Abstract
The bacterium Sinorhizobium morelense S-30.7.5 was isolated by a microbial screening using the sugar 1,5-anhydro-d-fructose (AF) as the sole carbon source. This strain metabolized AF by a novel pathway involving its reduction to 1,5-anhydro-d-mannitol (AM) and the further conversion of AM to d-mannose by C-1 oxygenation. Growth studies showed that the AF metabolizing capability is not confined to S. morelense S-30.7.5 but is a more common feature among the Rhizobiaceae. The AF reducing enzyme was purified and characterized as a new NADPH-dependent monomeric reductase (AFR, EC 1.1.1.-) of 35.1 kDa. It catalyzed the stereoselective reduction of AF to AM and also the conversion of a number of 2-keto aldoses (osones) to the corresponding manno-configurated aldoses. In contrast, common aldoses and ketoses, as well as nonsugar aldehydes and ketones, were not reduced. A database search using the N-terminal AFR sequence retrieved a putative 35-kDa oxidoreductase encoded by the open reading frame Smc04400 localized on the chromosome of Sinorhizobium meliloti 1021. Based on sequence information for this locus, the afr gene was cloned from S. morelense S-30.7.5 and overexpressed in Escherichia coli. In addition to the oxidoreductase of S. meliloti 1021, AFR showed high sequence similarities to putative oxidoreductases of Mesorhizobium loti, Brucella suis, and B. melitensis but not to any oxidoreductase with known functions. AFR could be assigned to the GFO/IDH/MocA family on the basis of highly conserved common structural features. His6-tagged AFR was used to demonstrate the utility of this enzyme for AF analysis and synthesis of AM, as well as related derivatives.
The rare sugar 1,5-anhydro-d-fructose (AF) was first prepared by a multistep chemical synthesis as a versatile chiral building block (28). Unlike common hexopyranoses AF lacks an anomeric carbon rendering it to a cyclic ether with a prochiral carbon-2 (4). AF can now be produced more efficiently from starch by a biocatalytic process using recombinant α-(1,4)-glucan lyase (EC 4.2.2.13; GLase) (50), which catalyzes the release of AF from the nonreducing end of α-(1,4)-glucans (26). The enhanced availability of AF from renewable resources promoted research to utilize AF as a starting material for various syntheses (4, 15, 31, 48). In biological systems AF was first detected in fungi, where it originated from glycogen through degradation by GLase (6, 13). In ascomycetes, discomycetes, and red algae AF is the metabolic precursor to antimicrobial secondary products such as ascopyrones, microthecin, and epipentenomycin I (7, 13), for which the complete pathway was elucidated only recently in the fungus Anthracobia melaloma (51). Although occurring in very small amounts, AF was found in bacteria (37), algae (9), higher plants (22), animal tissues (19), and human cell lines (46), suggesting the ubiquitous distribution of AF in living organisms. In Escherichia coli, as well as in plant and mammalian tissues AF is an intermediate in the formation of 1,5-anhydro-d-glucitol (AG) catalyzed by a specific NADPH-dependent anhydrofructose reductase (EC 1.1.1.263) (19, 22, 37). The widespread occurrence of the lytic—next to the established hydrolytic—and phosphorolytic glycogen degradation raised questions as to its metabolic role (19). In mammals, AF or AG, respectively, seems to stimulate insulin secretion (1, 46), and in humans differences in the AG serum concentrations between healthy and diabetic individuals were observed, having rendered AG a glycemic marker in diabetic control (10). Based on observations in various organisms that only a small fraction of glycogen is degraded to AF, it was assumed that AF and/or AG might play regulatory roles in the glycogen metabolism (19, 37). In E. coli C600 physiological evidence pointed to the possibility that AG promotes glycogenolysis, presumably by intervening with a signal pathway (36).
Here, we report on a microbial screening on AF yielding the bacterial strain Sinorhizobium morelense S-30.7.5, and we describe a new pathway by which AF is converted into the metabolizable sugar d-mannose. The first enzyme of this pathway was characterized as a new AFR with a unique substrate specificity and stereospecificity. We could assign AFR to the GFO/IDH/MocA family with an EC number of 1.1.1.- and demonstrated its general occurrence among the Rhizobiaceae. The AFR gene cloned from S. morelense S-30.7.5 was identified as the chromosomal locus SMc04400 in S. meliloti 1021, which now has a concrete function. Finally, we emphasize the biotechnological potential of AFR for use in the enzymatic analysis of AF and as a new biocatalyst for the efficient synthesis of rare sugars.
MATERIALS AND METHODS
Chemicals.
1,5-Anhydro-d-fructose (1,5-anhydro-d-arabino-hex-2-ulose [i.e., AF]) and ascopyrone P (1,5-anhydro-4-deoxy-d-glycero-hex-en-3-ulose) were prepared as described before (51); AG and 1,5-anhydro-d-mannitol (AM) were obtained from I. Lundt (3), Technical University of Denmark. The 2-keto aldoses d-glucosone, 6-deoxy-d-glucosone, d-allosone, d-galactosone, d-xylosone, and 3-keto-AF were prepared from the corresponding aldoses and AF, respectively, by selective oxidation with engineered pyranose-2-oxidase (EC 1.1.3.10) (5). Other biochemicals were purchased from Roche (Mannheim, Germany) and Sigma (Deisenhofen, Germany).
Strains, growth conditions, and screening.
S. morelense S-30.7.5, an isolate of this laboratory, was deposited as a patent strain (DSM 15760) at the German Culture Collection (Deutsche Sammlung von Mikroorganismen und Zellkulturen [DSMZ], Braunschweig, Germany). The reference strains Sinorhizobium meliloti DSM 1981, Ensifer fredii DSM 5851, Ensifer arboris DSM 13375, Rhizobium leguminosarum DSM 30132, Rhizobium trifolii DSM 30141, Mesorhizobium lotiDSM 2626, Mesorhizobium tianshanense DSM 11417, Bradyrhizobium japonicum DSM 30131, and Azorhizobium caulinodans DSM 5975 were obtained from DSMZ. The bacterial strains were grown at 28°C in Erlenmeyer flasks with shaking (142 rpm) in a mineral medium (pH 6.8) (29), supplemented with AF, d-mannose, or d-glucose (each 10 mM). Solid media contained 1.5% (wt/vol) of agar. For screening of microorganisms, soil samples were collected from the botanic garden of the Saarland University (Saarbrücken, Germany), suspended in 0.9% (wt/vol) saline solution and, after appropriate dilution, spread onto agar plates containing the medium described above with 1.8 mM AF. After 3 to 15 days of aerobic incubation at 28°C, the colonies formed were purified by standard techniques and grown in 5 ml of mineral medium. Larger batches were grown in 1.5-liter mineral medium at 28°C using a 2-liter bioreactor (Biolab CP; Braun, Melsungen, Germany) with pH regulation, aeration (4-liter air/min) and agitation (500 rpm). Escherichia coli TOP10 (Invitrogen, Karlsruhe, Germany) and E. coli BL21(DE3) (Novagen, Madison, Wis.) were recipients of cloning vectors and expression plasmids, respectively. They were grown on Luria-Bertani (LB) medium under appropriate selective conditions (33). Recombinant E. coli BL21(DE3) cells harboring plasmid pA6Ch1 and pA6Ch1HIS (see below), respectively, were grown in the bioreactor on LB medium at 37°C supplemented with 25 μg of kanamycin per ml. At an optical density at 600 nm of about 1, IPTG (isopropyl-β-d-thiogalactopyranoside) was added to a final concentration of 100 μM to initiate enzyme induction. After another 18 h of growth, cells were harvested by centrifugation.
Strain characterization.
The phenotypic characterization of the bacterial strain was performed according to keys to identify Sinorhizobium sp. (17). Antibiotic resistance was determined by the disk plate method (12, 38). The 16S rRNA gene of strain S-30.7.5 was amplified from genomic DNA (33) as a 1.5-kb DNA fragment using the primers fD1 and rD1 (43). The sequence of the amplified 16S rRNA gene was aligned with published sequences obtained from the GenBank and EMBL databases by using the BLAST program (NCBI, http://www.ncbi.nlm.nih.gov).
DNA techniques, cloning, and His6 tag fusion.
Standard and recombinant DNA techniques were applied according to an established manual (33). Plasmids were prepared as described previously (8). Transformation of E. coli was performed with an electroporator Gene Pulser II (Bio-Rad Laboratories, Munich, Germany). DNA sequencing was performed on a LI-COR 4200 DNA sequencer (MWG Biotech, Ebersberg, Germany) using a Sequenase fluorescence-labeled primer cycle sequencing kit (Amersham Biosciences, Freiburg, Germany). DNA sequences were determined with the program DNASTAR (Lasergene, Madison, WI) and aligned with published sequences from the GenBank and EMBL databases by using the BLAST program (see above). The derived amino acid sequences were aligned by using the CLUSTAL W algorithm (40). All PCR DNA amplifications were performed in gradient thermocycler (Biometra, Göttingen, Germany) using the Proofstart PCR Kit (QIAGEN, Hilden, Germany). The afr gene was amplified from genomic DNA (1 μg) of S. morlense S-30.7.5 by using the primers 5′-ATGAA(CT)CGCTGGGGACTGATCGGCGCGAGCACGAT-3′ and 5′-TCAAAGTCCCGTTTCGATCTCGAC-3′ derived from DNA sequences flanking the SMc04400 locus of the sequenced S. meliloti 1021 genome (11). The PCR product was separated in a 1% (wt/vol) agarose gel from which it was purified by using a MinElute gel extraction kit (QIAGEN) and then cloned into the vector pCRII-TOPO to yield plasmid pPS18 containing the 1-kb afr gene. Plasmid pPS18 was used as a template for subcloning afr into the expression vector pET24a(+) (Novagen, Darmstadt, Germany) via the NdeI and BamHI restriction sites (underlined) of the primers 5′-TCTGCAGAATTCGCCCATATGAATCGCTGGGGACTGATC-3′ and 5′-AGTGTGCTGGAATTCGGATCCTCAAAGTCCCGTTTCGAT-3′ to yield plasmid pA6Ch1, which was transformed into E. coli BL21(DE3). To provide AFR with a C-terminal His6 tag, plasmid pPS18 and the primers 5′-TCTGCAGAATTCGCCCATATGAATCGCTGGGGACTGATC-3′ and 5′-GGATCCTCA(GTG)6AAGTCCCGTTTCGATCTCGGC-3′ were used to introduce the afr fusion into pET24a(+), yielding plasmid pA6Ch1HIS for transformation into E. coli BL21(DE3). All inserts of the expression plasmids were verified by DNA sequence analysis.
Anhydrofructose reductase purification.
For AFR purification each 1 g of wet cells of S. morelense S-30.7.5 was suspended in 3 ml of standard buffer (50 mM Bistris-HCl [pH 7.0]) containing DNase I (0.5 mg/ml) and then disrupted by sonification with a MSE Soniprep 150 (Curtin Matheson Scientific, Inc., Houston, TX). The cell debris was removed by centrifugation (5,000 × g for 15 min at 4°C), and the supernatant was filtered through a 0.2-μm-pore-size membrane filter. The filtrate was applied to a Q-Sepharose HP column (2 cm2 by 7 cm) equilibrated with the standard buffer, and AFR was eluted with a linear KCl gradient (0.07 to 1.5 M) in the same buffer. Fractions containing high AF-reductase activities (see assay below) were concentrated by ultrafiltration and then further purified on a Superdex 200 HR10 30 column (2 cm2 by 12 cm) equilibrated with the standard buffer. The eluted fractions with AFR were adsorbed to a Red Sepharose CL-6B affinity column (3 cm2 by 7 cm) and then eluted by 1.5 M KCl in the standard buffer. The eluate with AFR was concentrated by ultrafiltration, desalted on a Sephadex G25-SF column (2 cm2 by 10 cm) equilibrated with the standard buffer, and stored at 4°C and −20°C, respectively.
AFR-His6 was purified from E. coli BL21(DE3)/(pA6Ch1HIS). By using cells suspended in 50 mM potassium phosphate (pH 7.0), cell extracts were prepared as described above and then applied to a HiTrap Chelating Sepharose column (5 ml) equilibrated with 20 mM sodium phosphate (pH 7.4) containing 0.5 M NaCl. The column was washed with 5 volumes of equilibration buffer containing 0.05 M imidazole, followed by elution of AFR-His6 with 10 volumes of the same buffer containing 0.5 M imidazole. Eluted AFR was desalted and finally purified on a Q-Sepharose HP column as described above.
Enzyme assays.
AFR and monooxygenase activities were determined spectrophotometrically at 365 nm by recording the change in NADPH absorbance (ɛ = 3.5 ml μmol−1 cm−1) at 30°C. The AFR assay contained in 1 ml of 100 μmol of Bistris-HCl (pH 6.5), 0.28 μmol of NADPH, 10 to 100 mU of AFR, and 30 to 100 μmol of substrate. The oxidation assay contained in 1 ml of 100 μmol of Tris-HCl (pH 9.0), 1.8 μmol of NADP+, 5 U of AFR, and 30 to 100 μmol of substrate. The reaction was started by substrate addition. The monooxygenase assay (18) contained in 1 ml of 100 μmol of Tris-HCl (pH 7.5) 0.28 μmol of NADPH, 0.1 μmol of FMN or FAD, and 100 μl of crude extract (1 to 3 mg of protein). The reaction was started by the addition of 20 μmol of AM. One unit of enzyme activity was defined as the amount of enzyme required to oxidize/reduce 1 μmol of NADPH/NADP+ min−1 under standard assay conditions.
The pH optimum of AFR was determined using the following buffers (each 100 mM): sodium citrate and sodium acetate (pH 3.5-6.0), potassium phosphate and Bistris-HCl (pH 5.5 to 7.5), and Tris-HCl (pH 7.0 to 9.0). The effects of metal salts and EDTA were tested each at a concentration of 1 mM in the standard assay. The temperature stability of AFR was tested in 20 mM Bistris-HCl (pH 7.0) in the range 20 to 50°C, and the storage stability in the same buffer in the presence of 1 mM dithiothreitol or 0.5 mM NADP, respectively, at −20, 4, 25, and 30°C.
Analytical methods.
Protein concentration was determined with the bicinchoninic acid assay kit (Sigma-Aldrich, Munich, Germany). Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (25) was carried out in 9% (wt/vol) slab gels (10 by 7 by 0.1 cm) using the protein standard (10 to 200 kDa) from MBI Fermentas (St. Leon-Roth, Germany). The gels were stained with Coomassie brilliant blue R-250.
The molecular mass of AF reductase was determined by matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF-MS) with a Bruker Reflex III spectrometer (Bruker-Daltonics, Bremen, Germany) as described recently (5). The relative molecular mass of AFR was determined by gel filtration on Superdex 200 HR10 30 equilibrated with the standard buffer. Protein standards were from Serva (Heidelberg, Germany). The N-terminal amino acid sequence of AFR was determined with the Procise Protein Sequencing System (Applied Biosystems, Foster City, CA). Isoelectric focusing of AFR was performed on Servalyt Precotes gels (pH 3 to 5 and pH 3 to 10) according to the manufacturer's instruction manual (Serva, Heidelberg, Germany).
Sugars and keto sugars were analyzed by high-pressure liquid chromatography (HPLC) equipped with a refractive index detector. An Aminex HPX-87H H+ column (300 by 7.8 mm; Bio-Rad) was used with 0.5 mM H2SO4 as the mobile phase at 60°C and a flow rate of 0.5 ml/min. For comparisons, authentic sugars were used as references for all analytical analyses. Thin-layer chromatography (TLC) was performed with silica gel 60 plates (Merck, Darmstadt, Germany) using acetone-butanol-H2O (4:5:1) as the mobile phase and 2,4-dinitrophenylhydrazine-sulfuric acid reagent for staining.
1H nuclear magnetic resonance (NMR) spectra were recorded in D2O on an Avance 500 spectrometer (Bruker, Rheinstetten, Germany) at 500 MHz and 300K, using the standard pulse program provided by the manufacturer. The analytes were either lyophilized and then dissolved in D2O or directly analyzed in 10% (vol/vol) D2O.
Metabolic studies of 1,5-anhydro-d-fructose.
Late-log-phase cells grown as described above were harvested, washed, and resuspended in 20 mM Tris-HCl (pH 7.5) to give a suspension of approximately 100 mg of wet cells per ml. To 1 ml of the cell suspension, 40 μmol AF or AM, respectively, was added, followed by incubation at 28°C with shaking (142 rpm). Samples were analyzed by HPLC after removal of the cells by ultrafiltration.
Cell extracts of S. morelense S-30.7.5 late-log-phase cells were used to study the conversion of AM. The reaction mixture contained at 28°C in 1.4 ml of the following: 50 μmol of Tris-HCl (pH 7.5), cell extract (30 mg of protein), 20 μmol of AM, 4.2 μmol of NADPH, and 1.0 μmol of FMN. Samples were removed and analyzed by HPLC after deproteinization.
Bioconversions using anhydrofructose reductase.
Conversions with cosubstrate regeneration were run in 50-ml Erlenmeyer flasks with gentle stirring at 28°C. The reaction mixture contained in 10 ml (final volume) of 20 mM Bistris-HCl (pH 6.5) the following: a 60 mM concentration of substrate, 60 mM glucose 6-phosphate, 0.03 mM NADPH, 1.2 mM NADP+, 50 U of glucose 6-phosphate dehydrogenase (EC 1.1.1.49; Serva), and 50 U of AFR. When the reaction was complete, the products were recovered by ultrafiltration, followed by anion (Dowex AG 1×8, formiate, 200 to 400 mesh; Serva)- and cation (Dowex 50 WX8 H+, 200 to 400 mesh)-exchange chromatography.
GenBank accession numbers.
The anhydrofructose reductase gene (afr) and the Sinorhizobium morelense S-30.7.5 16S rRNA gene sequences were deposited in GenBank under accession numbers DQ140417 and DQ140416, respectively.
RESULTS
Screening of AF-utilizing microorganisms.
The microbial screening yielded 50 bacterial strains, 14 of which grew well on liquid mineral medium with AF. From these strains extracts were prepared and analyzed for AF-converting enzyme activities. Remarkably, all strains able to grow on AF contained AF-reducing enzyme activities that required NADPH and were inactive with NADH. A specific AF-reducing enzyme activity (now called anhydrofructose reductase [AFR]) of >0.1 U/mg of protein, and a preliminary Km value for AF of <10 mM were the criteria for the selection of seven strains for closer examinations. When these strains were grown on mineral medium containing d-glucose, d-mannose, or LB medium, respectively, AFR activity was not detectable, suggesting that AFR was only induced in the presence of AF. One strain, S-30.7.5, exhibiting the highest specific AFR activity was selected for further and detailed characterization.
Characterization of the bacterial isolate S-30.7.5 as S. morelense.
The strain S-30.7.5 was characterized as an aerobic gram-negative rod with a polar flagellum capable of growing in complex (LB) and mineral medium with a wide range of carbohydrate substrates. Sequence analysis of 1,436 bp of the amplified 16S rRNA gene of strain S-30.7.5, and database alignments (40) over the total sequence length revealed the highest consensus to members of the genus Sinorhizobium (recently, a renaming of the genus to Ensifer was suggested [49]) and a global identity of 99% to the species S. morelense. Accordingly, phenotypic characterizations of strain S-30.7.5 also led to the genus Sinorhizobium species (17) and finally to the species S. morelense S-30.7.5, based on its multiple antibiotic resistance against kanamycin (1 mg ml−1), erythromycin (600 μg ml−1), chloramphenicol (250 μg ml−1), penicillin G (50 μg ml−1), and streptomycin (20 μg ml−1), which is a distinctive feature of S. morelense (42).
Growth of S. morelense S-30.7.5 and catabolism of AF.
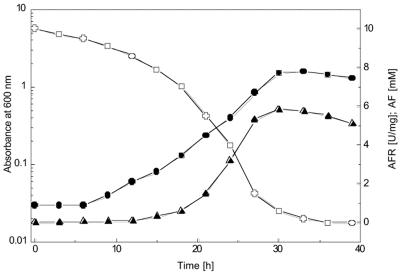
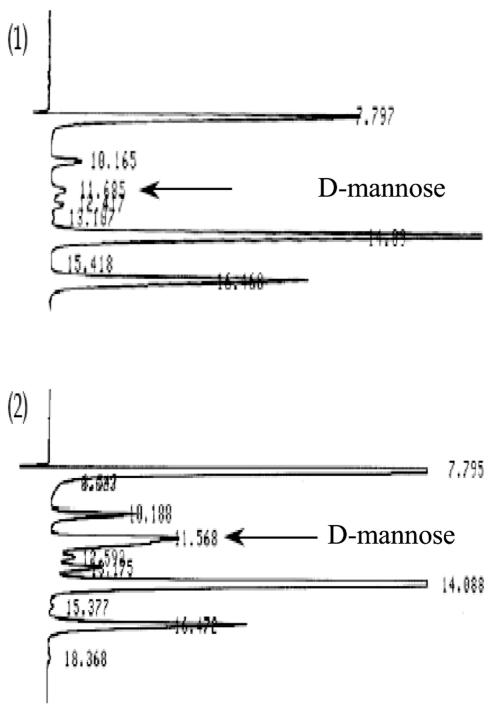
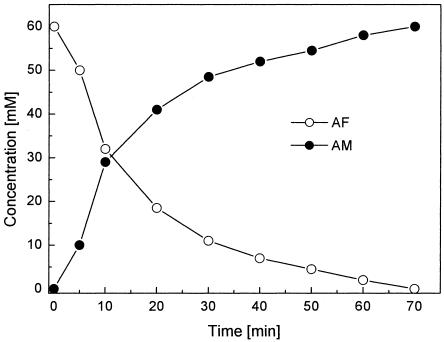
Fig. 1 illustrates the growth of S. morelense S-30.7.5 in a 2-liter bioreactor on a mineral medium containing 10 mM AF as a carbon source. After a lag period, the cells grew exponentially with a specific growth rate of μ = 0.17 h−1 up to an absorbance (A600) of about 1.5. AFR was formed, along with growth reaching a high maximum activity of 5.8 U/mg of protein in the late exponential growth phase (Fig. 1). To determine how the cyclic ether AF was metabolized by S. morelense S-30.7.5, cell extracts were prepared and incubated with AF and equimolar amounts of NADPH. MS analysis showed that a cyclic polyol was formed but did not discriminate between the two enantiomeric forms AM or AG (data not shown). In contrast, HPLC and TLC afforded the identification of a single product with a retention time of 13.7 min and an Rf value of 0.45 that exactly matched those of authentic AM (Table 1). Furthermore, the AM formed from AF was found to be further metabolized in S. morelense S-30.7.5 to d-mannose (Fig. 2), a well-metabolizable substrate for bacterial cells. In accordance with this, specific monooxygenase activities in a range of 0.04 to 0.3 U/mg of protein capable of converting AM to d-mannose were detected in cell extracts of S. morelense S-30.7.5. Figure 2 shows two HPLC runs of samples removed from the reaction mixture containing the cell extract, AF, and NADPH after 10 and 25 min. The peak at 11.6 min, which had increased during prolonged incubation, corresponded to the retention time of authentic d-mannose. Since AF has a similar retention time as d-mannose the possibility existed that a fraction of AM was reoxidized to AF by the cell extract. However, an AM oxidizing activity was not detectable in the standard assay in the presence of NADP or NAD, which therefore excluded this possibility. When intact cells were incubated with AF the sugar was incorporated, and AM was found as the sole product in the supernatant, whereas d-mannose was not detectable, suggesting that d-mannose was phosphorylated upon its formation and therefore retained within the cells.
FIG. 1.
Growth of S. morelense S-30.7.5 on AF and formation of AFR. A culture of 50 ml grown on d-glucose was used to inoculate 1.5 liter of mineral medium containing 10 mM AF in a 2-liter bioreactor. The culture was grown at 28°C with aeration (4-liter air/min) and agitation (500 rpm). Growth (•), AF concentration (○), and specific AFR activity (U/mg of protein) in cell extracts (⧫) were determined. All values represent means of double determinations.
TABLE 1.
HPLC and TLC characteristics of various sugarsa
| Authentic standard or sample | Retention time (min) | Rf |
|---|---|---|
| 1,5-Anhydro-d-fructose | 11.5 | 0.67 |
| 1,5-Anhydro-d-glucitol | 12.2 | 0.57 |
| 1,5-Anhydro-d-mannitol | 13.7 | 0.45 |
| d-Glucose | 10.8 | 0.36 |
| d-Mannose | 11.7 | 0.40 |
| Product of AF reduction (AM) | 13.7 | 0.45 |
| Product of AM conversion (d-mannose) | 11.7 | 0.40 |
The experimental conditions are described in Materials and Methods.
FIG. 2.
HPLC diagram of the conversion of AM to d-mannose by cell extracts of S. morelense S-30.7.5. For details, see Materials and Methods. The arrow indicates the elution peak of d-mannose at 11.6 min as the conversion product of AM at 14.1 min. 1, after 10 min of incubation; 2, after 25 min of incubation.
1,5-Anhydro-d-fructose utilization and demonstration of anhydrofructose reductase activities in members of the Rhizobiaceae.
AF utilization was not restricted to S. morelense S-30.7.5 since we found growth on AF and AFR activities (in U/mg of protein) in strains (for DSM numbers, see Materials and Methods) of Sinorhizobium meliloti (0.4), Rhizobium leguminosarum (0.3), Mesorhizobium loti (0.1), Ensifer fredii (0.04), Mesorhizobium tianshanense (0.04), and Rhizobium trifolii (0.01). In contrast, Ensifer arboris, Bradyrhizobium japonicum, and Azorhizobium caulinodans did not grow on AF and therefore were assumed not to form AFR. Altogether, these findings indicate that AF utilization is common among the Rhizobiaceae but not a feature of the whole family.
Purification and molecular properties of anhydrofructose reductase from S. morelense S-30.7.5.
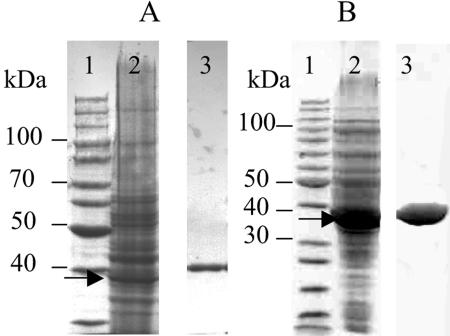
AFR was purified from the cell extracts made from 3.5 g of wet biomass of S. morelense S-30.7.5. The purification procedure consisted of three chromatographic steps (Table 2) resulting in a homogeneous AFR preparation (Fig. 3A) with a specific activity of 489 U/mg of protein and a yield of 8%. AFR is a monomer since the relative molecular mass determined by SDS-PAGE (Mr of ∼40,000) was similar to that obtained by gel filtration (Mr of ∼38,200). A precise molecular mass of 35,100 ± 130 Da was determined by MALDI-TOF-MS, which is in agreement with value derived from the AFR gene (see below). AFR has a pI of pH 4.3 as determined by isoelectric focusing. When AFR was stored at −20 or at 0°C the enzyme lost 50% of its initial activity within 50 days.
TABLE 2.
Purification of the native AFR from S. morelense S-30.7.5 and the His6-tagged AFR variant from E. coli BL21(DE3)/(pA6Ch1)
| Purification step | Total activity (U) | Total protein (mg) | Sp act (U/mg of protein) | Purification (fold) | Yield (%) |
|---|---|---|---|---|---|
| Native AFR | |||||
| Cell extract | 1,100 | 169.4 | 6.5 | 1 | 100 |
| Q-Sepharose HP | 777 | 11.4 | 68.2 | 11 | 71 |
| Superdex 200 HR | 595 | 1.9 | 313.2 | 48 | 54 |
| Red Sepharose CL-6B | 88 | 0.2 | 488.9 | 75 | 8 |
| AFR-His6 | |||||
| Cell extract | 5,586 | 44.9 | 124.4 | 1 | 100 |
| HiTrap chelating | |||||
| Sepharose | 4,900 | 12.5 | 392 | 3.4 | 88 |
| Q-Sepharose HP | 3,631 | 7.5 | 484 | 4 | 65 |
FIG. 3.
SDS-PAGE of AFR preparations. (A) AFR isolated from S. morelense S-30.7.5. Lanes: 1, protein standards; 2, cell extract (50 μg of protein), AFR indicated by arrow; 3, final preparation after Red Sepharose (5 μg of protein) on separate gel. (B) Recombinant His6-tagged AFR from E. coli. Lanes: 1, protein standards; 2, cell extract (50 μg of protein), AFR indicated by arrow; 3, final preparation after Q-Sepharose (15 μg of protein) on separate gel.
Catalytic properties and substrate specificity of anhydrofructose reductase.
At 30°C the highest activity of AFR was measured in 100 mM Bistris-HCl at pH 6.5, with 50% remaining activity at pH 5.2 (in citrate buffer) and pH 8.8 (in Tris-HCl buffer). Under optimum conditions the vmax of AFR was approximately 500 U/mg of protein, depending on the purification. The activity of AFR was not influenced by 1 mM EDTA or by any of the given metal salts NaCl, KCl, MgCl2, CaCl2, MnCl2, FeCl3, and ZnCl2 (each 1 mM), indicating that AFR does not require a metal ion for activity. AFR is specific for the cosubstrate NADPH and inactive toward NADH. The determination of the substrate specificity of AFR showed (Table 3) that the enzyme only acted on AF, 3-keto-AF, ascopyrone P and 2-keto-aldoses (called osones) that are structurally related with AF. In contrast, AFR was inactive toward common aldoses and ketoses, nonsugar aldehydes and ketones which are listed in the footnote of Table 3. It should be pointed out that AFR of S. morelense S-30.7.5 was also inactive toward pyridine-3-aldehyde, 2,3-butanedione, glucuronic acid, acetaldehyde, and formaldehyde, which were reported to be substrates of the NADPH-dependent AFR from porcine liver (32). The oxidation reaction of AFR at pH 9.0 was marginal, since AFR was 8,800 times less active toward AG and 53,000 times less active toward AM, respectively, than toward AF at pH 6.5 (Table 3). Other cyclic and acyclic polyols were not oxidized by AFR (see Table 3, footnote a). AFR followed Michaelis-Menten kinetics for AF, d-glucosone, and NADPH, with apparent Km values of 8.4 ± 0.15 mM, 11.0 ± 0.10 mM, and 0.2 ± 0.03 mM, respectively. The apparent kcat values for AF and d-glucosone were 286.7 and 63.2 s−1, and the corresponding catalytic efficiencies (kcat/Km) were 34,100 and 5,700 M−1 s−1.
TABLE 3.
Substrates and products of AFR from S. morelense S-30.7.5
| Substratea | Sp actb (U/mg of protein) | Relative activity (%) | Productc |
|---|---|---|---|
| Reduction | |||
| 1,5-Anhydro-d-fructose | 530 | 100 | 1,5-Anhydro-d-mannitol (100) |
| 3-Keto-1,5-anhydro-d-fructose | 191 | 36 | 1,5-Anhydro-d-mannitol (100) |
| d-Glucosone | 118 | 22 | d-Mannose (100) |
| 6-Deoxy-d-glucosone | 118 | 22 | 6-Deoxy-d-mannose (45) |
| d-xylosone | 88 | 17 | d-Lyxose (70) |
| d-Allosone | 53 | 10 | d-Altrose (90) |
| Ascopyrone P | 21 | 5 | ND |
| d-Galactosone | 21 | 4 | d-Talose (20) |
| Oxidation | |||
| 1,5-Anhydro-d-mannitol | 0.06 | 100 | ND |
| 1,5-Anhydro-d-glucitol | 0.01 | 23 | ND |
Substrate providers and substrate preparations are given in Materials and Methods. The following compounds were inactive as substrates in the reduction and oxidation assays. Reduction: d-glucose, d-mannose, d-fructose, l-sorbose, d-tagatose, d-xylose, d-lyxose, d-ribose, maltose, d-gluconolactone, 2-deoxy-d-glucose, glucose 6-phosphate. Oxidation: 1,5-anhydro-d-galactitol, 2-amino,2-deoxy-1,5-anhydro-d-mannitol, d-glucitol, d-mannitol, dulcitol, inositol.
Values represent means of double determinations of activity and protein concentration.
The percentage of aldoses with an axial OH-group (manno-configuration) at C-2 (in parentheses). ND, not determined.
Stereo- and regiospecificity of anhydrofructose reductase of S. morelense S-30.7.5.
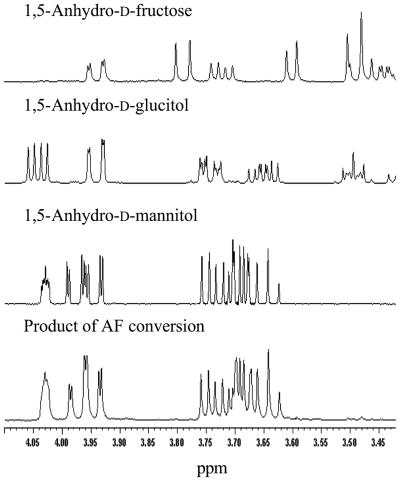
The preliminary results obtained with cell extracts with respect to the stereospecificity of AFR from S. morelense S-30.7.5 were confirmed with the purified enzyme. For this purpose, preparative conversions with 20 mM AF were performed by using a cosubstrate regeneration system. Analysis by HPLC indicated a single peak with the retention time (13.7 min) of authentic AM (Table 1), whereas based on 1H-NMR the product of AFR was identical to that of authentic AM and clearly distinct from that of AG (Fig. 4). This result showed that AFR from S. morelense S-30.7.5 catalyzed the stereoselective reduction of AF to AM according to the reaction equation in Fig. 5, thereby differing from the hepatic AFR which yields AG (32). To evaluate the stereospecificity of AFR with regard to 3-keto AF and the osones, similar conversions were performed. The products were analyzed by HPLC in comparison to the corresponding authentic aldose enantiomers. As a result, it was noted that AFR from S. morelense S-30.7.5 did not keep its strict stereospecificity toward all of these substrates (Table 3). Except for d-glucosone, which is stereospecifically converted to d-mannose, the osones yielded product mixtures with different proportions of manno- and gluco-configurated aldoses (Table 3), whereas 3-keto AF was completely reduced to AM. However, none of the osones was reduced at C-1, distinguishing AFR clearly from aldose reductase catalyzing the reduction of certain osones at C-1 to the corresponding ketoses (27).
FIG. 4.
1H-NMR spectra of authentic standards of AF, 1,5-anhydro-d-glucitol, and AM and of the product (bottom) of an AF bioconversion with AFR from S. morelense S-30.7.5.
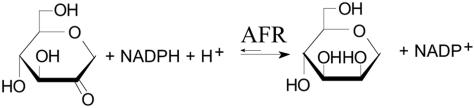
FIG. 5.
Reaction equation of AFR showing the stereoselective conversion of AF to AM.
Cloning of the anhydrofructose reductase gene from S. morelense S-30.7. 5 and its functional overexpression in E. coli.
We used the N-terminal sequence MNRWGLIGASTIAREWVIGAIRATG of AFR from S. morelense S-30.7.5 for a BLASTP database search and found 92% identity to a peptide sequence derived from a 1-kb open reading frame (ORF; locus SMc04400) localized on the sequenced 3.65 Mb main chromosome of S. meliloti 1021 (11). This ORF coded for a 333-amino-acid peptide with a calculated molecular mass of 34.9 kDa that was similar to that determined for AFR. Therefore, we assumed that the OFR SMc04400 represented the corresponding afr gene in S. meliloti 1021. By PCR using flanking primers derived from SMc04400 and a DNA template from S. morelense S-30.7.5 the afr gene was cloned and characterized as a 1,002-bp nucleotide sequence (including a stop codon) with a G+C content of 64.8 mol%. The sequence was deposited in GenBank under accession number DQ140417. The afr gene codes for a peptide of 333 amino acid residues of 35.1 kDa, corresponding exactly to the value experimentally determined for AFR. The afr gene revealed 80% identity to Smc04400 of S. meliloti 1021, whereas the derived polypeptides showed 86% identity over the total sequence length. The afr gene was subcloned into the expression vector pET24a(+) to give plasmid pA6Ch1, which was used to transform E. coli BL21(DE3). The recombinant E. coli BL21(DE3)/(pA6Ch1) grown in a 2-liter bioreactor produced AFR as an intracellularly active enzyme with a maximum yield of 100 kU per liter of culture after 15 h of growth. The specific activity of AFR in the soluble fraction of E. coli cell extracts was 130 U/mg of protein, which was approximately 20 times higher than in the original host S. morelense S-30.7.5. Recombinant AFR was purified to homogeneity from cell extracts of E. coli as wild-type AFR (Table 2) to a specific activity of 450 U/mg of protein. The subsequent characterization of the recombinant AFR revealed no significant changes compared to the wild-type enzyme.
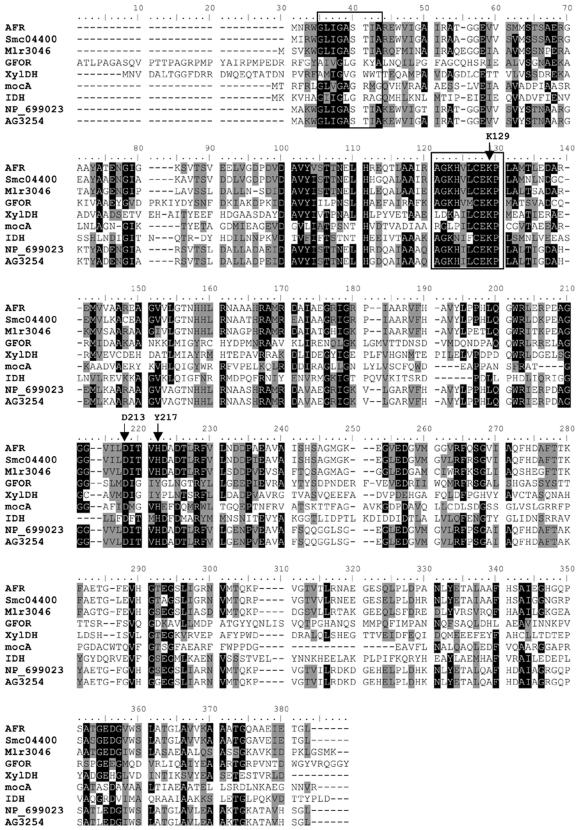
Anhydrofructose reductase primary structure analysis and assignment to the GFO/IDH/MocA protein family.
Using the afr-derived amino acid sequence for a BLASTP search in the NCBI database, we found no oxidoreductase with known function, but we retrieved a group of putative oxidoreductases with high sequence identities (73 to 86%) to AFR assigned to the bacteria S. meliloti, Mesorhizobium loti, Brucella suis, and B. melitensis (see legend to Fig. 6). Also, another prominent group of proteins, but with low sequence identities to AFR (<30%), was retrieved that belonged to the GFO/IDH/MocA family with glucose-fructose oxidoreductase (GFOR) from Zymomonas mobilis as a representative member (52). The sequence alignments (Fig. 6) showed that AFR shares highly conserved sequence motifs with GFOR and related members of the group. These include the characteristic NADP binding motif (-G5XXGXSXXA13-), the conserved substrate-binding motif (-A86GKHVLCEK94-), and the catalytic triad comprising two invariant residues (Lys94 and Asp176) and His180, which is Tyr217 in GFOR (21, 45). Based on these distinctive features that AFR has in common with the GFO/IDH/MocA proteins and in particular with GFOR of Z. mobilis, we assigned AFR of S. morelense S-30.7.5 as a new member of this family, and confirmed it by three-dimensional structure analysis of the AFR and site-directed mutagenesis (T. Dambe, A. Kühn, F. Giffhorn, and A. Scheidig, submitted for publication).
FIG. 6.
Multiple sequence alignment of the deduced amino acid sequences of anhydrofructose reductase (AFR) from S. morelense S-30.7.5 with GFO/IDH/MocA oxidoreductases, xylose dehydrogenase of H. marismortui, and putative oxidoreductases from S. meliloti 1021, M. loti, B. suis, and B. melitensis. The alignment was generated by means of CLUSTAL W with the Blosum62 matrix (40). NCBI/GenBank accession numbers: S. morelense S-30.7.5 AFR (DQ140417); E. meliloti 1021 Smc04400 (NP_387411); M. loti Mlr3046 (NP_104243); Z. mobilis GFOR (CAB_02496); H. marismortui XylDH (YP_137464); E. meliloti MocA, scyllo-inosamin/myoinositol 2-dehydrogenase (CAA55269); L. plantarum IDH, myo-inositol 2-dehydrogenase (NP_786809); B. suis oxidoreductase (NP_699023); B. melitensis oxidoreductase AG3254 (NP_538938). Identical residues among the polypeptides are underlined in black; variable residues are shaded. Framed boxes indicate the highly conserved motifs of cosubstrate and substrate binding. Arrows highlight catalytic residues (see the text).
Application of His6-tagged anhydrofructose reductase in bioconversions and enzymatic analysis of AF.
For convenient purification, the recombinant AFR was fused with a His6 tag at the C terminus, and AFR-His6 was produced in E. coli with yields of about 100 kU per liter of culture. AFR-His6 was purified in two steps to apparent homogeneity (Fig. 3B) with a 65% yield using chelating Sepharose affinity chromatography and a final polishing on Q-Sepharose (Table 2). The final preparation had a specific activity of 484 U/mg of protein and revealed no significant changes with respect to the kinetic properties and stereospecificity of the reaction compared to the native enzyme. AFR-His6 was used with cosubstrate regeneration for the preparative conversion of 60 mM AF to AM, which was complete after 70 min (Fig. 7). The AM formed was purified from the reaction mixture after protein removal by ion-exchange chromatography and obtained as a solid by evaporation. A yield of 100 mg of AM was recovered from 97 mg (0.6 mmol) of AF in a chromatographically pure form. Similarly, each 20 mM concentration of osones was converted to the corresponding aldoses (see above).
FIG. 7.
Bioconversion of AF to AM with cosubstrate regeneration. The conversion was performed in a final volume 10 ml of 20 mM Bistris (pH 6.5) at 28°C. The reaction mixture contained 60 mM AF, 60 mM glucose-6-phosphate, 1.2 mM NADP, 0.03 mM NADPH, 50 U of AFR-His6, and 50 U of glucose-6-phosphate dehydrogenase. AF and AM concentrations were determined by HPLC. The values given represent means of double determinations.
Because of the unique specificity of AFR and based on the fact that 2-keto sugars are rare in nature, we developed an enzymatic assay for AF affording the determination of this sugar in complex materials such as food, fruit juices, or animal body fluids. Using the standard assay containing 10 U of AFR, various amounts of AF (5, 15, 28, 40, and 50 nmol) were completely converted within 12 min (data not shown). The differences in the extinctions (ΔE) were directly proportional to the amounts of AF added to the assay. Conversely, the concentrations of AF could be calculated on the basis of ΔE and the extinction coefficient for NADPH at 340 nm. In the given assay, 5 nmol of AF could be determined with sufficient accuracy.
DISCUSSION
Catabolism of AF in Rhizobiaceae.
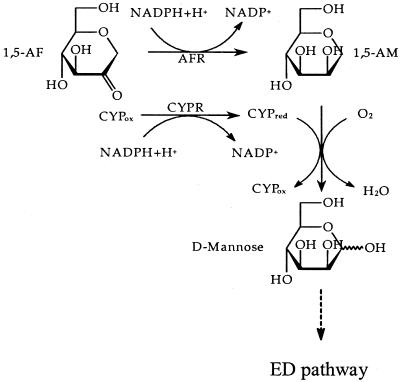
The present study reveals that the ability to grow on AF and to produce AFR is not confined to the newly isolated strain S. morelense S-30.7.5 but is a more common feature among species of the family Rhizobiaceae, in particular among the fast-growing species S. meliloti, R.leguminasorum, and M. loti that are phylogenetically closely related to S. morelense (34). In nature, AF concentrations are exceedingly low and are mostly found in the microgram range per gram of fresh cells or tissue (4), so that it appears unlikely that free-living microorganisms will be ever exposed to AF in substrate amounts. In this regard, it is questionable whether a specific AF catabolic pathway has evolved in S. morelense S-30.7.5. Rather, it seems that by analogy to E. coli C-600 (36, 37) the reduction of AF to AM may regulate the carbohydrate supply in symbiotic-living communities with the plant host where AF may arise from the lytic glucan degradation (22). If cells of laboratory cultures are exposed to artificially high AF concentrations, it may cause a sudden increase in AM with harmful consequences to the cells if it is not instantly eliminated by a CYP-mediated “detoxification” reaction. Why AF is reduced to AM in S. morelense S-30.7.5 and not to AG and whether AM is a communicating signal between host and symbiont warrants further investigation. The further conversion of AM by a CYP-catalyzed oxygenation at the O-linked C-1 was reasonable (35, 44) since the resulting hemiacetal yields d-mannose, which can enter the Entner-Doudoroff pathway (39). This may also apply to S. meliloti 1021, which possesses high AFR activities and the genetic equipment for two CYP systems (20). Altogether, the complete catabolic pathway for AF in S. morelense S-30.7.5 can be described (Fig. 8) based on these findings.
FIG. 8.
Pathway of the AF catabolism in S. morelense S-30.7.5. AFR, anhydrofructose reductase; CYP, cytochrome P450; CYPR, CYP reductase; ED-pathway, Entner-Doudoroff pathway.
New enzyme anhydrofructose reductase.
The AFR from S. morelense S-30.7.5 reported here is new since no similar enzyme could be found in the database. It is distinct from the only reported AFR from porcine liver that converts AF to AG (32). Indeed, both enzymes are similar in size and structure, but they possess essentially different peptide sequences. Based on the sequence data and amino acid substitutions (24) S. morelense S-30.7.5 AFR is a member of the GFO/IDH/MocA family, whereas the hepatic AFR is likely a member of one of the aldose reductase families (32). Also, there are qualitative and quantitative differences between both AFRs with regard to cosubstrate and substrate specificity and in particular with respect to the converse stereoselectivity of the AF reduction. The abundance and high specific activity of AFR in cells of S. morelense S-30.7.5 may reflect the importance of the reduction of AF to AM to the cells and the necessity to compensate for the relatively poor Km for AF. That even very low AF concentrations, as they were assumed to occur in nature, are quantitatively converted by AFR to AM was demonstrated by our in vitro conversions. In the case of hepatic AFR the poor specific activity is compensated for by a low Km for AF, affording its complete conversion to AG (32). Both AFRs are different from a homotetrameric 1,5-anhydro-d-glucitol dehydrogenase of the fungus Trichoderma longibrachiatum strain 11-3 that resembles typical dehydrogenases and/or reductases and catalyzes the NAD-dependent oxidation of AG (47).
Interestingly, in S. meliloti 1021 the afr gene (locus SMc04400) is localized on the 3.65-Mb chromosome, which contains the majority of genes coding for core metabolic functions (17). This finding may suggest a superior metabolic role of AFR, in particular since the corresponding genes were also found in the intracellular pathogens Brucella suis and B. melitensis, which have striking metabolic similarities with plant symbionts such as S. meliloti and M. loti (14, 30). Unfortunately, analysis of the gene region adjacent to SMc04400 provided no further information on its true function. The ORF flanking upstream of SMc04400 codes for a putative transcription regulator of the LacI-type, whereas the ORFs downstream code for putative enzymes of fatty acid synthesis and an l-sorbosone dehydrogenase.
Application of anhydrofructose reductase.
Due to structural similarities between AF and the osones listed in Table 3, these compounds were accepted as substrates by AFR. One requirement seems to be the pyran ring structure with the C-2 carbonyl group, as in AF (16, 23). The best structural match to AF (1-deoxy-d-glucosone) is obviously d-glucosone, which explains the high stereoselectivity of its conversion to d-mannose by AFR. Since d-mannose is still manufactured from birch and beech wood hydrolysates, we highlight a biocatalytic route of how it can be conveniently prepared from low-cost d-glucose (Fig. 9). The chemical synthesis of AM and d-mannose from AF and d-glucosone by catalytic hydrogenation is also possible (3, 16) but less effective than the synthesis with AFR. Although d-mannose is a well-established sugar on the market, uses for AM are just emerging, e.g., as a chiral building block in antivirals (41) and as a potential agent for the treatment of type II diabetes (2).
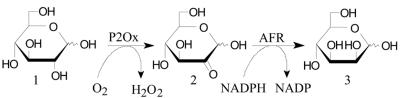
FIG. 9.
Principle of the biocatalytic synthesis of d-mannose from d-glucose. d-glucose 1 is oxidized by pyranose 2-oxidase (P2Ox) to d-glucosone 2 (5), which can be reduced to d-mannose 3 by anhydrofructose reductase (AFR).
In view of the need for the quantitative determination of AF in complex materials, we have developed the first enzymatic assay for AF. Various methods have been used for the determination of AF, but all are unspecific or else laborious and require sophisticated instrumentation (reference 4 and references therein). In contrast, the assay with AFR is specific, sensitive, and not affected by common sugars. Since osones practically do not appear in nature, their interference with the assay is unlikely.
Acknowledgments
We thank K. Hollemeyer for MALDI-TOF MS analysis, J. Zapp for MS and NMR analyses, W. Nastainczyk for N-terminal sequencing, and C. Zimmer for expert technical advice.
This study was financially supported by the European Union within the 5th Framework Programme NEPSA under contract no. QLK3-CT-2001-02400.
REFERENCES
- 1.Ahren, B., J. J. Holst, and S. Yu. 2000. 1,5-Anhydro-d-fructose increases glucose tolerance by increasing glucagon-like peptide-1 and insulin in mice. Eur. J. Pharmacol. 397:219-225. [DOI] [PubMed] [Google Scholar]
- 2.Ahren, B., and S. Yu. December 2004. Use of a cyclic ether for the preparation of medicaments affecting glucose tolerance. U.S. patent appl. 20,020,198,158.
- 3.Andersen, S. M., I. Lundt, and J. Marcussen. 2000. 1,5-Anhydro-d-fructose: stereoselective conversions to 1,5-anhydroalditols and deoxy/amino substituted analogues. J. Carbohydr. Chem. 19:717-725. [Google Scholar]
- 4.Andersen, S. M., I. Lundt, J. Marcussen, and S. Yu. 2002. 1,5-Anhydro-d-fructose, a versatile chiral building block: biochemistry and chemistry. Carbohydr. Res. 337:873-890. [DOI] [PubMed] [Google Scholar]
- 5.Bastian, S., M. J. Rekowski, K. Witte, D. M. Heckmann-Pohl, and F. Giffhorn. 2005. Engineering of pyranose 2-oxidase from Peniophora gigantea toward improved thermostability and catalytic efficiency. Appl. Microbiol. Biotechnol. 67:654-663. [DOI] [PubMed] [Google Scholar]
- 6.Baute, M. A., R. Baute, and G. Deffieux. 1988. Fungal enzymatic activity degrading 1,4-α-d-glucans to 1,5-d-anhydrofructose. Phytochemistry 27:3401-3403. [Google Scholar]
- 7.Baute, M. A., G. Deffieux, R. Baute, A. Badoc, J. Vercauteren, J. M. Léger, and A. Neveu. 1991. Fungal enzymic activity degrading 1,4-α-d-glucans to echinosporin (5-Epipentenomycin I). Phytochemistry 5:1419-1423. [Google Scholar]
- 8.Birnboim, H. C. 1983. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 100:243-255. [DOI] [PubMed] [Google Scholar]
- 9.Broberg, A., L. Kenne, and M. Pedersen. 1996. Presence of microthecin in the red alga Gracilariopsis lemaneiformis and its formation from 1,5-anhydrofructose. Phytochemistry 41:151-154. [Google Scholar]
- 10.Buse, J. B., J. L. Freeman, S. V. Edelman, L. Jovanovic, and J. B. McGill. 2003. Serum 1,5-anhydroglucitol (GlycoMark): a short-term glycemic marker. Diabetes Technol. Ther. 5:355-363. [DOI] [PubMed] [Google Scholar]
- 11.Capela, D., F. Barloy-Hubler, J. Gouzy, G. Bothe, F. Ampe, J. Batut, P. Boistard, A. Becker, M. Boutry, E. Cadieu, S. Dreano, S. Gloux, T. Godrie, A. Goffeau, D. Kahn, E. Kiss, V. Lelaure, D. Masuy, T. Pohl, D. Portetelle, A. Puhler, B. Purnelle, U. Ramsperger, C. Renard, P. Thebault, M. Vandenbol, S. Weidner, and F. Galibert. 2001. Analysis of the chromosome sequence of the legume symbiont Sinorhizobium meliloti strain 1021. Proc. Natl. Acad. Sci. USA 98:9877-9882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Davis, W. W., and T. R. Stout. 1971. Disc plate method of microbiological antibiotic assay. I. Factors influencing variability and error. Appl. Microbiol. 22:659-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Deffieux, G., R. Bauté, M. A. Baute, M. Atfani, and A. Carpy. 1986. 1,5-Anhydrofructose, the precursor of the pyrone microthecin in Morchella vulgaris. Phytochemistry 26:1391-1393. [Google Scholar]
- 14.DelVecchio, V. G., V. Kapatral, R. J. Redkar, G. Patra, C. Mujer, T. Los, N. Ivanova, I. Anderson, A. Bhattacharyya, A. Lykidis, G. Reznik, L. Jablonski, N. Larsen, M. D'Souza, A. Bernal, M. Mazur, E. Goltsman, E. Selkov, P. H. Elzer, S. Hagius, D. O'Callaghan, J. J. Letesson, R. Haselkorn, N. Kyrpides, and R. Overbeek. 2002. The genome sequence of the facultative intracellular pathogen Brucella melitensis. Proc. Natl. Acad. Sci. USA 99:443-448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deppe, O., A. Glumer, S. Yu, and K. Buchholz. 2004. Synthesis and co-polymerization of an unsaturated 1,5-anhydro-d-fructose derivative. Carbohydr. Res. 339:2077-2082. [DOI] [PubMed] [Google Scholar]
- 16.Freimund, S., A. Huwig, F. Giffhorn, and S. Köpper. 1998. Rare keto-aldoses from enzymatic oxidation: substrates and oxidation products of pyranose 2-oxidase. Chem. Eur. J. 4:2442-2455. [Google Scholar]
- 17.Holt, J. G., N. R. Krieg, P. H. A. Sneath, J. T. Staley, and S. T. Williams. 1994. Group 4: gram-negative aerobic/microaerophilic rods and cocci, p. 71-101. In J. G. Holt et al. (ed.), Bergey's manual of determinative bacteriology. The Williams & Wilkins Co., Baltimore, Md.
- 18.Kadiyala, V., and J. C. Spain. 1998. A two-component monooxygenase catalyzes both the hydroxylation of p-nitrophenol and the oxidative release of nitrite from 4-nitrocatechol in Bacillus sphaericus JS905. Appl. Environ. Microbiol. 64:2479-2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kametani, S., Y. Shiga, and H. Akanuma. 1996. Hepatic production of 1,5-anhydrofructose and 1,5-anhydroglucitol in rat by the third glycogenolytic pathway. Eur. J. Biochem. 242:832-838. [DOI] [PubMed] [Google Scholar]
- 20.Kelly, S. L., D. C. Lamb, C. J. Jackson, A. G. Warrilow, and D. E. Kelly. 2003. The biodiversity of microbial cytochromes P450. Adv. Microb. Physiol. 47:131-186. [DOI] [PubMed] [Google Scholar]
- 21.Kingston, R. L., R. K. Scopes, and E. N. Baker. 1996. The structure of glucose-fructose oxidoreductase from Zymomonas mobilis: an osmoprotective periplasmic enzyme containing non-dissociable NADP. Structure 4:1413-1428. [DOI] [PubMed] [Google Scholar]
- 22.Konishi, Y., K. Hashima, and K. Kishida. 2000. Increases in 1,5-anhydroglucitol levels in germinating amaranth seeds and in ripening banana. Biosci. Biotechnol. Biochem. 64:2462-2465. [DOI] [PubMed] [Google Scholar]
- 23.Köpper, S., and S. Freimund. 2003. The composition of keto aldoses in aqueous solution as determined by NMR spectroscopy. Helv. Chim. Acta 86:827-843. [Google Scholar]
- 24.Kühn, A. 2004. Ph.D. thesis. Universität des Saarlandes, Saarbrücken, Germany.
- 25.Laemmli, U. K. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 26.Lee, S. S., S. Yu, and S. G. Withers. 2003. Detailed dissection of a new mechanism for glycoside cleavage of α-1,4-glucan lyase. Biochemistry 42:13081-13090. [DOI] [PubMed] [Google Scholar]
- 27.Leitner, C., W. Neuhauser, J. Volc, K. D. Kulbe, B. Nidetzky, and D. Haltrich. 1998. The cetus process revisited: a novel enzymatic alternative for the production of aldose-free d-fructose. Biocatal. Biotrans. 16:365-382. [Google Scholar]
- 28.Lichtenthaler, F. W., E. S. H. El Ashry, and V. H. Göckel. 1980. Sugar enolones, XIV: a convenient access to 1,5-anhydroketoses. Tetrahedron Lett. 21:1429-1432. [Google Scholar]
- 29.Mayers-Küntzer, H., A. Reichert, K. H. Schneider, and F. Giffhorn. 1994. Isolation and characterization of a l-glucitol dehydrogenase from the newly isolated bacterium Pseudomonas sp. Ac. J. Biotechnol. 36:157-164. [Google Scholar]
- 30.Paulsen, I. T., R. Seshadri, K. E. Nelson, J. A. Eisen, J. F. Heidelberg, T. D. Read, R. J. Dodson, L. Umayam, L. M. Brinkac, M. J. Beanan, S. C. Daugherty, R. T. Deboy, A. S. Durkin, J. F. Kolonay, R. Madupu, W. C. Nelson, B. Ayodeji, M. Kraul, J. Shetty, J. Malek, S. E. Van Aken, S. Riedmuller, H. Tettelin, S. R. Gill, O. White, S. L. Salzberg, D. L. Hoover, L. E. Lindler, S. M. Halling, S. M. Boyle, and C. M. Fraser. 2002. The Brucella suis genome reveals fundamental similarities between animal and plant pathogens and symbionts. Proc. Natl. Acad. Sci. USA 99:13148-13153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Richard, G., S. Yu, P. Monsan, M. Remaud-Simeon, and S. Morel. 2005. A novel family of glucosyl 1,5-anhydro-d-fructose derivatives synthesized by transglycosylation with dextransucrase from Leuconostoc mesenteroides NRRL B-512F. Carbohydr. Res. 340:395-401. [DOI] [PubMed] [Google Scholar]
- 32.Sakuma, M., S. Kametani, and H. Akanuma. 1998. Purification and some properties of a hepatic NADPH-dependent reductase that specifically acts on 1,5-anhydro-d-fructose. J. Biochem. 123:189-193. [DOI] [PubMed] [Google Scholar]
- 33.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1987. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 34.Sawada, H., L. D. Kuykendall, and J. M. Young. 2003. Changing concepts in the systematics of bacterial nitrogen-fixing legume symbionts. J. Gen. Appl. Microbiol. 49:155-179. [DOI] [PubMed] [Google Scholar]
- 35.Sevrioukova, I. F., and J. A. Peterson. 1995. NADPH-P-450 reductase: structural and functional comparisons of the eukaryotic and prokaryotic isoforms. Biochimie 77:562-572. [DOI] [PubMed] [Google Scholar]
- 36.Shiga, Y., S. Kametani, T. Kadokura, and H. Akanuma. 1999. 1,5-Anhydroglucitol promotes glycogenolysis in Escherichia coli. J. Biochem. 125:166-172. [DOI] [PubMed] [Google Scholar]
- 37.Shiga, Y., H. Mizuno, and H. Akanuma. 1993. Conditional synthesis and utilization of 1,5-anhydroglucitol in Escherichia coli. J. Bacteriol. 175:7138-7141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Skinner, F. A., and D. W. Lovelock. 1972. Identification methods for microbiologists. Academic Press, London, England.
- 39.Stowers, M. D. 1985. Carbon metabolism in Rhizobium species. Annu. Rev. Microbiol. 39:89-108. [DOI] [PubMed] [Google Scholar]
- 40.Thompson, J. D., D. G. Higgins, and T. J. Gibson. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22:4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Van Aerschot, A. February 2004. Alkylated hexitol nucleoside analogues and oligomers thereof. U.S. patent appl. 20,040,033,967.
- 42.Wang, E. T., Z. Y. Tan, A. Willems, M. Fernandez-Lopez, B. Reinhold-Hurek, and E. Martinez-Romero. 2002. Sinorhizobium morelense sp. nov., a Leucaena leucocephala-associated bacterium that is highly resistant to multiple antibiotics. Int. J. Syst. Evol. Microbiol. 52:1687-1693. [DOI] [PubMed] [Google Scholar]
- 43.Weisburg, W. G., S. M. Barns, D. A. Pelletier, and D. J. Lane. 1991. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173:697-703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.White, G. F., N. J. Russell, and E. C. Tidswell. 1996. Bacterial scission of ether bonds. Microbiol. Rev. 60:216-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiegert, T., H. Sahm, and G. A. Sprenger. 1997. The substitution of a single amino acid residue (Ser-116 → Asp) alters NADP-containing glucose-fructose oxidoreductase of Zymomonas mobilis into a glucose dehydrogenase with dual coenzyme specificity. J. Biol. Chem. 272:13126-13133. [DOI] [PubMed] [Google Scholar]
- 46.Yamanouchi, T., T. Inoue, K. Ichiyanagi, T. Sakai, and N. Ogata. 2003. 1,5-Anhydroglucitol stimulates insulin release in insulinoma cell lines. Biochim. Biophys. Acta 1623:82-87. [DOI] [PubMed] [Google Scholar]
- 47.Yoshida, N., E. Uchida, T. Katsuragi, and Y. Tani. 2003. A novel NAD-dependent dehydrogenase, highly specific for 1,5-anhydro-d-glucitol, from Trichoderma longibrachiatum strain 11-3. Appl. Environ. Microbiol. 69:2603-2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yoshinaga, K., J. Abe, T. Tanimoto, K. Koizumi, and S. Hizukuri. 2003. Preparation and reactivity of a novel disaccharide, glucosyl 1,5-anhydro-d-fructose (1,5-anhydro-3-O-α-glucopyranosyl-d-fructose). Carbohydr. Res. 338:2221-2225. [DOI] [PubMed] [Google Scholar]
- 49.Young, J. M. 2003. The genus name Ensifer Casida 1982 takes priority over Sinorhizobium Chen et al. 1988, and Sinorhizobium morelense Wang et al. 2002 is a later synonym of Ensifer adhaerens Casida 1982. Is the combination “Sinorhizobium adhaerens” (Casida 1982) Willems et al. 2003 legitimate? Request for an Opinion. Int. J. Syst. Evol. Microbiol. 53:2107-2110. [DOI] [PubMed] [Google Scholar]
- 50.Yu, S., K. Bojsen, B. Svensson, and J. Marcussen. 1999. α-1,4-Glucan lyases producing 1,5-anhydro-d-fructose from starch and glycogen have sequence similarity to α-glucosidases. Biochim. Biophys. Acta 1433:1-15. [DOI] [PubMed] [Google Scholar]
- 51.Yu, S., C. Refdahl, and I. Lundt. 2004. Enzymatic description of the anhydrofructose pathway of glycogen degradation. I. Identification and purification of anhydrofructose dehydratase, ascopyrone tautomerase and α-1,4-glucan lyase in the fungus Anthracobia melaloma. Biochim. Biophys. Acta 1672:120-129. [DOI] [PubMed] [Google Scholar]
- 52.Zachariou, M., and R. K. Scopes. 1986. Glucose-fructose oxidoreductase, a new enzyme isolated from Zymomonas mobilis that is responsible for sorbitol production. J. Bacteriol. 167:863-869. [DOI] [PMC free article] [PubMed] [Google Scholar]