decidua | Ascomycota | Sordariomycetes | 684 | AF443851 | 90.6 |
| 1.51 | Ms41Mb20 | AJ875382 | Hymenogaster griseus | Basidiomycota | Homobasidiomycetes | 1,203 | AF325636 | 97.8 |
| 1.46 | Ms5Mb7g | AJ875344 | Cryptococcus carnescens | Basidiomycota | Heterobasidiomycetes | 914 | AB105438 | 97.8 |
| 1.41 | Ms20Mb11f | AJ875368 | Cylindrocarpon sp. | Ascomycota | Sordariomycetes | 963 | AY295332 | 99.8 |
| 1.36 | Ms25Mb24 | AJ875375 | Sebacina sp. | Basidiomycota | Heterobasidiomycetes | 452 | AF284135 | 88.4 |
| 1.36 | Ms4Mb4b | AJ875342 | Colletotrichum sublineolum | Ascomycota | Sordariomycetes | 325 | AJ301978 | 74.2 |
| 1.20 | Ms48Mb21 | AJ875390 | Sporobolomyces roseus | Basidiomycota | Urediniomycetes | 1,191 | AY015438 | 99.8 |
| 1.09 | Ms28Mb4b | AJ875377 | Podospora intestinacea | Ascomycota | Sordariomycetes | 549 | AY515363 | 87.5 |
| 1.09 | Ms9Mb28 | AJ875356 | Dactylaria junci | Ascomycota | Orbiliomycetes | 379 | AY265321 | 73.3 |
| 1.04 | Ms17Mb11f | AJ875366 | Cylindrocarpon sp. strain 5/97-12 | Ascomycota | Sordariomycetes | 1,142 | AJ279482 | 99.7 |
| 0.99 | Ms13Mb4 | AJ875362 | Choiromyces aboriginum | Ascomycota | Pezizomycetes | 1,067 | AF501259 | 100 |
| 0.89 | Ms9Mb7 | AJ875354 | Cistella grevillei | Ascomycota | Leotiomycetes | 702 | U57089 | 90.2 |
| 0.78 | Ms24Mb18 | AJ875372 | Tuber maculatum | Ascomycota | Pezizomycetes | 1,215 | AF106889 | 99.2 |
| 0.63 | Ms2Mb64 | AJ875339 | Bullera sp. | Basidiomycota | Heterobasidiomycetes | 145 | AY313032 | 31.6 |
| 0.57 | Ms13Mb9 | AJ875363 | Cladosporium sp. | Ascomycota | Dothideomycetes | 347 | AJ222807 | 75.9 |
| 0.57 | Ms15Mb7g | AJ875364 | Uncultured Phialophora | Ascomycota | Sordariomycetes | 793 | AY578279 | 98.6 |
| 0.57 | Ms23Mb25 | AJ875371 | Kabatiella caulivora | Ascomycota | Dothideomycetes | 351 | AJ244251 | 68.0 |
| 0.57 | Ms7Mb28a | AJ875350 | Phaeosphaeria eustoma | Ascomycota | Dothideomycetes | 1,092 | AJ496629 | 98.0 |
| 0.57 | Ms9Mb43c | AJ875359 | Cladosporium sp. | Ascomycota | Dothideomycetes | 381 | AJ222807 | 79.0 |
| 0.52 | Ms25Mb5 | AJ875373 | Calyptrozyma arxii | Ascomycota | Eurotiomycetes | 244 | AJ133432 | 60.7 |
| 0.52 | Ms28Mb91 | AJ875378 | Sporobolomyces gracilis | Basidiomycota | Urediniomycetes | 1,183 | AF444578 | 100 |
| 0.52 | Ms39Mb5 | AJ875380 | Stachybotrys elegans | Ascomycota | Sordariomycetes | 1,207 | AF081481 | 100 |
| 0.52 | Ms8Mb20 | AJ875352 | Hebeloma collariatum | Basidiomycota | Homobasidiomycetes | 1,259 | AY309962 | 98.7 |
| 0.47 | Ms43Mb21g | AJ875386 | Unidentified ericoid mycorrhiza | Ascomycota | Undefined | 307 | AF149079 | 72.0 |
| 0.47 | Ms7Mb5 | AJ875348 | Rhizopus sexualis | Zygomycota | Mucorales | 137 | AB113011 | 54.8 |
| 0.42 | Ms43Mb9 | AJ875384 | Scuellinia scutellata | Ascomycota | Pezizomycetes | 618 | AF072091 | 91.1 |
| 0.36 | Ms88Mb20 | AJ875397 | Agrocybe erebia | Basidiomycota | Homobasidiomycetes | 1,209 | AY168831 | 97.9 |
| 0.36 | Ms9Mb18 | AJ875355 | Stenella araguata | Ascomycota | Dothideomycetes | 369 | AF362066 | 70.0 |
| 0.26 | Ms43Mb11 | AJ875385 | Cylindrocarpon sp. | Ascomycota | Sordariomycetes | 597 | AY295335 | 89.3 |
| 0.26 | Ms45Mb46 | AJ875389 | Cryptococcus chernovii | Basidiomycota | Heterobasidiomycetes | 1,233 | AF444354 | 99.4 |
| 0.26 | Ms63Mb4 | AJ875393 | Phaeoacremonium inflatipes | Ascomycota | Sordariomycetes | 331 | AF118140 | 71.0 |
| 0.26 | Ms88Mb26 | AJ875398 | Thelephoraceae sp. | Basidiomycota | Homobasidiomycetes | 866 | U83471 | 91.1 |
| 0.21 | Ms9Mb105 | AJ875360 | Dactylaria junci | Ascomycota | Orbiliomycetes | 626 | AY265320 | 89.2 |
| 0.16 | Ms20Mb5 | AJ875367 | Dioszegia crocea | Basidiomycota | Heterobasidiomycetes | 646 | AF444406 | 91.8 |
| 0.16 | Ms44Mb20 | AJ875387 | Hebeloma pusillum | Basidiomycota | Homobasidiomycetes | 1,181 | AF124702 | 99.3 |
| 0.16 | Ms44Mb67 | AJ875388 | Psathyrella gracilis | Basidiomycota | Homobasidiomycetes | 1,031 | AY228352 | 92.8 |
| 0.16 | Ms56Mb4 | AJ875392 | Unidentified isolate | Ascomycota | Undefined | 781 | AY559361 | 96.0 |
| 0.16 | Ms88Mb6 | AJ875395 | Uncultured ectomycorrhiza | Basidiomycota | Homobasidiomycetes | 414 | AY310829 | 79.8 |
| 0.10 | Ms30Mb28 | AJ875379 | Phaeococcomyces nigricans | Ascomycota | Chaetothyriomycetes | 337 | AF050278 | 71.1 |
| 0.10 | Ms3Mb3 | AJ875340 | Glomus sp. | Glomeromycota | Glomeromycetes | 944 | AY174689 | 97.7 |
| 0.10 | Ms6Mb21 | AJ875346 | Phaeoacremonium aleophilum | Ascomycota | Sordariomycetes | 359 | AY159787 | 71.3 |
| 0.10 | Ms77Mb41 | AJ875394 | Candida castellii | Ascomycota | Saccharomycetes | 218 | AY046196 | 44.2 |
| 0.10 | Ms88Mb19 | AJ875396 | Sebacina vermifera | Basidiomycota | Heterobasidiomycetes | 337 | AF202728 | 73.9 |
| 0.05 | Ms10Mb19 | AJ875361 | Uncinula septata | Ascomycota | Leotiomycetes | 163 | AB022421 | 56.5 |
| 0.05 | Ms16Mb14 | AJ875365 | Exophiala salmonis | Ascomycota | Chaetothyriomycetes | 955 | AF050274 | 91.4 |
| 0.05 | Ms1Mb17 | AJ875338 | Phialophora sp. | Ascomycota | Sordariomycetes | 511 | U31845 | 86.0 |
| 0.05 | Ms23Mb21 | AJ875370 | Phaeoacremonium aleophilum | Ascomycota | Sordariomycetes | 373 | AY159787 | 71.5 |
| 0.05 | Ms41Mb91 | AJ875383 | Agrocybe praecox | Basidiomycota | Homobasidiomycetes | 1,156 | AF124713 | 98.4 |
| 0.05 | Ms7Mb20 | AJ875349 | Sporobolomyces yunnanensis | Basidiomycota | Urediniomycetes | 1,164 | AB030353 | 99.3 |
OTUs were characterized by MspI and MboI digestion. The suffixes a, b, and c indicate cases in which a third enzyme, PalI, was used for further differentiation.
Relative abundance for the combined libraries, which was used to sort the entries.
BLASTN score.
Accession number of the closest database match.
Level of similarity for pairwise alignments with the closest match, using the Martinez-Needleman-Wunsch algorithm.
The OTU is >97% similar to a fungal isolate that was recovered previously from reeds.
Related sequences diverge up to 3%. The sequence most similar to the closest database hit was used in the analysis.
Identification of undocumented fungi.
The entire lengths of the 61 nonredundant sequences resulting from the analyses described above were used as queries in BLASTN searches to find related sequences annotated in current gene databases (Table 1). The majority of the closest database matches found represented fungal species that are known to interact with plants as either pathogens, ectomycorrhizas, arbuscular mycorrhizas, ericoid mycorrhizas, epiphytes, or endophytes. A threshold of 97% similarity was used as an approximation to differentiate the sequences at a level close to the species level, by analogy to the practice used to distinguish bacterial species by their 18S rRNA sequences, since it was reported that in the Basidiomycota the level of intraspecies variation commonly ranges from 0 to 3% (43). Using the 97% threshold, we identified five sequences that were the best database matches with our previous isolates from reeds that were obtained by conventional cultivation (9, 41, 42). The two most common OTUs found in this study, Ms9Mb4 and Ms4Mb4a, thus corresponded to a Cladosporium sp. (accession no. AJ279487) and an unclassified ascomycete (accession no. AJ279484), respectively. The other three belonged to two species of Cylindrocarpon (accession no. AY295332; AJ279482) and one species of Stagonospora (accession no. AJ496627). Eighteen sequences represented fungi having levels of similarity of at least 97% to database entries from other, unrelated studies. At this level, 62% of the sequences were novel. The degree of novelty covered a wide range, as indicated by the BLAST scores and similarity values for pairwise alignments. In some cases, similarity to database entries was limited to the conserved parts of the sequence analyzed. In these cases, there were not significant matches for the ITS1 and ITS2 boxes when they were used individually for BLAST searches. Thus, some of the sequences must have originated from fungi that were only distantly related to currently annotated taxa.
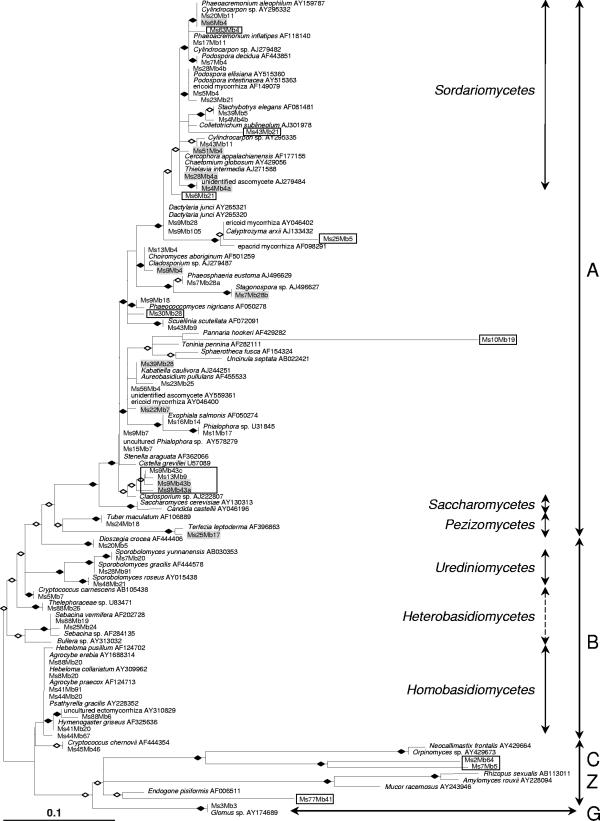
A phylogenetic analysis was performed in order to insert the sequences into a molecular taxonomic framework provided by the closest matches in the databases. The highly divergent ITS1 and ITS2 boxes prohibited us from generating consistent alignments that included the entire lengths of all sequences. Therefore, we restricted this analysis to the conserved 5.8S rRNA, which previously had been shown to have variation appropriate for examining relationships within the Mycota at higher taxonomic levels (6, 24, 27). The phylogram shown in Fig. 1 was obtained by a state of the art maximum-likelihood approach using a GTR+G8+I model. The fungal phyla were correctly recovered; however, the limited amount of information contained in the 5.8S rRNA (156 positions) is reflected by the inability to group sequences belonging to the same class or order. This was notably the case for the Ascomycota. Furthermore, it is likely that the high rate of evolution within the Ascomycota resulted in the nonmonophyly of the Basidiomycota in the tree. The fastest-evolving basidiomycete lineages are obviously attracted by the Ascomycota, resulting in a long-branch attraction tree reconstruction artifact. Most of the OTUs grouped closely with their database matches. However, OTUs Ms2Mb64, Ms7Mb5, and Ms77Mb41 were not located in a context anticipated from the BLAST searches (Table 1) and were placed in the Chytridiomycota and Zygomycota. Their BLAST scores, as well as the similarity values resulting from pairwise alignments with the closest matches, were particularly low, which indicated their great phylogenetic distance from currently annotated fungi. OTUs Ms7Mb5 and Ms2Mb64 formed a sister group to the Neocallimastigales, an order of the Chytridiomycota comprising the only obligate anaerobic fungi currently known (4). OTU Ms77Mb41 was connected to the Endogonales of the Zygomycota, whose members associate with plants as a distinct type of mycorrhizae. The Ascomycota group comprised several novel sequences as well. Ms63Mb4, Ms43Mb21, Ms6Mb21, Ms25Mb5, and Ms30Mb28 were attached to branches without their database matches, and Ms10Mb19 was connected to a particularly long branch (Fig. 1). In addition, there was a distinct cluster of four OTUs (Ms13Mb9, Ms9Mb43a, Ms9Mb43b, and Ms9Mb43c) that represented novelty at a lower taxonomic level. Affiliation with the Ascomycota was supported in all these cases by BLAST searches. Within the Ascomycota many of the OTUs sequenced were connected to the classes Sordariomycetes (17 OTUs) and Dothideomycetes (10 OTUs), which also included all OTUs except two whose total abundance was 2% or more (Table 1). Most of the sequences of OTUs that were attached to long branches were all confirmed with two or more independent clones; the only exception was Ms10Mb19, which was recovered only once. In contrast to the other phyla, OTUs belonging to the Basidiomycota were located mostly on short branches, relatively close to their database matches (Fig. 1). The taxonomic novelty for this phylum was therefore restricted to lower levels. Within the Basidiomycota eight OTUs belonged to the class Homobasidiomycetes, which includes many ectomycorrhizal fungi, five OTUs belonged to the Heterobasidiomycetes, and three OTUs belonged to the Urediniomycetes. Members belonging to the last class were closely related to ballistosporous yeasts inhabiting the phyllosphere (19). The Sebacinales of the Heterobasidiomycetes comprise species that form ectomycorrhizas, orchid mycorrhizas, ericoid mycorrhizas, and endophytes (38).
FIG. 1.
Molecular phylogeny of 5.8S rRNAs from reed-associated fungi. The tree is based on a total of 129 sequences and 156 unambiguous nucleotide positions and was obtained by a maximum-likelihood analysis using the program Treefinder with a GTR+G8+I model. It shows the relationships between 61 sequences originating from reed fungi, their closest database matches, additional reference sequences, and several outgroup sequences. Reed fungi are referred to as OTUs. Reference sequences are shown as they are annotated in the database, including their accession numbers. OTUs in boxes represent potential new lineages. OTUs that are highlighted with gray have a total abundance of more than 2%. Open diamonds indicate branches receiving more than 50% LRSH-RELL bootstrap support, and solid diamonds indicate branches receiving more than 80% LRSH-RELL bootstrap support. A, Ascomycota; B, Basidiomycota; C, Chytridiomycota; Z, Zygomycota; G, Glomeromycota.
Structure of fungal communities associated with the common reed.
Between 94 and 217 clones per library were analyzed to determine their RFLP patterns, and this analysis yielded between 15 and 83 different types per library, which was referred to as the observed richness (Table 2). Initially, we intended to saturate the RFLP analysis, but this could not be achieved by reasonable efforts, as indicated by the accumulation curves obtained (data not shown). Therefore, we used several approximation methods to estimate the “true” richness. For most libraries, the predicted values were considerably higher than the observed values. We used two different nonparametric methods to derive 38 to 224 different OTUs per library (ACE) and, respectively, 28 to 198 different OTUs per library (Chao1) (Table 2). Alternatively, use of a parametric approach implemented in the SDR software that assumed a truncated lognormal model for species abundance distribution resulted in a prediction that there were 49 to 177 OTUs per library. Roots growing in nonflooded soil were apparently associated with the richest fungal community, and stems were associated with the poorest fungal community. Flooded habitat conditions seemed to reduce the number of OTUs that could be recovered from roots. However, statistical analyses by the Kruskal-Wallis test (JMP software) did not prove that the differences were significant (P = 0.05).
TABLE 2.
Community analysis of reed-associated mycofloras
| Parameter | Library(ies)a
|
|||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | 15 | 1-4 | 5-8 | 9-12 | 1,5,9 | 2,6,10 | 3,7,11 | 4,8,12 | 13-15 | 1-15 | |
| Plant | 1 | 1 | 1 | 1 | 2 | 2 | 2 | 2 | 3 | 3 | 3 | 3 | 4 | 5 | 6 | 1 | 2 | 3 | 1-3 | 1-3 | 1-3 | 1-3 | 4-6 | 1-6 |
| Organ | l | s | rh | ro | l | s | rh | ro | l | s | rh | ro | ro | ro | ro | all | all | all | l | s | rh | ro | ro | All |
| Site | d | d | d | d | d | d | d | d | d | d | d | d | f | f | f | d | d | d | d | d | d | d | f | d,f |
| Sizeb | 100 | 103 | 103 | 217 | 113 | 105 | 134 | 153 | 113 | 150 | 124 | 115 | 94 | 152 | 142 | 523 | 505 | 502 | 326 | 358 | 361 | 485 | 388 | 1,918 |
| Observedc | 36 | 29 | 15 | 83 | 54 | 26 | 45 | 52 | 26 | 45 | 62 | 36 | 15 | 39 | 67 | 133 | 134 | 135 | 93 | 82 | 105 | 141 | 103 | 345 |
| ACEd | 146 | 107 | 38 | 219 | 224 | 76 | 126 | 135 | 58 | 95 | 122 | 103 | 57 | 97 | 172 | 355 | 420 | 275 | 285 | 187 | 230 | 353 | 263 | 753 |
| Chao1d | 164 | 77 | 28 | 193 | 198 | 57 | 92 | 118 | 53 | 116 | 99 | 82 | 38 | 71 | 177 | 327 | 401 | 267 | 277 | 165 | 169 | 297 | 242 | 757 |
| Lognormale | 98 | 81 | 177 | 116 | 79 | 139 | 69 | 78 | 49 | 66 | 66 | 68 | n.a. | 77 | 79 | 289 | 333 | 194 | 201 | 153 | 148 | 230 | 198 | 726 |
| Hf | 2.6 | 2.4 | 1.4 | 3.8 | 3.3 | 2.1 | 3.2 | 3.3 | 2.4 | 3.1 | 3.8 | 2.5 | 0.9 | 2.7 | 3.8 | 3.9 | 3.9 | 4.0 | 3.4 | 3.5 | 3.8 | 3.9 | 3.5 | 4.6 |
| Df | 7.1 | 6.1 | 2.5 | 24.1 | 14.4 | 4.1 | 19.3 | 16.7 | 7.1 | 13.0 | 42.1 | 5.0 | 1.5 | 8.2 | 33.7 | 23.2 | 26.2 | 25.8 | 11.2 | 19.4 | 21.9 | 20.4 | 13.7 | 39.3 |
| Alphaf | 20.2 | 13.4 | 4.8 | 49.1 | 40.6 | 11.1 | 23.8 | 27.8 | 10.6 | 21.8 | 49.3 | 18.0 | 5.0 | 17.0 | 49.6 | 57.5 | 59.6 | 60.6 | 43.5 | 33.3 | 49.7 | 66.8 | 45.8 | 122.7 |
| Modelg | s,n | s,n | s,n | s,n | s,n | n | g,s,n | s,n | s,n | s,n | g,s,n | s,n | None | s,n | g,s,n | None | None | n | n | n | None | n | n | None |
The libraries are characterized by the samples used to construct them (i.e., the individual plants [plants 1 to 6]), the organ (leaf [l], stem [s], rhizome [rh], or root [ro]), and the site, which was dry (d) or flooded (f).
Number of clones analyzed per library.
Number of OTUs observed or richness.
Richness estimated by nonparametric models.
Richness estimated by lognormal model.
H, Shannon diversity index; D, Simpson diversity index; Alpha, Fisher diversity index.
Fit of abundance distribution to geometric (g), log series (s), and truncated lognormal (n) models. The best fit is indicated by boldface type. The first 15 columns of data show the results of analyses for individual libraries; the next eight columns show the results for combined data sets prior to analysis; and the last column shows the combined data for libraries 1 to 15.
Diversity indices were calculated by using the SDR software (Table 2). The Shannon-Wiener index varied from 0.9 to 3.8 for single libraries, with an average of 2.8, and it was 4.6 for all combined libraries for the entire study. The Simpson index and the Fisher alpha index have been used previously to evaluate the degree of dominance in microbial communities (44). In the study of Zhou et al., the upper limits for a dominance profile were indicated by values of 50 for the Simpson index and 100 for the alpha index. Based on these limits, all fungal communities reflected dominance. A randomization test implemented in the SDR software was used to test variation between diversity indices in a pairwise manner (29). When different host organs, host habitats, and host individuals were compared, the differences for leaves versus rhizomes and roots from the dry habitats were found to be significant (P = 0.05) with both the Shannon-Wiener and Simpson indices. In addition, the values for the former index were found to differ significantly for stems versus rhizomes and roots from the dry habitats and for roots from flooded habitats versus rhizomes and roots from dry habitats. The Simpson index values for leaves and stems were significantly different in addition to the differences mentioned above.
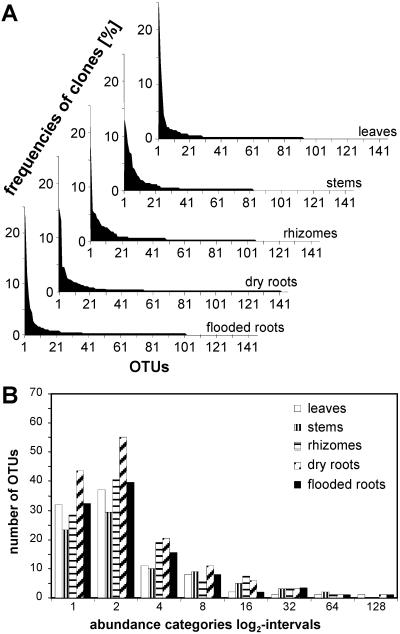
The relatively small surveys typically used for molecular analysis of microbial communities have the potential to recognize the underlying type of community structure (20). Data for the triplicate libraries from each host organ were combined and used to construct rank abundance plots (Fig. 2A). These plots showed that much of the high diversity that was observed was a result of the large proportion of OTUs recovered only once (56% of all OTUs). Few of the many OTUs present on each plant organ were prevalent. In addition, we used species abundance plots to assess community structure (Fig. 2B). The observed distributions were compared to the truncated lognormal, geometric series, log series, and broken stick models (15) by using the SDR software. The truncated lognormal model fit for all vegetative organs analyzed except rhizomes, and no alternative model fit. When the individual data sets were used, this model also fit the distributions from 14 of 15 libraries (Table 2). For 11 libraries, this was also the best matching model, whereas for two libraries the best match was found with a log series model, for one library the best match was found with a geometric series model, and for another library no model fit. The broken stick model did not fit any data set.
FIG. 2.
Community structures of reed-associated mycofloras. The plots are based on the combined triplicate libraries. (A) Rank abundance plots: relative abundance of OTUs sorted for each host organ by rank order size. (B) Species (OTU) abundance plot. The bars represent observed numbers of OTUs binned on a log2 scale of abundance categories. The first group of bars represents one-half of the OTUs having an abundance of 1, the second group represents one-half of the OTUs with an abundance of 1 and the OTUs with an abundance of 2, the third group represents all OTUs having an abundance of 3 and one-half of the OTUs having abundances of 2 and 4, and so forth (37).
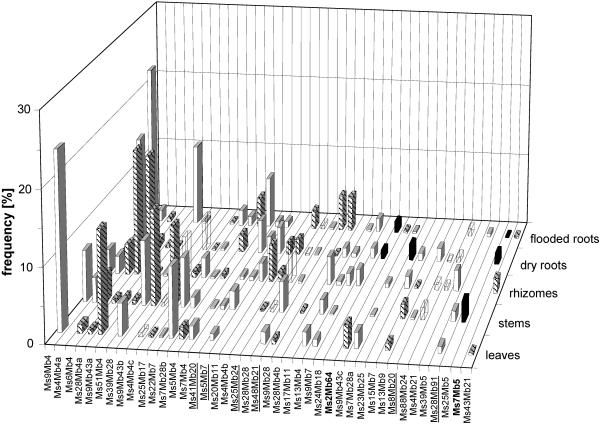
Data sets for the triplicate libraries were combined, and a plot was generated to show the distribution of sequence novelty and gross taxonomic affiliations of the OTUs. This plot included the 41 most prevalent OTUs with total abundance values down to and including 0.5%. For illustration, the similarity scores obtained by pairwise alignments with the closest database match (Table 1) were used to sort OTUs into three arbitrary classes for taxonomic novelty. The scores for these classes were 100 to 90% for lower taxonomic levels, 90 to 70% for intermediate taxonomic levels, and less than 70% for higher taxonomic levels (Fig. 3). OTUs that had less than 70% similarity to current database entries generally had low abundance values (at most 2.8% for any vegetative organ). All organs analyzed harbored unknown OTUs at similarity levels between 70% and 90%, and the maximal abundance was 17% for OTU Ms28Mb4a in rhizomes. Of the 41 most common OTUs, 6 belonged to the Basidiomycota (Fig. 3) and 2 were tentatively assigned to the Chytridiomycota (Ms7Mb5 and Ms2Mb64). The abundance of none of these OTUs was as high as 5% for any organ. The Ascomycota thus outweighed the other phyla with respect to numbers and abundances of OTUs and also as a source of sequence novelty.
FIG. 3.
Distribution and taxonomic novelty of OTUs retrieved from vegetative organs of P. australis. The data are data from combined triplicate libraries and include data for the 41 most prevalent OTUs. The relative frequencies are sorted by total abundance. The solid columns indicate similarity scores of less than 70% in alignments with the closest database match, the cross-hatched columns indicate similarity scores of 70% to 90%, and the open columns indicate similarity scores of more than 90%. Translucent columns represent OTUs that were not sequenced. OTUs identified as members of the Basidiomycota are underlined, OTUs that are members of the Chytridiomycota and Zygomycota are indicated by boldface type, and all other OTUs belong to the Ascomycota.
Variations in the distribution of OTUs with respect to host organ, host habitat, and host individual were analyzed by the Kruskal-Wallis test for statistical significance. OTUs Ms4Mb4b, Ms4Mb4c, and Ms15Mb7 were significantly more frequent in libraries from roots and rhizomes (P = 0.05). The frequency of Ms9Mb43a was significantly higher in leaves, the frequency of Ms20Mb11 was significantly higher in roots, and the frequency of Ms43Mb21 was significantly higher in rhizomes. When we compared dry and flooded roots, the frequencies of OTUs Ms4Mb4a, Ms4Mb4b, Ms4Mb4c, and Ms20Mb11 were significantly higher in dry roots. When we compared the three host individuals, OTU Ms22Mb7 was the only OTU that showed a significant increase (for plant 2).
Nested PCR confirmed niche differentiation.
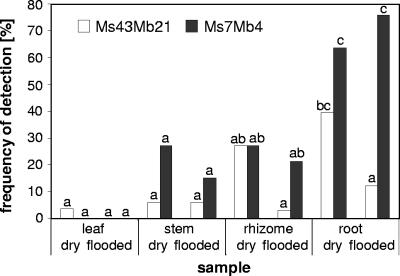
Although the analyses described above were based on just three replicate libraries, significant variation for host organ or host habitat was observed for some fungi. To investigate putative niche differentiation in depth, we developed nested PCR assays that specifically targeted two OTUs in order to assess their occurrence in numerous field samples. The targets were OTUs Ms7Mb4 and Ms43Mb21; the level of the latter was significantly enhanced in rhizomes (see above), and this OTU was only distantly related to currently annotated species in the Ascomycota. Specificity tests showed that these assays amplified only DNA from the targeted fungi (Fig. 4A to C). The occurrence of the targeted fungi was monitored in 252 tissue samples originating from 66 standing reed plants harvested at four sites at Lake Constance over a 3-year period. The authenticity of the assays was checked by sequencing three PCR products for each target. In each case, the sequence of the PCR product was the sequence determined previously. Most fungi were detected at all sites at each of 10 harvests; the only exception was Ms43Mb21, which was not detected on one occasion. When we examined the occurrence in different vegetative host organs and habitat types, the frequency of OTU Ms43Mb21 was significantly increased in rhizomes and roots, but only at dry sites (Fig. 5). In contrast, OTU Ms7Mb4 occurred significantly more frequently in roots than in the other organs, and there was no clear association with habitat.
FIG. 4.
Specificity of nested PCR assays for the detection of two OTUs. (A) First PCR step, using primers ITS1F and ITS4. Lanes M contained 100-bp ladders for reference. Other labels above the lanes indicate the OTUs that were the sources of plasmids used as templates. (B and C) Second PCR step, using the product of the first PCR and gene-specific primers targeting OTUs Ms7Mb4 and Ms43Mb21, respectively.
FIG. 5.
Monitoring of two OTUs in field samples by nested PCR: summary of data for 252 DNA preparations that originated from 66 P. australis plants harvested over a 3-year period at Lake Constance. The frequency of detection is the percentage of preparations that yielded a band after the second step of the nested PCR assays targeting OTUs Ms7Mb4 and Ms43Mb21. For each OTU for each type of sample, the same letter indicates samples for which the results did not differ significantly.
DISCUSSION
In conclusion, approximately 350 fungal OTUs were detected on just six individual reed plants, but the abundance was >2% for only 11 of these OTUs and only 3 of these OTUs could be isolated by conventional methods. Among the large number of the remaining, less abundant OTUs, there were only two that were previously cultivated from reeds. The very uneven structure of plant-associated fungal communities might complicate attempts to cultivate novel taxa that are rare. Such cultivation might require procedures that are currently not available for mycology and thus need to be developed.
The first outcome of this study is that the mycoflora associated with mature, healthy reeds is much more diverse than previous cultivation-based approaches that covered the same sampling sites had indicated (9, 41, 42). This was true for all vegetative organs analyzed. The total number of OTUs experimentally observed in this study was 345, whereas two nonparametric extrapolations, which did not assume a particular distribution, predicted a total of 753 (ACE) and 757 (Chao1) OTUs for theoretical, infinite clone libraries (Table 2). Interestingly, an extrapolation assuming an underlying truncated lognormal distribution provided almost the same result (726 OTUs). On average, a single reed plant hosted 134 different experimentally observed OTUs; the extrapolated total numbers were 350 OTUs (ACE), 332 OTUs (Chao1), and 272 OTUs (log normal).
Several considerations suggest that these extrapolations might still underestimate the total mycoflora diversity associated with P. australis. Our OTU definition relied on RFLP typing, which might not separate closely related species in a few instances. In addition, the common reed grows up to 4 m tall in Lake Constance and has an extensive root system that penetrates into the soil to depths ranging from about 50 cm at permanently flooded sites to up to 4 m at sites with severely fluctuating groundwater levels (25). Obviously, samples used for DNA extraction could cover this large biomass only to a limited degree. Many of the observed OTUs were singletons, and it is likely that more extensive sampling would have retrieved more such OTUs. Furthermore, neither the accumulation curves for the observed richness nor the predicted accumulation curves reached saturation, indicating that analyses of additional clones from the same libraries would have increased the numbers (data not shown).
Conversely, problems inherent with any PCR-based approach might have artificially increased the observed richness. Most of the point mutations introduced by PCR are not detected by RFLP analysis and thus do not influence OTU counts. Another problem is the generation of chimeras during PCR. Some chimeras were detected among the OTUs sequenced and were removed. Nevertheless, there would have been some chimeras among the OTUs that were not sequenced. A previous report of a study in which the workers used the same DNA polymerase that was used in our experiments indicated that 14% of the clones analyzed contained sequence artifacts ranging from 0.2% to 1.2% (30). In most cases, such PCR artifacts would not be detected by RFLP analysis and, therefore, would not considerably increase the OTU counts.
The diversity of plant-associated fungi observed here is within the range of diversities observed in some previous cultivation-based studies of samples from the tropics (1-3). Since our molecular approach is much more sensitive, it is plausible that hundreds of fungal species can colonize an individual plant also in temperate climates.
The second important outcome is that many of the reed-associated fungi belong to species that are currently not covered by the GenBank database. One might object that this could be just a result of poor representation of the molecular marker used. A database search looking for the presence of all segments of the fungal ITS region revealed 32,050 entries, whereas a search for fungal 18S rRNA and 28S rRNA retrieved 30,651 and 24,744 entries, respectively (as of 26 June 2005). Thus, the target chosen is the best-annotated locus for the Mycota. In particular, the Ascomycota is broadly covered, with 20,903 entries. Still, much of the novelty observed here is connected to just this phylum, hinting that there are many uncultivated or undescribed species. However, it is possible that a few branches of the Mycota are currently not well represented in the GenBank database despite the fact that fungi belonging to these branches were cultivated and described previously. This fraction will disappear in the future through the increased efforts of mycologists to annotate ITS sequences when they describe their specimens.
In spite of several previous studies on the reed mycoflora, for 62% of the sequences determined here there were not database matches at a threshold of 3% for sequence novelty, which was used as a proxy for species rank diversity. The many novel fungal sequences retrieved from reeds indicate the limitations of cultivation, which is reminiscent of reports investigating bacterial diversity. Our study confirmed the existence of many undocumented fungal species associated with roots (28, 36) and additionally showed that the same is true for other vegetative host organs. Several novel OTUs appear to be linked to intermediate taxonomic levels, whereas four hold the promise of being new at higher levels. Three of these OTUs were inserted at basal portions of the fungal pedigree, one within the Ascomycota. The taxonomic novelty within the Basidiomycota was limited to lower levels.
The third outcome of this study is that the mycofloras associated with P. australis assemble along dominance profiles. Furthermore, the Simpson and Fisher alpha indices determined from all libraries were clearly below the limits suggested previously as turning points for uniform distributions (44). In a recent report workers showed plots of the observed richness against the Simpson index for theoretical microbial communities following lognormal, geometric, and uniform model distributions and compared these data to experimental data for bacterial communities (20). The type of underlying distribution could be deduced over much of the range that these curves covered. In such a plot, the data from all our libraries would correspond to the lognormal model and would thus support the results obtained with the SDR software.
Recent work indicated that the diversity of root-associated fungi is high, but how the communities are structured remained uncertain (36). In addition to the diversity in roots, we found high fungal diversity in other vegetative host organs. However, our quantitative analyses showed that only a few fungi were able to attain higher abundance in a given organ. Our results suggest that competition and niche differentiation may shape the observed dominance profiles of plant-associated fungal communities. In contrast, uniform profiles have been found for bacterial communities under certain soil conditions (44). The lack of dominance was interpreted to be a result of reduced competition, and this interpretation was experimentally supported by microcosm experiments (35). The dominance profiles observed here fit a truncated lognormal model that reflected a species-rich community in equilibrium, in which species abundance was influenced by many independent factors (15, 20). The abiotic and biotic factors influencing fungal species abundance on or in a plant are manifold and often difficult to determine. Nevertheless, in the present study we succeeded in addressing two such factors. On the one hand, we compared mycofloras from various vegetative host organs. The greatest diversity was observed for roots, whereas leaves seemed to reflect more severe dominance. Several OTUs showed increased abundance in the libraries from distinct organs, which was substantiated for two OTUs by nested PCR assays. The significant variation indicates that different vegetative host organs offer such contrasting habitat conditions that fungi colonizing one organ might not be able to successfully spread to other organs. Whether specialization for resources, contrasting host defense reactions, competition by other fungi, and/or other factors can explain this finding remains to be determined. On the other hand, we compared different mycoflora samples from roots growing in dry and flooded sediments. These contrasting habitats differ drastically in terms of oxygen availability and, most likely, pH and chemical composition. Several fungi were detected significantly more often in the libraries originating from the dry sites. Nested PCR monitoring of the occurrence of OTU Ms43Mb21 over 3 years in a large collection of samples provided independent evidence that some properties of the flooded sites limited the competitiveness of this OTU in this habitat. Support for the hypothesis that sediment conditions have an effect on fungal persistence on the host has also come from studies showing that arbuscular mycorrhizal fungi do not colonize reed roots in flooded sediments (40) and that only certain fungi are able to infect roots under anaerobic conditions (8). Adaptation to a host organ and a host habitat leads to niche differentiation which separates putative competitors and expands the range of fungi associated with a host. Besides space, additional factors that were not examined here lead to further niche differentiation. One of these factors is seasonal variability, as previously observed for fungi in reeds (9) and in tundra soils (28). Furthermore, the carbohydrates available in or on a plant might influence the associated fungal community (12). In future investigations workers should examine whether some recently developed models could lead to a deeper understanding of the assembly and dynamics of plant-associated fungal communities (21, 32, 34, 37).
Acknowledgments
Financial support was provided by the Deutsche Forschungsgemeinschaft through the Collaborative Reseach Centre 454 (Littoral Zone of Lake Constance).
We thank Michael Ernst for the provision of reed DNA preparations and Willi Nagl for help with statistics (both at Universität Konstanz).
REFERENCES
- 1.Aptroot, A. 2001. Lichenized and saprobic fungal biodiversity of a single Elaeocarpus tree in Papua New Guinea, with the report of 200 species of ascomycetes associated with one tree. Fungal Divers. 6:1-11. [Google Scholar]
- 2.Arnold, A. E., Z. Maynard, and G. S. Gilbert. 2001. Fungal endophytes in dicotyledonous neotropical trees: patterns of abundance and diversity. Mycol. Res. 105:1502-1507. [Google Scholar]
- 3.Arnold, A. E., L. C. Mejia, D. Kyllo, E. I. Rojas, Z. Maynard, N. Robbins, and E. A. Herre. 2003. Fungal endophytes limit pathogen damage in a tropical tree. Proc. Natl. Acad. Sci. USA 100:15649-15654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brookman, J. L., G. Mennim, A. P. Trinci, M. K. Theodorou, and D. S. Tuckwell. 2000. Identification and characterization of anaerobic gut fungi using molecular methodologies based on ribosomal ITS1 and 185 rRNA. Microbiology 146:393-403. [DOI] [PubMed] [Google Scholar]
- 5.Castresana, J. 2000. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17:540-552. [DOI] [PubMed] [Google Scholar]
- 6.Cullings, K. W., and D. R. Vogler. 1998. A 5.8S nuclear ribosomal RNA gene sequence database: applications to ecology and evolution. Mol. Ecol. 7:919-923. [DOI] [PubMed] [Google Scholar]
- 7.Curtis, T. P., W. T. Sloan, and J. W. Scannell. 2002. Estimating prokaryotic diversity and its limits. Proc. Natl. Acad. Sci. USA 99:10494-10499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Damm, U., A. Brune, and K. Mendgen. 2003. In vivo observation of conidial germination at the oxic-anoxic interface and infection of submerged reed roots by Microdochium bolleyi. FEMS Microbiol. Ecol. 45:293-299. [DOI] [PubMed] [Google Scholar]
- 9.Ernst, M., K. W. Mendgen, and S. G. R. Wirsel. 2003. Endophytic fungal mutualists: seed-borne Stagonospora spp. enhance reed biomass production in axenic microcosms. Mol. Plant-Microbe Interact. 16:580-587. [DOI] [PubMed] [Google Scholar]
- 10.Ganley, R. J., S. J. Brunsfeld, and G. Newcombe. 2004. A community of unknown, endophytic fungi in western white pine. Proc. Natl. Acad. Sci. USA 101:10107-10112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gardes, M., and T. D. Bruns. 1993. ITS primers with enhanced specificity for Basidiomycetes—application to the identification of mycorrhizae and rusts. Mol. Ecol. 2:113-118. [DOI] [PubMed] [Google Scholar]
- 12.Hadacek, F., and G. F. Kraus. 2002. Plant root carbohydrates affect growth behaviour of endophytic microfungi. FEMS Microbiol. Ecol. 41:161-170. [DOI] [PubMed] [Google Scholar]
- 13.Hawksworth, D. L. 2004. Fungal diversity and its implication for genetic resource collections. Stud. Mycol. 50:9-18. [Google Scholar]
- 14.Hawksworth, D. L. 2001. The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycol. Res. 105:1422-1432. [Google Scholar]
- 15.Hill, T. C. J., K. A. Walsh, J. A. Harris, and B. F. Moffett. 2003. Using ecological diversity measures with bacterial communities. FEMS Microbiol. Ecol. 43:1-11. [DOI] [PubMed] [Google Scholar]
- 16.Hughes, J. B., J. J. Hellmann, T. H. Ricketts, and B. J. Bohannan. 2001. Counting the uncountable: statistical approaches to estimating microbial diversity. Appl. Environ. Microbiol. 67:4399-4406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jobb, G., A. von Haeseler, and K. Strimmer. 2004. TREEFINDER: a powerful graphical analysis environment for molecular phylogenetics. BMC Evol. Biol. 4:18. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 18.Kishino, H., and M. Hasegawa. 1989. Evaluation of the maximum likelihood estimate of the evolutionary tree topologies from DNA sequence data, and the branching order in Hominoidea. J. Mol. Evol. 29:170-179. [DOI] [PubMed] [Google Scholar]
- 19.Nakase, T. 2000. Expanding world of ballistosporous yeasts: distribution in the phyllosphere, systematics and phylogeny. J. Gen. Appl. Microbiol. 46:189-216. [DOI] [PubMed] [Google Scholar]
- 20.Narang, R., and J. Dunbar. 2004. Modeling bacterial species abundance from small community surveys. Microb. Ecol. 47:396-406. [DOI] [PubMed] [Google Scholar]
- 21.Pachepsky, E., J. W. Crawford, J. L. Bown, and G. Squire. 2001. Towards a general theory of biodiversity. Nature 410:923-926. [DOI] [PubMed] [Google Scholar]
- 22.Philippe, H. 1993. MUST, a computer package of management utilities for sequences and trees. Nucleic Acids Res. 21:5264-5272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rappe, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 24.Redecker, D., M. Hijri, H. Dulieu, and I. R. Sanders. 1999. Phylogenetic analysis of a dataset of fungal 5.8S rDNA sequences shows that highly divergent copies of internal transcribed spacers reported from Scutellospora castanea are of ascomycete origin. Fungal Genet. Biol. 28:238-244. [DOI] [PubMed] [Google Scholar]
- 25.Rodewald-Rudescu, L. (ed.). 1974. Das Schilfrohr Phragmites communis Trinius, vol. XXVII. Schweizerbart'sche Verlagsbuchhandlung, Stuttgart, Germany.
- 26.Ronquist, F., and J. P. Huelsenbeck. 2003. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572-1574. [DOI] [PubMed] [Google Scholar]
- 27.Roose-Amsaleg, C., Y. Brygoo, and M. Harry. 2004. Ascomycete diversity in soil-feeding termite nests and soils from a tropical rainforest. Environ. Microbiol. 6:462-469. [DOI] [PubMed] [Google Scholar]
- 28.Schadt, C. W., A. P. Martin, D. A. Lipson, and S. K. Schmidt. 2003. Seasonal dynamics of previously unknown fungal lineages in tundra soils. Science 301:1359-1361. [DOI] [PubMed] [Google Scholar]
- 29.Solow, A. R. 1993. A simple test for change in community structure. J. Anim. Ecol. 62:191-193. [Google Scholar]
- 30.Speksnijder, A. G., G. A. Kowalchuk, S. De Jong, E. Kline, J. R. Stephen, and H. J. Laanbroek. 2001. Microvariation artifacts introduced by PCR and cloning of closely related 16S rRNA gene sequences. Appl. Environ. Microbiol. 67:469-472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stach, J. E., L. A. Maldonado, D. G. Masson, A. C. Ward, M. Goodfellow, and A. T. Bull. 2003. Statistical approaches for estimating actinobacterial diversity in marine sediments. Appl. Environ. Microbiol. 69:6189-6200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugihara, G., L. F. Bersier, T. R. Southwood, S. L. Pimm, and R. M. May. 2003. Predicted correspondence between species abundances and dendrograms of niche similarities. Proc. Natl. Acad. Sci. USA 100:5246-5251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tilman, D. 2004. Niche tradeoffs, neutrality, and community structure: a stochastic theory of resource competition, invasion, and community assembly. Proc. Natl. Acad. Sci. USA 101:10854-10861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Treves, D. S., B. Xia, J. Zhou, and J. M. Tiedje. 2003. A two-species test of the hypothesis that spatial isolation influences microbial diversity in soil. Microb. Ecol. 45:20-28. [DOI] [PubMed] [Google Scholar]
- 36.Vandenkoornhuyse, P., S. L. Baldauf, C. Leyval, J. Straczek, and J. P. Young. 2002. Extensive fungal diversity in plant roots. Science 295:2051. [DOI] [PubMed] [Google Scholar]
- 37.Volkov, I., J. R. Banavar, S. P. Hubbell, and A. Maritan. 2003. Neutral theory and relative species abundance in ecology. Nature 424:1035-1037. [DOI] [PubMed] [Google Scholar]
- 38.Weiss, M., M. A. Selosse, K. H. Rexer, A. Urban, and F. Oberwinkler. 2004. Sebacinales: a hitherto overlooked cosm of heterobasidiomycetes with a broad mycorrhizal potential. Mycol. Res. 108:1003-1010. [DOI] [PubMed] [Google Scholar]
- 39.White, T. J., T. Bruns, S. Lee, and J. Taylor. 1990. Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics, p. 315-322. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (ed.), PCR protocols: a guide to methods and applications. Academic Press, New York, N.Y.
- 40.Wirsel, S. G. R. 2004. Homogenous stands of a wetland grass harbour diverse consortia of arbuscular mycorrhizal fungi. FEMS Microbiol. Ecol. 48:129-138. [DOI] [PubMed] [Google Scholar]
- 41.Wirsel, S. G. R., W. Leibinger, M. Ernst, and K. Mendgen. 2001. Genetic diversity of fungi closely associated with common reed. New Phytol. 149:589-598. [DOI] [PubMed] [Google Scholar]
- 42.Wirsel, S. G. R., C. Runge-Froböse, D. G. Ahren, E. Kemen, R. P. Oliver, and K. W. Mendgen. 2002. Four or more species of Cladosporium sympatrically colonize Phragmites australis. Fungal Genet. Biol. 35:99-113. [DOI] [PubMed] [Google Scholar]
- 43.Zervakis, G. I., J. M. Moncalvo, and R. Vilgalys. 2004. Molecular phylogeny, biogeography and speciation of the mushroom species Pleurotus cystidiosus and allied taxa. Microbiology 150:715-726. [DOI] [PubMed] [Google Scholar]
- 44.Zhou, J., B. Xia, D. S. Treves, L. Y. Wu, T. L. Marsh, R. V. O'Neill, A. V. Palumbo, and J. M. Tiedje. 2002. Spatial and resource factors influencing high microbial diversity in soil. Appl. Environ. Microbiol. 68:326-334. [DOI] [PMC free article] [PubMed] [Google Scholar]