Abstract
KaiC from Synechococcus elongatus PCC 7942 (KaiC) is an essential circadian clock protein in cyanobacteria. Previous sequence analyses suggested its inclusion in the RecA/DnaB superfamily. A characteristic of the proteins of this superfamily is that they form homohexameric complexes that bind DNA. We show here that KaiC also forms ring complexes with a central pore that can be visualized by electron microscopy. A combination of analytical ultracentrifugation and chromatographic analyses demonstrates that these complexes are hexameric. The association of KaiC molecules into hexamers depends on the presence of ATP. The KaiC sequence does not include the obvious DNA-binding motifs found in RecA or DnaB. Nevertheless, KaiC binds forked DNA substrates. These data support the inclusion of KaiC into the RecA/DnaB superfamily and have important implications for enzymatic activity of KaiC in the circadian clock mechanism that regulates global changes in gene expression patterns.
Keywords: DnaB, RecA, cyanobacteria, Synechococcus
Circadian rhythms are endogenous biological programs that “free-run” with a period close to 24 h in constant conditions but will entrain to appropriate environmental cycles of light/dark or temperature. Before 1985, it was believed that circadian programs were exclusively found among eukaryotes; however, it is now clearly documented that eubacterial cyanobacteria exhibit circadian rhythms (1, 2). In cyanobacteria, this circadian clockwork orchestrates rhythmic changes in the expression of nearly every gene in the organism (1, 3). A mutational analysis in the genetically tractable cyanobacterium, Synechococcus elongatus PCC 7942, pinpointed a cluster of three genes, kaiA, kaiB, and kaiC, that were essential circadian clock genes but were not required for viability (4, 5). The kaiC gene is the largest of the three, and most of the mutations that have been isolated by mutational screening were mapped to kaiC.
On the basis of a genomic survey, it was proposed that full-length KaiC from S. elongatus PCC 7942 (KaiC) is a member of the bacterial RecA/DnaB family (6). RecA is an ATP-dependent DNA recombinase, and DnaB is the replication fork helicase in bacteria. The kaiC gene appears to be an internally duplicated version of a RecA/DnaB-like gene; it has two parts that are very similar (7). In each half of the kaiC gene, there is a Walker A motif that binds ATP; when the Walker A motifs are mutated, nucleotide binding is abolished, and rhythmicity is severely disrupted or abolished (3, 8). At this time, the enzymatic function (if any) of KaiC is unknown, but its membership in the RecA/DnaB superfamily suggests a function relating to DNA (3).
We undertook a study using biophysical methods and electron microscopy (EM) to ascertain whether the sequence similarity between the structures of KaiC and RecA/DnaB was manifested in a structural relationship. We used KaiC from S. elongatus (optimal growth temperature 30–35°C), herein called “KaiC,” and also from the mildly thermophilic cyanobacterium Synechococcus lividus (optimal growth temperature 50–55°C), herein called “KaiC-P2.” We found that both of these KaiCs associate as homohexamers, dependent on ATP. Finally, both KaiCs bind forked DNA substrates despite the fact that neither of their sequences includes the DNA-binding motifs found in other members of the RecA/DnaB gene family.
Experimental Procedures
Cloning of KaiC-P2.
To isolate the kaiC gene from a thermophilic cyanobacterium, PCR was performed with the genomic DNA from S. lividus strain P2, which was isolated from a 50–55°C site within the Octopus Spring microbial mat at Yellowstone National Park (Wyoming, MT). Primers were designed to amplify a 389-bp conserved region inside the kaiC-P2 gene (5′-CTY GAT GCT TCM CCC GAT CC-3′ and 5′-GGA TAT TCC CCY TTC ATG TGG-3′). One major PCR product was obtained and sequenced. On the basis of the sequence of the PCR product obtained, oligonucleotide primers were designed for PCR-based chromosome walking. The entire ORF of the kaiC-P2 gene on the chromosome was progressively sequenced by inverse PCR (9) and adaptor ligation PCR (10) methods. The nucleic acid sequence for the kaiC-P2 gene has been deposited in GenBank (accession no. AF497977).
Purification of KaiC, KaiCI, and KaiC-P2.
The kaiC ORF (from S. elongatus PCC 7942) was amplified by PCR using the following primers: 5′-TAT ACA TAT GAC TTC CGC TGA GAT GAC TAG C-3′ (NdeI site underlined) and 5′-CAT GCT AGCCTA ATG ATG ATG ATG ATG ATG GCT CTC CGG CCC TTT TTC TTG AAC-3′ (NheI site underlined, hexahistidine sequence in italics). The amplified DNA fragment was cleaved with NdeI and NheI and inserted into the NdeI-NheI site of pRSET-B (Invitrogen). Escherichia coli BL21(DE3) cells were transformed with the construct, and His-6-tagged KaiC protein was expressed under the control of the T7 promoter. The first “half” of KaiC from S. elongatus PCC 7942 (KaiCI) is the N-terminal half of KaiC (residues 1–268 of KaiC). For KaiCI, a His-6-tag fusion protein expression construct was made as described above except the oligonucleotide 5′-CTG CTA GCC TTA ATG ATG ATG ATG ATG ATG GAC GAC ACC AGA TGA AAC ACG CAC-3′ (NheI site underlined, hexahistidine sequence in italics) was used as a reverse primer. The recombinant His-6-tagged KaiC and KaiCI proteins were purified by essentially the same method used for HisP (11) to the point of elution from the TALON resin (BD Biosciences CLONTECH); thereafter, our protocol differed slightly: in the case of KaiC and KaiCI, the resin was washed on the column with buffer containing 40 mM imidazole and then eluted with 100 mM imidazole. For the gel shift assays depicted in Fig. 5B, KaiC was further purified on MonoQ resin and eluted with a NaCl gradient. For purification of KaiC and KaiCI, 5 mM ATP was included in all buffers. If ATP was not included in the elution buffer, KaiC precipitated within hours after elution; if ATP was included in the elution buffer, KaiC did not precipitate in the eluate for many days at 4°C.
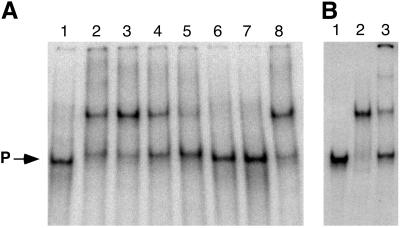
Fig 5.
EMSA of KaiC-P2 and KaiC binding to DNA. (A) Binding of KaiC-P2 to DNA. Lanes 1–8, radiolabeled forked DNA probe (≈5 fmol/20-μl reaction volume). Lanes 2–8 include purified KaiC-P2 (0.46 μg = 7.26 pmol)/20-μl reaction volume) and 1 mM AMP-PNP. Lanes 3–7 include unlabeled forked DNA probe at the following concentrations per 20-μl reaction volume: 5 fmol (lane 3), 10 fmol (lane 4), 25 fmol (lane 5), 250 fmol (lane 6), and 2.5 pmol (lane 7). Lane 8 includes 2.5 pmol unlabeled 60-mer poly dN random oligonucleotide (single-stranded DNA). Incubation at 53°C was for 30 min before loading on the gel. “P” marks the position of the unbound probe. (B) Binding of KaiC and KaiC-P2. Lanes 1–3, radiolabeled forked DNA probe (≈3 fmol/20-μl reaction volume) and 1 mM AMP-PNP. Lane 2 includes purified KaiC-P2 (0.46 μg = 7.3 pmol/20-μl reaction volume). Lane 3 includes purified KaiC (0.52 μg = 8.8 pmol/20-μl reaction volume). There was a 30-min incubation before loading on the gel at 53°C (lane 2) or at 30°C (lane 3). “P” marks the position of the unbound probe.
The kaiC-P2 gene from S. lividus P2 was amplified by genomic PCR by using the primers 5′-ATG GCT AGC TAC GAC GAT GAC GAT AAG ATG AAC CAG TCA TTG GGG CCT TCT-3′ (NheI site underlined, enterokinase recognition site in italics) and 5′-ATG CTA GCT CAC AAC CCT TCT TCC TCG AG-3′ (NheI site underlined). The amplified DNA fragment was cleaved with NheI and inserted into the NheI site of pRSET-B. This construct allows KaiC-P2 to be expressed under the control of the T7 promoter and adds 19 residues (MRGSHHHHHHGMASDDDDK) to its N terminus (Fig. 1). KaiC-P2 was purified in the absence of ATP by essentially the same procedure as for KaiC and KaiCI, except that the E. coli homogenate was heat treated (55°C for 20 min) and partially purified by an ammonium sulfate precipitation. The pellet from the 50% saturated ammonium sulfate precipitation was resuspended in TALON column buffer, and KaiC-P2 was purified on TALON metal affinity resin followed by gel filtration on a Sephacryl S-300 HR column equilibrated with buffer A (0.1 M NaCl/20 mM Tris⋅HCl, pH 8) plus 10% glycerol. Peak fractions of KaiC, KaiCI, and KaiC-P2 were pooled and concentrated by ultrafiltration, snap-frozen in liquid nitrogen, and stored at −80°C.
Fig 1.
Alignments of the sequences of KaiC, KaiC-P2, and KaiCI. Black backgrounds indicate identical residues. Shaded backgrounds indicate conserved substitutions (criteria defined by CLUSTAL X). His-6 tags for protein purification are underlined. Hyphens indicate gaps.
Analytical Ultracentrifugation Analysis.
Sedimentation velocity analysis was carried out in a Beckman Coulter optima XL-I analytical ultracentrifuge by using interference optics at 20°C. Medium was buffer A plus 10% glycerol in the presence or absence of 5 mM ATP. The concentration profiles were analyzed by time-derivative methods with dcdt (12) and by fitting with sedanal (13). sedanal is a Windows program that fits concentration time difference data to eliminate systematic errors in the interference data using numerical solutions of the Lamm equation according to the method of Todd and Haschemeyer (14). Molar mass was also determined from time derivative sedimentation velocity analysis by fitting the boundary profiles to a Gaussian to obtain both the sedimentation and diffusion coefficients (15) by using the following equations:
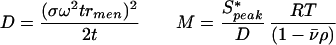 |
where σ is the standard deviation of the Gaussian in units of svedbergs; ω is the angular velocity of the rotor; rmen is the radius of the meniscus; t is the time of sedimentation; v̄is the partial specific volume calculated from the amino acid composition; and ρ is the buffer density. Both methods gave comparable results. Other details have been reported previously (16). The calculated value of the partial specific volume was 0.7335 cc/g for KaiC and 0.7292 for KaiC-P2. The density of the buffer was 1.01443; the viscosity correction was 1.1422 calculated with the program sednterp (17).
Analytical Gel Filtration of KaiC.
Gel filtration was carried out on a 1.45 × 32.5 cm column filled with Sephacryl S-300 HR resin using buffer A plus 10% glycerol. Standards for the gel filtration column were blue dextran (void volume marker), apoferritin [molecular mass = 443 kDa; Stokes radius (Rs) = 6.1 nm], β-amylase (molecular mass = 200 kDa; Rs = unknown), BSA (molecular mass = 66 kDa, Rs = 3.55 nm), cytochrome c (molecular mass = 12.4 kDa; Rs = 1.70 nm), and nicotinamide (molecular mass = 122 Da; included volume marker). The gel filtration column was calibrated by using the Rs data and the inverse complement error function of the partition coefficients according to the method of Ackers (18). The partition coefficient, σ, is defined as σ = (Ve − Vo)/(Vx − Vo), where Ve is the elution volume of a particular protein, Vx is the included volume, and Vo is the void volume of the column. The Rs is related to the partition coefficient by Rs = a + b[erfc−1(σ)].
EM.
KaiC, KaiCI, and KaiC-P2 proteins were maintained in storage buffer (buffer A plus 100 mM imidazole/5% glycerol/5 mM ATP/0.1 mM EDTA/1 mM DTT, final pH 8.0). For all reactions and incubations, the protein was diluted at least 5-fold into a standard buffer containing 20 mM Tris⋅HCl, pH 8.0, 5 mM Mg acetate, 5 mM ATP, 10 mM KCl, 0.1 M NaCl, 0.1 mM EDTA, and 5 mM 2-mercaptoethanol, to the indicated concentration. Incubations were carried out at 37°C.
Reaction mixtures were prepared for EM by one of two procedures: (i) Alcian method (positive staining): Samples were prepared by adsorption to Alcian-activated carbon-coated grids as previously described (19). An 8-μl sample of KaiC protein was diluted 100-fold with 200 mM ammonium acetate/Hepes (10 mM, pH 7.0)/10% glycerol, and adsorbed to the Alcian-activated carbon film for 3 min. After washing with the same buffer for 1 min, the sample was stained with 5% uranyl acetate followed by a very brief circular shadowing with platinum (19).
(ii) Negative staining: Carbon films were activated by glow discharge (20). Activated grids were used immediately. Undiluted protein (8 μl) was placed on the activated carbon surface and samples mounted and stained with 2% uranyl acetate as described previously (20).
Native Gel Analyses.
Nondenaturing PAGE was performed with a standard Tris-glycine buffer system from which SDS was excluded (7.5% T and 2.6% C polyacrylamide gels). Electrophoresis was performed at 10 V/cm at 4°C. For KaiC, 1 mM ATP was added to the gels and the electrode buffer. After the electrophoresis, gels were fixed with methanol-glacial acetic acid and stained with Coomassie brilliant blue R-250.
Electrophoretic Mobility-Shift Assay (EMSA) for DNA Binding.
Binding of KaiC and KaiC-P2 to DNA was tested using different DNA substrates: linear double- or single-stranded DNA and a forked DNA substrate (i.e., partially double-stranded with forks of single-stranded dT DNA). The design of the forked DNA substrate was suggested by Anthony Schwacha based on the work of Lee and Hurwitz (21). To prepare the forked DNA probe, the oligonucleotides (5′-TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TGG TTG GCC GAT CAA GTG CCC AGT CAC GAC GTT GTA AAA CGA GCC C-3′) and (5′-CAC TCG GGC TCG TTT TAC AAC GTC GTG ACT GGG CAC TTG ATC GGC CAA CCT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TT-3′) were radiolabeled by using [γ-32P]ATP (6,000 Ci/mmol; ICN) and T4 polynucleotide kinase, annealed, and purified by polyacrylamide gel electrophoresis. The KaiC or KaiC-P2 samples that were used for EMSA were purified to greater than 99% purity. Purified KaiC or KaiC-P2 was incubated with radiolabeled DNA substrates in 20 μl of binding buffer (25 mM Hepes⋅KOH, pH 7.5/100 mM KCl/10% glycerol/2 mM DTT/0.1 mg/ml acetylated BSA/5 mM magnesium acetate/5 mM ATP or 1 mM AMP-PNP) for 30 min at 30°C (for KaiC) or 53°C (for KaiC-P2). Reactions were loaded onto polyacrylamide gels (8% T and 2.6% C) containing 5 mM magnesium acetate and 10 μM AMP-PNP and run at 10 V/cm in 0.5 × TBE buffer (25 mM Tris⋅HCl, pH 8.5/25 mM boric acid/0.5 mM EDTA) containing 5 mM magnesium acetate and 10 μM AMP-PNP at 4°C for 8 h. The gels were fixed, dried, and exposed to a storage phosphor screen (Molecular Dynamics, Amersham Pharmacia Biosciences) for 24≈72 h.
Results and Discussion
Sequences of KaiC Proteins.
The sequence of the KaiC protein from S. elongatus PCC 7942 is aligned in Fig. 1 with KaiC-P2 from S. lividus strain P2 that was isolated from a 50–55°C hot spring in Yellowstone National Park. There is clear homology between these two proteins except for an N-terminal sequence present in KaiC-P2 that is absent from KaiC. Fig. 1 also shows the sequence of KaiCI, which is the N-terminal half of the internally duplicated KaiC (7).
KaiC Forms Hexameric Complexes in the Presence of ATP.
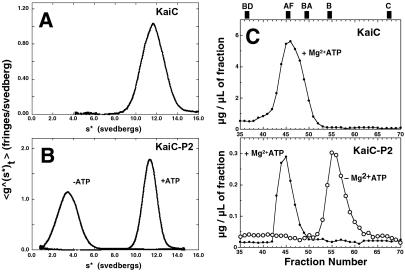
We found that KaiC and KaiC-P2 form hexameric complexes in the presence of ATP. We used two different methods to determine the size of these KaiC complexes: analytical ultracentrifugation and gel filtration chromatography. Analytical ultracentrifugation of KaiC was performed in the presence of ATP. As shown in Fig. 2A, time-derivative sedimentation velocity analysis and fitting with sedanal indicated that KaiC sedimented with a sedimentation coefficient s20,w of 11.7 S and a diffusion coefficient of D20,w of 3.17 F [1 fick (F) = 10−7 cm2 sec−1], giving a molar mass of 336 kg/mol, which is close to that predicted for a hexamer of KaiC [6 × 58.8 = 353 kg/mol (353 kDa)]. From the diffusion coefficient, we calculated a Rs of 6.8 nm for the KaiC hexamer. For KaiC-P2 in the absence of ATP, we observed 3.61 S, 5.57 F, Rs = 3.8 nm, giving a molar mass of 58.0 kg/mol for KaiC-P2 (expected mass for the monomer from its sequence would be 64.7 kg/mol). For KaiC-P2 in the presence of ATP, we observed 11.4 S, 2.77 F, Rs = 7.8 nm, giving a molar mass of 368 kg/mol (Fig. 2B). Again, this is as expected for a hexamer of KaiC-P2 (6 × 64.7 kDa = 388 kg/mol).
Fig 2.
KaiC and KaiC-P2 form homohexameric complexes. (A and B) Time-derivative sedimentation velocity analysis of KaiC and KaiC-P2. Plots of g(s*) vs. s*. The peak value of s* gives the sedimentation coefficient, and the standard deviation gives the diffusion coefficient, both corrected to water at 20°C. The combination of s and D gives an estimate of molecular weight. (A) Analytical ultracentrifugation of KaiC in the presence of 5 mM ATP, 40,000 rpm. s20,w = 11.7S; D20,w = 3.17F; Rs = 6.8 nm; M = 336 kg/mol. (B) Analytical ultracentrifugation of KaiC-P2 in the absence and presence of 5 mM ATP, 50,000 rpm. In the presence of ATP: s20,w = 11.4S; D20,w = 2.77F; Rs = 7.8 nm; M = 368 kg/mol, whereas in the absence of ATP: s20,w = 3.61S; D20,w = 5.57F; Rs = 3.8 nm; M = 58.0 kg/mol. (C) Gel filtration of KaiC (Upper) and KaiC-P2 (Lower) complexes on a Sephacryl S-300 HR column. Positions of standards are shown at the top of Upper (standards: BD = blue dextran, AF = apoferritin, BA = β-amylase, B = BSA, C = cytochrome c).
Determination of the Rs of KaiC by analytical gel filtration chromatography on Sephacryl S-300 HR of these samples (Fig. 2C) corroborated the Rs measurements determined by sedimentation–diffusion analysis. Gel filtration analysis for KaiC gave a value of 6.0 nm for the Rs, which agrees well with the Rs value of 6.8 nm obtained by sedimentation–diffusion analysis. For KaiC-P2, gel filtration analyses gave values of 6.2 nm for the hexamer in the presence of ATP and of 3.6 nm for the monomer in the absence of ATP. These values agree reasonably well with the values of 7.8 and 3.8 nm, respectively, derived from the diffusion coefficients measured by sedimentation analysis. Comparison of the Rs measurements from sedimentation and gel filtration indicates that there is no significant heterogeneity in the presence of ATP. Heterogeneity would have resulted not only in poor fits of the sedimentation velocity data but also in much smaller values for the Rs derived from the fitted apparent diffusion coefficients. Heterogeneity in sedimentation velocity experiments is manifested in apparent diffusion coefficients that increase with time. This type of behavior was not observed with either KaiC or KaiC-P2 in the presence of ATP indicating that the 11S peak is indeed a homogeneous hexamer. Taken together, the analytical ultracentrifugation and gel filtration data indicate that KaiC and KaiC-P2 form complexes in the presence of ATP that are hexamers.
Formation of KaiC and KaiC-P2 Complexes Depends on ATP.
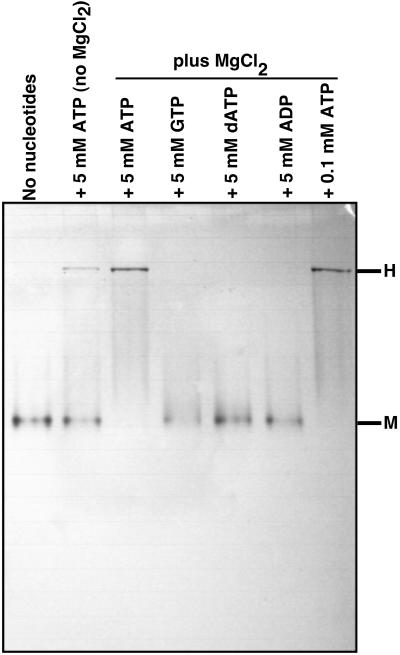
When recombinant His-6-tagged KaiC was purified in the absence of ATP, it precipitated rapidly after elution from the metal affinity column. On the other hand, if ATP was included during the purification and in the elution buffer, KaiC remained in solution as a hexamer for days. Unlike KaiC, KaiC-P2 could be purified in the absence of ATP and remained soluble. As shown in Fig. 2 B and C, KaiC-P2 appears to convert from a monomeric to a hexameric form after ATP is added. We used native gel electrophoresis to further analyze this phenomenon. KaiC-P2 in the absence of ATP migrates on native gels to a position consistent with a monomeric form (Fig. 3, “no nucleotide”). Conversion of KaiC-P2 monomers to hexamers in the presence of ATP and Mg2+ could be observed and monitored by the large decrease in mobility seen by native gel electrophoresis. ATP alone will partially allow this conversion, but Mg2+ alone will not. This association is specific for ATP: neither GTP, dATP, nor ADP will substitute for ATP in the formation of the hexamers (Fig. 3).
Fig 3.
Formation of hexameric rings by KaiC-P2 specifically depends on ATP. Purified KaiC-P2 was incubated with the indicated nucleotides in 25 mM Hepes⋅KOH, pH 7.5/50 mM KCl/1 mM DTT/0.5 mM EDTA/±5 mM MgCl2 at 4°C for 2 h, and analyzed by nondenaturing PAGE (the gel did not contain nucleotides). Lane 1, no nucleotides (includes 5 mM MgCl2); lane 2, 5 mM ATP excluding MgCl2; lane 3, 5 mM ATP; lane 4, 5 mM GTP; lane 5, 5 mM dATP; lane 6, 5 mM ADP; lane 7, 0.1 mM ATP (lanes 3–7 include 5 mM MgCl2). H, migration position of hexameric complexes; M, migration position of monomers.
EM of KaiC Proteins.
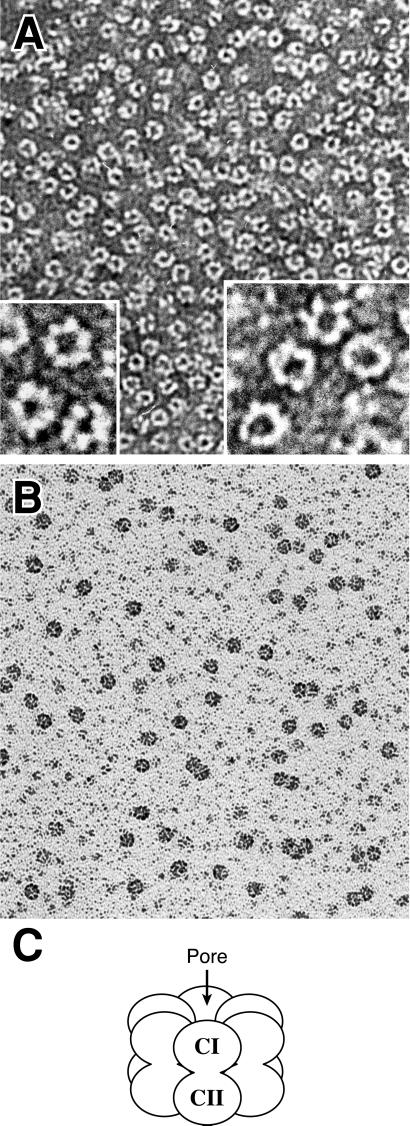
Purified recombinant KaiC, KaiCI, and KaiC-P2 were examined directly by EM. The EM visualizations shown in Fig. 4 are of KaiC. A standard reaction containing 1.7 μM wild-type KaiC protein was incubated at 37°C for 30 min, and the results are shown in Fig. 4. The negatively stained sample shows an abundance of hexagonal ring-like structures (Fig. 4A). The diameter of the rings was estimated to approach the diameter of a DNA-bound RecA filament (≈10 nm). A hole in the center of many rings is clearly evident, although the hole is not seen in all of the views provided within the field. Fig. 4A Insets show two enlargements of several of these structures. On the basis of the data shown in Figs. 2 and 3, this ring-like structure corresponds to a KaiC hexamer.
Fig 4.
EM of ATP-dependent protein complexes formed by the KaiC protein. A and B show the KaiC protein from S. elongatus PCC 7942 in an ATP-containing solution. (A) Sample viewed with the negative staining procedure. Enlarged sections of this image are shown in Insets. (B) A more diluted sample viewed after preparation by positive staining. (C) Model for KaiC hexamers. Each monomer consists of an N-terminal (CI) and a C-terminal (CII) half. These monomers associate into a hexamer in which the monomers surround a central pore.
KaiC samples spread by using the Alcian procedure followed by positive staining and very light shadowing revealed structures of similar size, indicating that the negatively stained images are not an artifact (Fig. 4B). Internal structure can be observed in these positively stained samples because the shadowing was very light, but the central depression was not observed in most objects by this procedure. This very likely arises because positively stained objects tend to present a number of orientations in three dimensions, whereas negatively stained objects tend to align so that internal holes are vertical to the substrate. Direct dilution of the KaiC stock solution, followed by immediate spreading without a 37°C incubation, revealed ring structures that were as plentiful as in the incubated sample, suggesting that the rings were present in the stock solution. The structures we observed were clearly proteinaceous in nature. When KaiC was digested with 0.5 mg/ml proteinase K in 0.1 M NaCl, 0.2% SDS at 37°C for 30 min, EM of the digested sample demonstrated that the rings disappeared (data not shown).
Consistent with the data of Figs. 2 and 3, EM analyses of KaiC and KaiC-P2 confirmed that ring formation depended on the presence of ATP. Three KaiC samples were prepared, treated in different ways, and the circular complexes counted in each. The first was the concentrated KaiC stock solution, diluted 100,000 times into buffer A plus ATP (5 mM), Mg acetate (10 mM), and 2-mercaptoethanol (5 mM). In a survey of 48 fields, an average of 53 KaiC complexes were observed (53 ± 11.6 SD). The sample was diluted to the same extent after the following treatments to allow direct comparison. The ATP was removed from the KaiC stock solution by dialysis overnight at 4°C against buffer A plus 10 mM KCl/0.3 mM EDTA/5 mM 2-mercaptoethanol (pH 8.0). EM of the dialyzed sample showed that the number of the rings in the solution decreased substantially (3.4 ± 1.5 SD complexes per field; 48 fields examined). When ATP (5 mM), Mg acetate (10 mM), and 2-mercaptoethanol (5 mM) were added back to this dialyzed sample and incubated overnight at 37°C, the rings reappeared (24.3 ± 7.6 SD complexes per field; 60 fields counted). This demonstrates that KaiC ring formation is ATP-dependent and reversible. We noted that ring formation in the presence of ATP appeared to be slow. The ATP-depleted sample, examined 30 min after reintroduction of ATP, showed no significant increase in KaiC ring structures (data not shown).
KaiCI protein is the N-terminal half of KaiC that was truncated so that it contains only one of the RecA-like domains (Fig. 1). KaiCI also forms ring structures. Stock protein solutions of KaiCI were examined by EM directly without dilution or incubation at 37°C. Ring structures were abundant in these samples (data not shown). Finally, reversible and ATP-dependent formation of ring structures was also demonstrated with the KaiC-P2 in the EM (data not shown).
On the basis of the data of Figs. 2–4, we envision that the KaiC ring complex is like that shown in Fig. 4C. Based on the internal sequence duplication (7), each KaiC monomer is depicted as a bipartite “dumbbell-like” molecule. These molecules associate into a hexameric ring with a central pore that is obvious in the negatively stained KaiC EM images (Fig. 4A). Because KaiCI molecules also associate into a ring that can be visualized in the EM, we predict that there are intermolecular interaction domains in the N-terminal half of KaiC that are sufficient to mediate ring formation. Some of the complexes shown in the Fig. 4 Insets clearly look like hexamers, whereas others suggest that there might be more than six KaiC monomers in the ring complex. Remembering, however, that KaiC (and KaiC-P2) is an internally duplicated molecule, we expect that a KaiC monomer might look like a dumbbell (Fig. 4C). Therefore, if six dumbbell-like molecules were to splay apart from a ring complex, the appearance would be predicted to be similar to that seen in a few of the complexes depicted in the Fig. 4A Insets that appear to have more than six densities.
KaiC Binds Forked DNA Substrates.
Our initial attempts to demonstrate binding of KaiC or KaiC-P2 to DNA used linear DNA substrates and were unsuccessful. For a single-stranded linear substrate, we tested a random 60-mer single-stranded DNA oligonucleotide. For a linear double-stranded substrate, we tested binding to the kaiBC promoter (5). However, when we changed the substrate to a forked DNA substrate, our EMSA detected significant band shifts for both KaiC-P2 and KaiC (Fig. 5). As shown in Fig. 5A, the inclusion of KaiC-P2 with the labeled forked substrate caused a significant band shift that was competed by higher concentrations of the specific unlabeled forked substrate (Fig. 5A, lanes 2–7). Nonspecific unlabeled single-stranded DNA (using the random 60 mer) did not compete with the forked substrate for KaiC-P2 binding (Fig. 5A, lane 8) nor did an unlabeled dsDNA [poly(dI-dC)⋅poly(dI-dC), not shown]. At equivalent protein/DNA ratios, essentially the same band shift was obtained with KaiC, although we sometimes obtained a second, lower mobility band shift as well (Fig. 5B). The KaiC band shift was also competed by unlabeled forked substrate, but not by linear substrates, in a manner comparable to that for KaiC-P2 (not shown).
Implications of Hexameric Structure.
KaiC is the first circadian clock protein for which structural information about the full-length protein has been visualized. The data described herein indicate that in the presence of ATP and Mg2+, KaiC monomers form hexameric complexes with a central pore. In this respect, KaiC is similar to proteins in the RecA/DnaB superfamily with some of which it shares sequence similarities (6). Unlike RecA and DnaB, however, the KaiC sequence does not include an obvious DNA-binding motif. It is therefore intriguing that KaiC binds forked DNA substrates, implying a direct action of KaiC on DNA metabolism and bolstering its similarity to the proteins of the RecA/DnaB superfamily. That KaiC binds forked substrates suggests a function relating to DNA forks, as a helicase activity, etc. (21).
The sequence similarity (6) and now the structural similarity to the RecA/DnaB superfamily and DNA-binding capability represent important steps in elucidating function. The RecA/DnaB protein superfamily includes a large number of proteins that act on DNA. In addition to RecA and its functional homologues such as the eukaryotic Rad51 protein, these include a range of DNA helicases (22) and DNA pumps (23). However, the RecA/DnaB superfamily also includes structurally related proteins that do not act on DNA, such as the F1-ATPase (24) and adenosylcobinamide kinase/adenosylcobinamide phosphate guanylyltransferase (CobU protein) from Salmonella typhimurium (25). The data on DNA binding of KaiC (Fig. 5) imply that the KaiC protein is a member of the subset of the RecA superfamily that acts on DNA. Because the KaiC sequence does not include an obvious DNA-binding motif, it is possible that further study of KaiC will elucidate a novel motif for protein–DNA interaction.
Our attempts to identify a specific activity of KaiC other than DNA binding have not yet succeeded, but it is likely that KaiC acts in a complex with other partners that have not been identified. Other factors are often necessary to allow enzymatic activity of other members of the RecA superfamily, as is also the case for DnaB (26). KaiC is known to interact with other cyanobacterial proteins, in particular the clock proteins KaiA and KaiB (7) and the histidine kinase SasA (27). It is tempting to consider that these proteins (or others) interact with KaiC to allow KaiC to perform a helicase or other DNA-related activity. If so, KaiC might be a critical factor mediating global regulation of circadian gene expression in cyanobacteria as previously suggested (3). Of course, it is also possible that the hexameric rings formed by KaiC have another function not yet considered. Whether the DNA-action hypothesis is true, the hexameric structure of KaiC ring complexes and its DNA-binding capability provide further support for the inclusion of KaiC in the DnaB/RecA superfamily (6) and tantalizing clues to its enzymatic function.
Acknowledgments
We thank Dr. Michael Ferris and Yellowstone National Park for provision of S. lividus strain P2 and for suggestions on the design of primers to amplify the internal region of kaiC-P2. We also thank Dr. Anthony Schwacha for the suggestion to try forked DNA substrates for KaiC binding, and Drs. Schwacha, Vladimir Podust, and Gisela Mosig for advice about gel shift assays (and, in the case of Dr. Podust, for providing KaiC purified on MonoQ resin). We are grateful for support from the National Science Foundation (MCB-9874371 to C.H.J.; BIR-9513060 to W.F.S.) and the National Institutes of Health (MH01179 and MH43836 to C.H.J.; GM32335 to M.M.C.).
Abbreviations
KaiC, full-length KaiC from Synechococcus elongatus PCC 7942
KaiCI, the first “half” of KaiC from S. elongatus PCC 7942
KaiC-P2, full-length KaiC from the thermophilic cyanobacterium Synechococcus lividus
EM, electron microscope or electron microscopy
EMSA, electrophoretic mobility-shift assay
Rs, Stokes radius
AMP-PNP, adenosine 5′-(β,γ-imido)-triphosphate
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF497977).
References
- 1.Johnson C. H., Golden, S. S., Ishiura, M. & Kondo, T. (1996) Mol. Microbiol. 21, 5-11. [DOI] [PubMed] [Google Scholar]
- 2.Johnson C. H. (2001) Annu. Rev. Physiol. 63, 695-728. [DOI] [PubMed] [Google Scholar]
- 3.Mori T. & Johnson, C. H. (2001) Semin. Cell Dev. Biol. 12, 271-278. [DOI] [PubMed] [Google Scholar]
- 4.Kondo T., Tsinoremas, N. F., Golden, S. S., Johnson, C. H., Kutsuna, S. & Ishiura, M. (1994) Science 266, 1233-1236. [DOI] [PubMed] [Google Scholar]
- 5.Ishiura M., Kutsuna, S., Aoki, S., Iwasaki, H., Andersson, C. R., Tanabe, A., Golden, S. S., Johnson, C. H. & Kondo, T. (1998) Science 281, 1519-1523. [DOI] [PubMed] [Google Scholar]
- 6.Leipe D. D., Aravind, L., Grishin, N. V. & Koonin, E. V. (2000) Genome Res. 10, 5-16. [PubMed] [Google Scholar]
- 7.Iwasaki H., Taniguchi, Y., Kondo, T. & Ishiura, M. (1999) EMBO J. 18, 1137-1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nishiwaki T., Iwasaki, H., Ishiura, M. & Kondo, T. (2000) Proc. Natl. Acad. Sci. USA 97, 495-499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ochman H., Gerber, A. S. & Hartl, D. L. (1988) Genetics 120, 621-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Siebert P. D., Chenchik, A., Kellogg, D. E., Lukyanov, K. A. & Lukyanov, S. A. (1995) Nucleic Acid Res. 23, 1087-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nikaido K., Liu, P.-Q. & Ames, G. F.-L. (1997) J. Biol. Chem. 272, 27745-27752. [DOI] [PubMed] [Google Scholar]
- 12.Stafford W. F. (1992) Anal. Biochem. 203, 295-301. [DOI] [PubMed] [Google Scholar]
- 13.Stafford, W. (2003) Biophys. Soc. Abstracts, in press.
- 14.Todd G. P. & Haschemeyer, R. H. (1981) Proc. Natl. Acad. Sci. USA 78, 6739-6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stafford W. F. (1997) Curr. Opin. Biotechnol. 8, 14-24. [DOI] [PubMed] [Google Scholar]
- 16.Stafford W. F., Jacobsen, M. P., Woodhead, J., Craig, R., O'Neall-Hennessey, E. & Szent-Gyorgyi, A. G. (2001) J. Mol. Biol. 307, 137-147. [DOI] [PubMed] [Google Scholar]
- 17.Laue T. M., Shah, B. D., Ridgeway, T. M. & Pelletier, S. L. (1992) in Analytical Ultracentrifugation in Biochemistry and Polymer Science, eds. Harding, S. E., Rowe, A. J. & Horton, J. C. (Royal Society of Chemistry, Cambridge, U.K.), pp. 90–125.
- 18.Ackers G. K. (1967) J. Biol. Chem. 242, 3237-3241. [PubMed] [Google Scholar]
- 19.Webb B. L., Cox, M. M. & Inman, R. B. (1995) J. Biol. Chem. 270, 31397-3140. [DOI] [PubMed] [Google Scholar]
- 20.Schnos M. & Inman, R. B. (1999) Methods Mol. Biol. 117, 229-243. [DOI] [PubMed] [Google Scholar]
- 21.Lee J.-K. & Hurwitz, J. (2001) Proc. Natl. Acad. Sci. USA 98, 54-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Egelman E. H. (1998) J. Struct. Biol. 124, 123-128. [DOI] [PubMed] [Google Scholar]
- 23.Gomis-Ruth F. X., Moncalian, G., Perez-Luque, R., Gonzalez, A., Cabezon, E., de la Cruz, F. & Coll, M. (2001) Nature 409, 637-641. [DOI] [PubMed] [Google Scholar]
- 24.Abrahams J. P., Leslie, A. G., Lutter, R. & Walker, J. E. (1994) Nature 370, 621-628. [DOI] [PubMed] [Google Scholar]
- 25.Thompson T. B., Thomas, M. G., Escalante-Semerena, J. C. & Rayment, I. R. (1998) Biochemistry 37, 686-695. [DOI] [PubMed] [Google Scholar]
- 26.Learn B. A., Um, S.-J., Huang, L. & McMacken, R. (1997) Proc. Natl. Acad. Sci. USA 94, 1154-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Iwasaki H., Williams, S. B., Kitayama, Y., Ishiura, M., Golden, S. S. & Kondo, T. (2000) Cell 101, 223-233. [DOI] [PubMed] [Google Scholar]