Abstract
Gibberella zeae (anamorph: Fusarium graminearum) is an important pathogen of maize, wheat, and rice. Colonies of G. zeae produce yellow-to-tan mycelia with the white-to-carmine red margins. In this study, we focused on nine putative open reading frames (ORFs) closely linked to PKS12 and GIP1, which are required for aurofusarin biosynthesis in G. zeae. Among them is an ORF designated GIP2 (for Gibberella zeae pigment gene 2), which encodes a putative protein of 398 amino acids that carries a Zn(II)2Cys6 binuclear cluster DNA-binding domain commonly found in transcription factors of yeasts and filamentous fungi. Targeted gene deletion and complementation analyses confirmed that GIP2 is required for aurofusarin biosynthesis. Expression of GIP2 in carrot medium correlated with aurofusarin production by G. zeae and was restricted to vegetative mycelia. Inactivation of the 10 contiguous genes in the ΔGIP2 strain delineates an aurofusarin biosynthetic gene cluster. Overexpression of GIP2 in both the ΔGIP2 and the wild-type strains increases aurofusarin production and reduces mycelial growth. Thus, GIP2 is a putative positive regulator of the aurofusarin biosynthetic gene cluster, and aurofusarin production is negatively correlated with vegetative growth by G. zeae.
Fusarium head blight is caused by several species of Fusarium, including Fusarium graminearum (teleomorph: Gibberella zeae), F. culmorum, and F. crookwellense. These fungi are distributed worldwide and produce mycotoxins that cause economic losses in terms of crop and animal production. They also produce pigments that range from yellow to tan to carmine red (23). Two of the pigments produced by G. zeae and F. culmorum are naphthoquinones: aurofusarin and rubrofusarin (1, 8, 28). Aurofusarin is toxic to poultry and can reduce the nutritional quality of quail eggs (5, 6). Rubrofusarin is antimycobacterial, antiallergenic, and phytotoxic (7, 14, 15, 20).
We previously identified a type I polyketide synthase gene (PKS12) and a putative laccase gene (GIP1) that are required for aurofusarin biosynthesis in G. zeae (13). Thus, aurofusarin is synthesized in G. zeae in a manner similar to that used for other fungal polyketide pigments. Agrobacterium-mediated transformation of F. pseudograminearum also results in aurofusarin-deficient mutants, and targeted mutagenesis in G. zeae confirmed the function of PKS12 (21). However, the role(s) of aurofusarin production in the physiology of G. zeae is not well understood. It may play a role in vegetative growth and zearalenone production in G. zeae based on the phenotype of aurofusarin-deficient mutants (21).
Genes involved in the biosynthesis of secondary metabolites are often clustered in filamentous fungi (11). For example, clusters for biosynthetic genes of trichothecenes (9), fumonisins (27), and gibberellins (31) have been identified in Fusarium species that produce these metabolites. These clusters include genes encoding metabolic enzymes, transcription factors, and transporters. Thus the genes encoding the protein products for aurofusarin biosynthesis might also be clustered. Previously, putative open reading frames (ORFs) were found near the PKS12 gene in G. zeae (13, 21); one of them, which was similar to a fungal transcription factor, was involved in expression of PKS12 (21). However, the role of this putative transcription factor for aurofusarin biosynthesis is unproven and the limits of the aurofusarin gene cluster are unknown.
Our objectives in this study were (i) to determine whether GIP2 is required for aurofusarin biosynthesis, (ii) to identify the genes in the aurofusarin biosynthetic gene cluster, and (iii) to determine whether overproduction or underproduction of aurofusarin affects growth and colony morphology of G. zeae. The results from this study and further characterization of the aurofusarin biosynthetic genes will be used to determine the biological significance of aurofusarin in the growth and physiological activities of G. zeae.
MATERIALS AND METHODS
Strains and media.
Strains SCKO4 and Z03643 were used as wild-type strains of G. zeae (13). SCKO4 is a lineage 6 strain that produces nivalenol and zearalenone. Z03643 is a lineage 7 strain that produces deoxynivalenol and zearalenone (24) and is more pigmented than SCKO4 (13). Fungal strains were stored as spore suspensions in 20% glycerol at −80°C. For inoculum and pigment production, the strains were grown on potato dextrose agar (PDA; Difco Laboratories, Detroit, MI) at 25°C. For DNA extraction, the strains were inoculated in 100 ml of complete medium in 250-ml Erlenmeyer flasks (4) and grown for 3 days at 25°C on a rotary shaker at 150 rpm. For RNA extraction and induction of sexual development, fungi were grown on carrot medium as previously described (16).
Nucleic acid manipulations and PCR primers.
Fungal genomic DNA was extracted as described previously (12). Escherichia coli colonies carrying recombinant plasmids were screened by using a single-tube miniprep method (19). For fungal transformation, plasmids were purified from 5 ml of E. coli culture by using a plasmid purification kit (NucleoGen Biotech, Siheung, Korea). Total RNA was extracted from mycelia (0.1 to 0.2 g) or lawns of perithecia ground in liquid N2 with 1 ml of TRIZOL reagent (Invitrogen, Carlsbad, CA) following the manufacturer's instructions. DNA and RNA gel blot hybridizations with 32P-labeled probes were made as previously described (26). DNA fragments used as probes were amplified from genomic DNAs of Z03643 and SCKO4 with proper primer pairs (Table 1) as described previously (18). PCR primers were obtained from the Bioneer oligonucleotide synthesis facility (Bioneer Corporation, Chungwon, Korea), resuspended at 100 μM in sterile water, and stored at −20°C.
TABLE 1.
Primers used in this study
| Primer | Sequencea (5′ to 3′) | Positionb (bp at contig) or description |
|---|---|---|
| G2-5′f | TTTGCGGGATGATATGACTGAG | 123,112-123,091 at 1.116 |
| G2-5′r | CTCCACTAGCTCCAGCCAAGC | 121,978-121,999 at 1.116 |
| AAACGCAGCTAACAGAGGAGA | ||
| G2-3′f | TGAAAATTCCGTCACCAGCCTC | 120,415-120,394 at 1.116 |
| TCGCGGCAGCTTGTAATCAT | ||
| G2-3′r | GCCATGCTAGCCCAACTCTCC | 118,949-118,969 at 1.116 |
| NG2-5′f | GCTTGCATCGGACCGAAGGAATGA | 122,880-122,856 at 1.116 |
| NG2-3′r | GCCCTGAGCTTGCAGCAGAGTGTCTT | 119,164-119,189 at 1.116 |
| P12-for | TATAGGGGATTGTGCTTC | 136,498-136,481 at 1.116 |
| P12-rev | AATACACAAACAGCCCTCTC | 129,906-129,925 at 1.116 |
| G1-for | AGGAGCCCTCCAAGTAAT | 144,157-144,140 at 1.116 |
| G1-rev | TCAACGCAGTCGATTGTA | 141,752-141,769 at 1.116 |
| G2-for | ACGACCTTCCTAAACTGCACCTATCA | 122,182-122,157 at 1.116 |
| G2-rev | GGATGCCTGCACCCACCTA | 120,235-120,253 at 1.116 |
| G3-for | GGCTTAACGGCTGAGGACCAAT | 122,940-122,961 at 1.116 |
| G3-rev | GCACGGCCGATCTCATCAAG | 124,669-124,650 at 1.116 |
| G4-for | GCATCACCATTCAATCCT | 127,093-127,076 at 1.116 |
| G4-rev | CCTTTTTATTGCATTGCA | 125,277-125,294 at 1.116 |
| G5-for | TCTCGTCAATACCAACCC | 127,538-127,555 at 1.116 |
| G5-rev | GCAGATTGCCTTCATTCT | 129,844-129,827 at 1.116 |
| G6-for | CGCCTCATAGTGATACCCAAGAAA | 139,925-136,948 at 1.116 |
| G6-rev | AAACGCGGATCTGCCTTCAT | 137,264-137,245 at 1.116 |
| G7-for | GAACACGGCATCCACTGTAAGAT | 139,307-139,285 at 1.116 |
| G7-rev | GCGATATCAGCGAGATCAAAAATA | 137,647-137,670 at 1.116 |
| G8-for | CACCCCGACCCGAAGAGC | 139,452-139,469 at 1.116 |
| G8-rev | CTCACAACAGTCAATCAGGAACCAC | 141,641-141,617 at 1.116 |
| G9-for | TCATCAATGTCAGCCAAG | 144,377-144,394 at 1.116 |
| G9-rev | CGAGTCGCACTGAGTATG | 145,843-145,826 at 1.116 |
| G10-for | GCTTGCCATCTCGAGTTTGAAT | 148,254-148,233 at 1.116 |
| G10-rev | CAGTGCGACTCGAATGAGGC | 145,832-145,851 at 1.116 |
| G2ov-p1 | CATTACATACAGCCGTCAATA | 121,713-121,692 at 1.116 |
| .......................................................... | ||
| TGAGTTCCACAGACCCCCTTC | ||
| G2ov-p2 | GTGTTGGTGCCGTTGCCC | 119,150-119,167 at 1.116 |
| G2ov-p3 | TGTTCGTCGTCGGCTTCGTTC | 123,273-123,293 at 1.116 |
| GCGGGGTCTACAATAAGC | ||
| G2ov-p4 | GCACGGCCGATCTCATCAAG | 124,669-124,650 at 1.116 |
| bpro-5′ | CGAAGCCGACGACGAACA | 49,310-49,327 at 1.393 |
| bpro-3′ | ATTGACGGCTGTAGATGTAATG | 51,455-51,434 at 1.393 |
| ................................................................. | ||
| NG2ov-5′f | ACCCAATACTATCGCCTGTCG | 124,493-124,473 at 1.116 |
| NG2ov-3′r | GGAACGCCTTGAAGAGAATGTC | 119,613-119,634 at 1.116 |
| nHygB-f | CTTGGCTGGAGCTAGTGGAGGT | For amplification of hygB cassette from pBCATPH |
| nHygB-r | GGCTGGTGACGGAATTTTCATA | For amplification of hygB cassette from pBCATPH |
Underlined sequences with the same patterns (e.g., light underline, heavy underline, double underline, dotted underline) are complementary and promote hybridization between the PCR products.
The positions indicated as bp at contig are from the F. graminearum database at http://www.broad.mit.edu/annotation/fungi/fusarium/index.html.
Construction of transforming vectors.
For PCR-based construction of fungal transforming vectors, a double-joint PCR method with appropriate primers (Table 1) was employed as previously described (33). To delete GIP2, DNA fragments corresponding to regions 5′ (1.1 kb) and 3′ (1.5 kb) of the GIP2 ORF were amplified from genomic DNA of either Z03643 or SCKO4 with primer pair G2-5′f and G2-5′r and primer pair G2-3′f and G2-3′r, respectively. A 1.8-kb fragment containing the hygB gene under the control of the Aspergillus nidulans trpC promoter and terminator was amplified from the vector pBCATPH (34) with primers nHygB-f and nHygB-r. Three amplicons (5′-flanking region of GIP2, the hygB cassette, and the 3′-flanking region of GIP2) were mixed in a 1:2:1 ratio, and a second round of PCR was carried out. Using a new nested primer pair (NG2-5′f and NG2-3′r, which are nested in G2-5′f and G2-3′r, respectively) and the second PCR product as the template, a 4.4-kb fusion PCR product was amplified.
For the GIP2 construct fused to a putative promoter of the G. zeae β-tubulin gene (GenBank accession no. AAP68979), a 2.6-kb region containing the GIP2 ORF and its 3′ flank (1.3 kb) and a 1.4-kb 5′-flanking region of GIP2 were amplified from Z03643 with primer pair G2ov-p1 and G2ov-p2 and primer pair G2ov-p3 and G2ov-p4, respectively. These amplicons and a 2.1-kb putative β-tubulin gene promoter amplified from Z03643 with primer pair bpro-5′ and bpro-3′ were fused each other in a PCR with a nested primer pair (NG2ov-5′f and NG2ov-3′r), resulting in a 5.5-kb fusion PCR product.
Fungal transformation.
Following phenol extraction and ethanol precipitation, ∼5 μg of the final PCR product alone, if it carried a selectable marker, was incorporated directly into fungal protoplasts or in combination with plasmid pSK660 which carries the geneticin resistance gene (gen) as a selectable marker as previously described (13).
RESULTS
Structural organization of a putative PKS12 gene cluster.
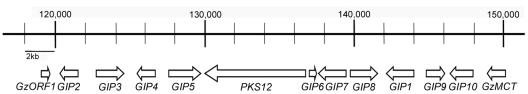
The 30 kb-region carrying PKS12 and GIP1, required for aurofusarin biosynthesis in G. zeae, contains several ORFs, including sequences that are similar to transcription factors, an efflux pump, and some metabolic enzymes (Fig. 1, Table 2). This putative PKS12 gene cluster was arbitrarily defined, because there is no significant sequence structure outside of this region for proximity of a gene cluster. A 1.2-kb ORF designated GIP2, which contains no putative intron, exhibited the highest amino acid identity (21%) to the transcription factor AFLR required for sterigmatocystin or aflatoxin production in Aspergillus species (32). However, significant identity (53%) with AFLR was mostly due to the conserved Zn(II)2Cys6-type DNA binding motif that frequently occurs in other transcription factors of yeasts and filamentous fungi (3).
FIG. 1.
Molecular organization of the putative aurofusarin biosynthesis cluster in G. zeae. This cluster was found in contig 1.116 from the F. graminearum genome databases. Numbers and arrows indicate possible ORFs and transcriptional directions, respectively. Nucleotide positions of the contig are indicated on the thick vertical bar.
TABLE 2.
Nucleotide sequence similarities of the genes located at the 30-kb region carrying PKS12
| Gene | Locusa | Similarity (accession no.) | Species | E value |
|---|---|---|---|---|
| PKS12 | FG02324.1 | Polyketide synthase (AF025541) | A. fumigatus | 0 |
| GIP1 | FG02328.1 | brown 2 (AF104823) | A. fumigatus | e−142 |
| GIP2 | FG02320.1 | AFLR (AY618557) | A. flavus | 8e−08 |
| GIP3 | FG02321.1 | FAD/FMN-containing dehydrogenases (NZ_AAED01000004) | Mesorhizobium sp. | 9e−29 |
| GIP4 | FG02322.1 | DHA14-like major facilitator (AF238225) | Botryotinia fuckeliana | e−108 |
| GIP5 | FG02323.1 | Putative transcriptional activator (NP_593170) | Schizosaccharomyces pombe | 6e−14 |
| GIP6 | FG02325.1 | Hypothetical protein | A. nidulans | 3e−29 |
| GIP7 | FG02326.1 | AFLJ (AY510453) | A. flavus | 3e−24 |
| GIP8 | FG02327.1 | Flavin-containing monooxygenase 5 (AAA67848) | Cavia porcellus | 1e−44 |
| GIP9 | FG02329.1 | Fasciclin I family protein, putative (BX649607) | A. fumigatus | 8e−27 |
| GIP10 | FG02330.1 | Ascorbate oxidase (AB010110) | Acremonium sp. | e−130 |
| GzORF1 | FG02319.1 | Predicted protein (EAA69859) | G. zeae | 2e−104 |
| GzMCT | FG02331.1 | Major superfacilitator superfamily monocarboxylate transporter, putative | A. fumigatus | 6e−29 |
The sources of each locus from the F. graminearum database can be found at http://www.broad.mit.edu/annotation/fungi/fusarium/index.html.
Targeted deletion of GIP2.
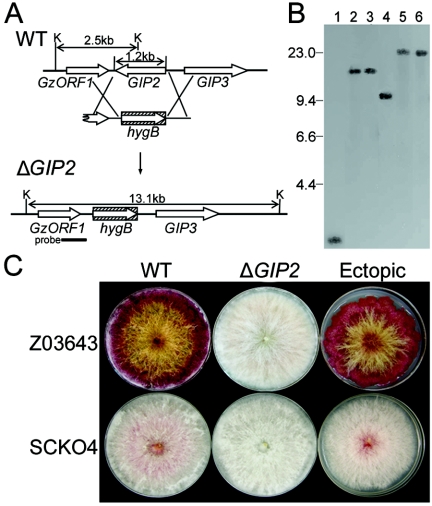
We deleted GIP2 from strains Z03643 and SCKO4. The entire ORF of GIP2 was replaced with the fungal selectable marker hygB via double homologous recombination between a PCR fragment carrying both the 5′ and 3′ regions of the GIP2 ORF fused to hygB and the corresponding genomic regions (Fig. 2A). Genomic DNAs of the ΔGIP2 strains derived from Z03643 and SCKO4 carried a 13.1-kb band and a ∼22.8-kb hybridizing band, when digested with KpnI, instead of the 2.5-kb and ∼9.7-kb bands found in the wild-type strains, respectively, suggesting that the 1.2-kb GIP2 ORF had been deleted and replaced with the hygB gene (Fig. 2B). The ΔGIP2 strains derived from both strains could not produce aurofusarin when grown on PDA, but a transgenic strain carrying the transforming DNA at an ectopic position had red wild-type pigmentation (Fig. 2C). Radial growth on PDA of the ΔGIP2 transgenic Z03643 strains increased ∼30% relative to that of its wild-type progenitor. Mycelial growth of the transgenic ΔGIP2 strain of SCKO4 was not significantly different from that of the untransformed parent (data not shown).
FIG. 2.
Targeted deletion of GIP2 from the genome of wild-type G. zeae strains Z03643 and SCKO4. (A) Deletion strategy. WT, genomic DNA of the wild-type strain Z03643; ΔGIP2, genomic DNA of the strain with GIP2 deleted; K, KpnI; hygB, hygromycin B resistance gene. The probe used for blot hybridization, which is amplified from genomic DNA of Z03643 with primers G2-3′f and G2-3′r (Table 1), is indicated by a thick bar. (B) Gel blot of KpnI-digested genomic DNAs from ΔGIP2 strains, hybridized with the probe. Lanes 1 and 4, Z03643 and SCKO4, respectively; lanes 2 and 3, the ΔGIP2 strains of Z03643 (Tzg2-1 and Tzg2-2, respectively); lanes 5 and 6, the ΔGIP2 strains of SCKO4 (Tsg2-1 and Tsg2-2, respectively). The sizes of λDNA standards (in kilobases) are indicated on the left of the blot. (C) Pigmentation of transformants of Z03643 and SCKO4. WT, wild-type strains; ΔGIP2, the GIP2-deleted strains Tzg2-1 and Tsg2-1; Ectopic, transformants carrying ectopic vector integrations (Tzg2-5 and Tsg2-8).
Complementation analyses.
Intact copies of GIP2 were introduced into a ΔGIP2 mutant by cotransformation (Fig. 3A). Of 16 geneticin-resistant transformants with GIP2, four produced as much aurofusarin as Z03643 (Fig. 3B). One of these transformants has integrated the GIP2 gene at its original genomic locus by homologous recombination (Fig. 3C). To determine whether the heterologous genes were expressed in these transformants, total RNA was extracted from colonies grown on carrot medium and hybridized with GIP2. In pigmented transformants carrying GIP2, GIP2 transcripts were present at a level similar to that found in the wild-type strain (Fig. 3D).
FIG. 3.
Complementation of pigmentation in the G. zeae ΔGIP2 strain. (A) Complementation strategy with GIP2 from G. zeae. ΔGIP2, genomic DNA from the GIP2-deleted strain of Z03643 (Tzg2-1); Rg2-3, genomic DNA of the transgenic Tzg2-1 strain carrying GIP2; K, KpnI; hygB, hygromycin resistance gene; gen and amp, genes conferring resistance to geneticin and ampicillin, respectively. GzORF1 and GIP3 are indicated by open arrows. (B) Pigmentation of the transgenic strains examined by DNA gel blot analysis. Left, Z03643; middle, Tzg2-1; right, Rg2-3. (C) DNA gel blots of transformants derived from the ΔGIP2 strain by using an intact copy of the GIP2 ORF. Genomic DNAs of these transformants were digested with KpnI and hybridized with the probes indicated in panel A. The probe for hybridization with GIP2 was amplified from genomic DNA of Z03643 with primers G2-for and G2-rev (Table 1). Lane 1, wild-type strain Z03643; lane 2, ΔGIP2 recipient strain Tzg2-1; lane 3, transformants (Rg2-3) carrying GIP2. The sizes of λDNA standards (in kilobase) are indicated on the left of the blot. (D) RNA gel blot of strain Z03643 and the transgenic ΔGIP2 strain probed with GIP2. Z03643 and ΔGIP2, total RNAs from wild-type strain and the ΔGIP2 recipient strain Tzg2-1, respectively; Rg2-3, total RNAs from transgenic Tzg2-1 strains carrying GIP2. The probe and the incubation time are indicated on the left and above, respectively. The ethidium bromide-stained rRNAs are indicated as a loading control.
Transcript analyses.
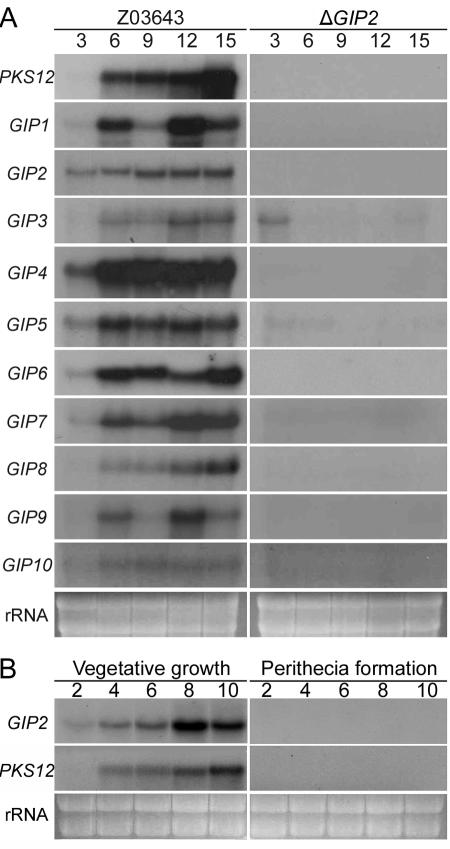
To determine whether the nine genes closely linked to both PKS12 and GIP1 are associated with aurofusarin biosynthesis, we analyzed their expression patterns on carrot medium, which is conducive to aurofusarin production by G. zeae (13). Total RNA was extracted from the mycelia of both Z03643 and the transgenic ΔGIP2 strain (Tzg2-1) grown for 3, 6, 9, 12, and 15 days. Transcripts of all 11 genes (GIP2 to GIP10; Fig. 1) were detected beginning either 3 or 6 days after inoculation in Z03643, but none of the genes except GIP3 and GIP5 were expressed in Tzg2-1 during the entire incubation period (Fig. 4A). The expression pattern for most of the genes paralleled the time course of fungal pigmentation, but signal intensities differed between individual genes. The transcripts of PKS12 and GIP8 began to accumulate on day 6 and increased gradually until day 15. Transcripts of GIP2, GIP4, GIP5, and GIP7 were first detected on day 3, and the accumulation levels remained high through day 15. No transcripts of the putative ORFs (GzORF1 and GzMCT) that were located 3′ of GIP2 and 5′ of GIP10, respectively, were detected in either Z03643 or Tzg2-1 (data not shown). Thus, the coregulated set of 11 contiguous genes probably defines the aurofusarin biosynthetic gene cluster regulated by GIP2. No transcripts of GIP2 or PKS12 were detected in Z03643 during sexual development, i.e., during perithecial formation on carrot agar medium (Fig. 4B).
FIG. 4.
Transcript analyses of the aurofusarin gene cluster. (A) RNA gel blots of Z03643 and its ΔGIP2 strain Tzg2-1, probed with each cluster gene. Z03643 and ΔGIP2, total RNAs from strains Z03643 and Tzg2-1 grown on carrot medium, respectively. The probe and the incubation time are indicated on the left and above, respectively. The probes used for hybridization were amplified from genomic DNA of Z03643 by use of the following primer pairs: P12-for and P12-rev for PKS12, G1-for and G1-rev for GIP1, G2-for and G2-rev for GIP2, G3-for and G3-rev for GIP3, G4-for and G4-rev for GIP4, G5-for and G5-rev for GIP5, G6-for and G6-rev for GIP6, G7-for and G7-rev for GIP7, G8-for and G8-rev for GIP8, G9-for and G9-rev for GIP9, and G10-for and G10-rev for GIP10 (Table 1). (B) RNA gel blots of Z03643 grown on carrot medium, probed with GIP2 and PKS12, respectively. The incubation times for vegetative growth and perithecia induction are indicated above the blots. The ethidium bromide-stained rRNAs are indicated as a loading control.
Overexpression of GIP2.
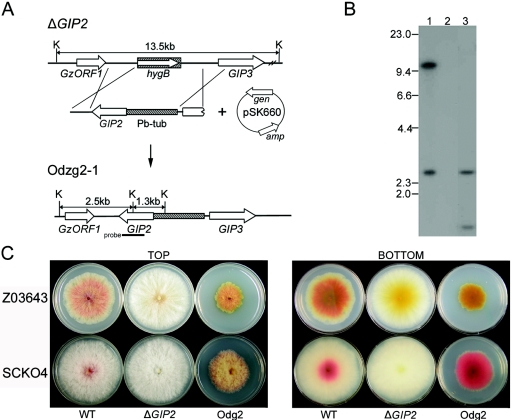
We cotransformed the ΔGIP2 mutant with a 5.5-kb DNA fragment (Fig. 5A) carrying the GIP2 ORF fused to the β-tubulin promoter of G. zeae to assess the effects of GIP2 overexpression. Of 15 geneticin-resistant transformants derived from Tzg2-1, four were sensitive to hygromycin B, which suggested that the heterologous GIP2 gene under the control of β-tubulin gene promoter was integrated into the original GIP2 genomic locus. The presence of hybridizing bands (2.5 kb and 1.3 kb) of the expected size confirmed this type of integration (Fig. 5B). All four hygBS genR transformants were more pigmented than was Z03643 (Fig. 5C). These transformants began to produce red pigments on PDA 2 days after inoculation, while strain Z03643 did not begin to produce pigments until day 4. Mycelial growth of all of four transformants was severely retarded relative to that of Z03643. A similar pattern was observed when GIP2 was overproduced in the ΔGIP2 SCKO4 strains, but the reduction in radial growth of these transformants was not as severe as was the reduction in the strains derived from Z03643 (Fig. 5C). The overexpressing transformants produced ∼50% less fungal mass but three times more aurofusarin in potato broth at 3 days after inoculation compared with wild-type strain results (data not shown).
FIG. 5.
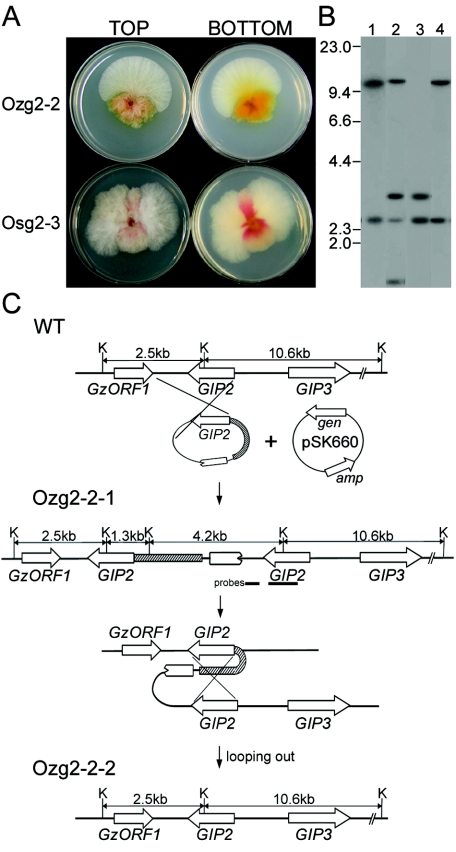
Overexpression of GIP2 in G. zeae ΔGIP2 strains. (A) Overexpression strategy. ΔGIP2, genomic DNA of Tzg2-1, the ΔGIP2 strain derived from Z03643; Odzg2-1, genomic DNA of Odzg2-1, the transgenic Tzg2-1 strain carrying the GIP2 gene under control of the G. zeae β-tubulin gene promoter; pSK660, a vector used for cotransformation; K, KpnI; Pb-tub, promoter region of the β-tubulin gene from Z03643; hygB, hygromycin resistance gene; gen and amp, genes conferring resistance to geneticin and ampicillin, respectively. The probe used for blot hybridization, which is amplified from genomic DNA of Z03643 with primers G2-for and G2-rev (Table 1), is indicated by a thick bar. GzORF1 and GIP3 are indicated by open arrows. (B) DNA gel blot of transformants derived from ΔGIP2 strains, hybridized with GIP2. Lane 1, Z03643; lanes 2, Tzg2-1, the ΔGIP2 recipient strains derived from Z03643; lanes 3, Odzg2-1, a transformant derived from Tzg2-1. The sizes of λDNA standards (in kilobases) are indicated on the left of the blot. (C) Pigmentation of transgenic strains. The upper and lower plates in each panel indicate the strains derived from Z03643 and SCKO4, respectively. WT, wild-type strains; ΔGIP2, the transgenic strains with GIP2 deleted (Tzg2-1 and Tsg2-2 derived from Z03643 and SCKO4, respectively); Odg2, the GIP2-overexpressing transformants from the ΔGIP2 strains (Odzg2-1 and Odsg2-2 derived from Tzg2-1 and Tsg2-2, respectively).
The fused β-tubulin/GIP2 construct also was introduced into the wild-type Z03643 and SCKO4 strains. These transformants had unstable mycelial morphology and produced sectored colonies with differing morphologies. Seventeen transformants of Z03643 were highly pigmented and had slow growth. Fifteen of these 17 transformants frequently produced fast-growing sectors on PDA plates (Fig. 6A). When the mycelia of the sector were transferred to a fresh PDA plate, they grew as fast as the original wild-type strain but were much less pigmented; this phenotype remained stable for five successive transfers on PDA. When mycelia of the highly pigmented original transformants were transferred to fresh media, the resulting colonies continued to form sectors.
FIG. 6.
Overexpression of GIP2 in wild-type G. zeae strains Z03643 and SCKO4. (A) Pigmentation of transgenic strains. The upper and lower plates in each panel indicate transgenic strains carrying the heterologous GIP2, Ozg2-2, and Osg2-3 derived from Z03643 and SCKO4, respectively. (B) DNA gel blot of transformants derived from wild-type strains. Lane 1, Z03643; lanes 2 and 3, Ozg2-2-1 probed with GIP2 and a 3′ flanking region of GIP2, respectively; lane 4, Ozg2-2-2, probed with GIP2. The sizes of λDNA standards (in kilobases) are indicated on the left of the blot. (C) Overexpression strategy. WT, genomic DNA of Z03643; Ozg2-2-1, genomic DNA from mycelia of the highly pigmented original transformant Ozg2-2; Ozg2-2-2, genomic DNA from the wild-type sector from Ozg2-2; pSK660, a vector used for cotransformation; K, KpnI; Pb-tub, promoter region of the β-tubulin gene from Z03643; gen and amp, genes conferring resistance to geneticin and ampicillin, respectively. The probes of GIP2 and the 3′ flank of GIP2, which are amplified from genomic DNA of Z03643 with primer pairs G2-for and G2-rev and G2-3′f and G2-3′r, respectively (Table 1), are indicated by thick bars. GzORF1 and GIP3 are indicated by open arrows.
The highly pigmented transformants carried the heterologous GIP2 gene, which was integrated into the genome by a single homologous recombination event between a circular PCR product and the corresponding genomic region (Fig. 6B). The hybridization patterns in transformant Ozg2-2-1 are consistent with this integration mechanism. The four expected hybridizing bands appeared when probed with GIP2, but only two of four bands were seen when probed with 3′ flanking region of GIP2 (Fig. 6B and C). The presence of a ∼3.0-kb band, instead of the expected 4.2-kb band, when probed with GIP2 may have resulted from loss of terminal portions of the PCR product during the circularization through self-ligation. Genomic DNA of the mycelia from the fast-growing, less-pigmented sector had the same hybridization pattern as that of Z03643 when probed with GIP2, indicating that the sector no longer carried the heterologous GIP2 sequence. These possible revertants could be generated by looping out and deleting the β-tubulin promoter from the chromosome through homologous recombination between adjacent two copies of GIP2 (carrying heterologous and endogenous promoters, respectively) in the transformant Ozg2-2-1 (Fig. 6C). The same sectoring phenotype also was observed in the transformants of SCKO4 that carried the heterologous GIP2 sequence (Fig. 6A).
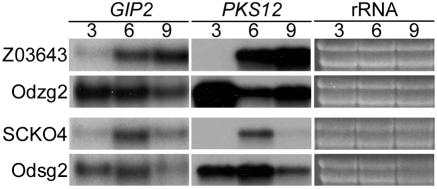
Northern blot analysis confirmed overexpression of both PKS12 and GIP2 in transformants carrying the β-tubulin/GIP2 constructs. The transcripts of both genes were present at high levels from day 3 in the GIP2-overexpressing strains, but little or no transcript was detected in the wild-type strains (Fig. 7).
FIG. 7.
RNA gel blots of the GIP2-overexpressing transformants derived from the ΔGIP2 strains, probed with GIP2 and PKS12, respectively. Z03643 and SCKO4, total RNAs from the wild-type strains; Odzg2-1 and Odsg2-2, total RNAs from the transgenic strains overexpressing GIP2, derived from the ΔGIP2 strains of Z03643 and SCKO4, respectively. The probe and the incubation time are indicated above each blot. The ethidium bromide-stained rRNAs are indicated as a loading control.
DISCUSSION
In filamentous fungi, genes responsible for the biosynthesis of secondary metabolites often are coordinately regulated and physically clustered in the genome. We previously described two aurofusarin biosynthetic genes, PKS12 and GIP1, which encode an unreduced polyketide synthase and a putative laccase, respectively (13). In this report, we identify an additional gene, designated GIP2, that is closely linked to these genes and that is essential for aurofusarin biosynthesis in G. zeae. The requirement for all of these closely linked genes for aurofusarin biosynthesis suggests that aurofusarin is produced in G. zeae through the coordinated activities of the proteins encoded by the genes in the cluster, as is known for other fungal secondary metabolites (9, 11, 27, 31).
Eight additional genes were closely spaced within the 30-kb region carrying the three genes known to be involved in aurofusarin biosynthesis. Sequence similarity of the putative GIP2 protein to fungal transcription factors, especially those associated with a number of metabolic pathways (29), suggests that GIP2 plays an important role in the transcription of the putative clustered genes. Malz et al. (21) reported that GIP2 is important for regulating PKS12 and other genes in the putative aurofusarin biosynthetic gene cluster. They provided reverse transcription-PCR data for altered expression of the genes in the G. zeae strains with insertional mutations in the promoter region of GIP2. In this study we generated RNA gel blots with ΔGIP2 and GIP2-overexpressing G. zeae strains to conclusively define the genes in the aurofusarin biosynthetic gene cluster in G. zeae and to show that GIP2 was required for transcription of the clustered genes.
Coexpression of the same set of genes (11 contiguous genes) in the wild type and their lack of expression in the ΔGIP2 strain under conditions favorable for pigmentation suggest that the gene cluster, coordinately induced by GIP2, is associated with aurofusarin biosynthesis in G. zeae. In addition, the high and early accumulation of PKS12 transcripts in the overexpressing G. zeae strain is consistent with the hypothesis that GIP2 is a transcriptional activator. Additional work remains to be done for the other eight genes in the aurofusarin biosynthetic cluster. We have not shown that GIP2 binds to specific sequences in the promoter regions of the aurofusarin genes. Neither has the role of two other putative transcription factors (GIP5 and GIP7) been determined. Furthermore, it is possible that GIP2 may regulate genes other than those involved exclusively in aurofusarin biosynthesis, as has been shown for a regulatory gene TRI10 in trichothecene biosynthesis in F. sporotrichioides (25).
Aurofusarin production can significantly affect the growth of G. zeae, especially the mycelial growth. The faster growth rate of the ΔGIP2 mutants of Z03643 suggests that aurofusarin accumulation may adversely affect mycelial growth of G. zeae. A similar result was obtained with the albino, ΔPKS12 mutants from G. zeae strain PH-1 (21). These results are consistent with previous reports that highly pigmented G. zeae isolates infected with double-stranded RNAs also have reduced growth (2). However, this phenotypic change is neither simple nor universal, since ΔGIP2 mutants from SCKO4 did not grow at a rate obviously different from that of the SCKO4 wild type. The different effects of aurofusarin deficiency on mycelial growth may be attributed to genetic differences between these three G. zeae strains. SCKO4, a lineage 6 strain obtained from barley in Korea (13), is less pigmented than are Z03643 and PH-1, which belong to lineage 7 and were collected in the United States (13, 24). The aurofusarin deficiency may have little effect on mycelial growth of SCKO4, because this strain has already adapted to growth with less pigmentation. To clearly evaluate the effect of aurofusarin accumulation on fungal growth, we generated aurofusarin-overproducing strains through the constitutive expression of heterologous GIP2 sequences in both nonpigmented ΔGIP2 mutants and pigmented wild-type strains. Severe growth reduction resulted in the highly pigmented transformants derived from both types of strains, which is consistent with the hypothesis that aurofusarin biosynthesis should be reduced or inhibited for normal early vegetative growth by G. zeae.
The effect of aurofusarin overproduction on mycelial growth also depended on the wild-type strain from which the GIP2-overexpressing mutants were derived. Mycelial growth of ΔGIP2 mutants from Z03643 was more increased than was that of the ΔGIP2 mutants from SCKO4, suggesting that proper aurofusarin biosynthesis is more important for hyphal growth of Z03643 than it is for SCKO4. The frequent revertants arising in cultures of aurofusarin-overproducing mutants also suggests that early accumulation of aurofusarin can alter the fungus's metabolism and favor genomic rearrangements that reduce the physiological stress. We do not know why the revertants of GIP2-overexpressing transformants derived from wild-type strains were less pigmented than their wild-type progenitors.
Aurofusarin biosynthesis may be associated with a particular developmental stage of G. zeae, as are other developmentally-regulated fungal pigments (10, 17, 22, 30). Neither GIP2 nor PKS12 is transcribed during sexual development, which is consistent with the hypothesis that aurofusarin biosynthesis is coordinated with mycelial growth, especially as the hyphae mature or age. Further analysis is needed of the regulatory mechanism(s), including those mediated by GIP2 that act on the aurofusarin gene cluster in response to environmental changes. Also, the possible utilization of aurofusarin by G. zeae for other purposes, e.g., for survival, for protection against adverse environmental conditions, or as an antimicrobial agent, also should be more carefully evaluated.
In conclusion, through functional studies of the GIP2 gene, which encodes a putative transcription factor, we identified the members of the aurofusarin biosynthetic gene cluster, whose products appear to have a role in the biosynthesis of the compound. We also determined that this pigment may have a significant impact on colony growth rate and morphology, although the mechanism(s) by which these unanticipated morphological changes are mediated remains to be elucidated. Thus, aurofusarin has a previously unanticipated biological role that is worthy of further investigation.
Acknowledgments
This study was supported by a grant (CG 1413) from the Crop Functional Genomics Center of the 21st Century Frontier Research Program funded by the Korean Ministry of Science and Technology and the Rural Development Administration of the Republic of Korea and by a grant (KRF-2004-005-F00013) from the Korea Research Foundation Grant funded by the Korean Government (MOEHRD). J.-E.K. and J.J. were supported by graduate and postdoctoral fellowships, respectively, from the Korean Ministry of Education through the Brain Korea 21 project.
We thank S. Kang, Department of Plant Pathology, Pennsylvania State University, for providing the vector pSK660 carrying the geneticin resistance gene.
REFERENCES
- 1.Ashley, J. N., B. C. Hobbs, and H. Raistrick. 1937. LV. Studies in the biochemistry of micro-organisms. LIII. The crystalline coloring matters of Fusarium culmorum (W. G. Smith) Sacc. and related forms. Biochem. J. 31:385-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chu, Y.-M., J.-J. Jeon, S. J. Yea, Y.-H. Kim, S.-H. Yun, Y.-W. Lee, and K.-H. Kim. 2002. Double-stranded RNA mycovirus in Fusarium graminearum. Appl. Environ. Microbiol. 68:2529-2534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coleman, J. E. 1992. Zinc proteins: enzymes, storage proteins, transcription factors, and replication proteins. Annu. Rev. Biochem. 61:897-946. [DOI] [PubMed] [Google Scholar]
- 4.Correll, J. C., C. J. R. Klittich, and J. F. Leslie. 1987. Nitrate nonutilizing mutants and their use in vegetative compatibility tests. Phytopathology 77:1640-1646. [Google Scholar]
- 5.Dvorska, J. E., and P. F. Surai. 2004. Protective effect of modified glucomannans against changes in antioxidant systems of quail egg and embryo due to aurofusarin consumption. Asian-Australas. J. Anim. 17:434-440. [Google Scholar]
- 6.Dvorska, J. E., P. F. Surai, B. K. Speake, and N. H. C. Sparks. 2001. Effect of the mycotoxin aurofusarin on the antioxidant composition and fatty acid profile of quail eggs. Br. Poultry Sci. 42:643-649. [DOI] [PubMed] [Google Scholar]
- 7.Graham, J. G., H. J. Zhang, S. L. Pendland, B. D. Santarsiero, A. D. Mesecar, F. Cabieses, and N. R. Farnsworth. 2004. Antimycobacterial naphthopyrones from Senna obliqua. J. Nat. Prod. 67:225-227. [DOI] [PubMed] [Google Scholar]
- 8.Gray, J. S., G. C. J. Martin, and W. Rigby. 1967. Aurofusarin. J. Chem. Soc. C 1967:2580-2587. [Google Scholar]
- 9.Hohn, T. M., R. Krishna, and R. H. Proctor. 1995. Characterization of a transcription activator controlling trichothecene toxin biosynthesis. Fungal Genet. Biol. 26:224-235. [DOI] [PubMed] [Google Scholar]
- 10.Kawamura, C., T. Tsujimoto, and T. Tsuge. 1999. Targeted disruption of a melanin biosynthesis gene affects conidial development and UV tolerance in the Japanese pear pathotype of Alternaria alternata. Mol. Plant-Microbe Interact. 12:59-63. [DOI] [PubMed] [Google Scholar]
- 11.Keller, N. P., and T. M. Hohn. 1997. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet. Biol. 21:17-29. [PubMed] [Google Scholar]
- 12.Kereyi, Z., K. Zeller, L. Hornok, and J. F. Leslie. 1999. Molecular standardization of mating type terminology in the Gibberella fujikuroi species complex. Appl. Environ. Microbiol. 65:4071-4076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, J.-E., K.-H. Han, J. Jin, H. Kim, J.-C. Kim, S.-H. Yun, and Y.-W. Lee. 2005. Putative polyketide synthase and laccase genes for biosynthesis of aurofusarin in Gibberella zeae. Appl. Environ. Microbiol. 71:1701-1708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kimura, Y., A. Shimada, H. Nakajima, and T. Hamasaki. 1988. Structures of naphthoquinones produced by the fungus, Fusarium sp., and their biological activity toward pollen germination. Agric. Biol. Chem. 52:1253-1259. [Google Scholar]
- 15.Kitanaka, S., T. Nakayama, T. Shibano, E. Ohkoshi, and M. Takido. 1998. Antiallergic agent from natural sources. Structures and inhibitory effect of histamine release of naphthoquinone glycosides from seeds of Cassia obtusifola L. Chem. Pharm. Bull. 46:1650-1652. [DOI] [PubMed] [Google Scholar]
- 16.Klittich, C. J. R., and J. F. Leslie. 1988. Nitrate reduction mutants of Fusarium moniliforme (Gibberella fujikuroi). Genetics 118:417-423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Langfelder, K., M. Streibel, B. Jahn, G. Hasse, and A. A. Brakhage. 2003. Biosynthesis of fungal melanins and their importance for human pathogenic fungi. Fungal Genet. Biol. 38:143-158. [DOI] [PubMed] [Google Scholar]
- 18.Lee, T., Y.-K. Han, K.-H. Kim, S.-H. Yun, and Y.-W. Lee. 2002. Tri13 and Tri7 determine deoxynivalenol- and nivalenol-producing chemotypes of Gibberella zeae. Appl. Environ. Microbiol. 68:2148-2154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu, Z., and N. C. Mishra. 1995. A single-tube method for plasmid mini-prep from large numbers of clones for direct screening by size or restriction digestion. BioTechniques 18:214-217. [PubMed] [Google Scholar]
- 20.Macias, M., M. Ulloa, A. Gamboa, and R. Mata. 2000. Phytotoxic compounds from the new coprophilous fungus from Guanomyces polydrix. J. Nat. Prod. 63:757-761. [DOI] [PubMed] [Google Scholar]
- 21.Malz, S., M. N. Grell, C. Thrane, F. J. Maier, P. Rosager, A. Felk, K. S. Albertsen, S. Salomon, L. Bohn, W. Schafer, and H. Giese. 2005. Identification of a gene cluster responsible for the biosynthesis of aurofusarin in the Fusarium graminearum species complex. Fungal Genet. Biol. 42:420-433. [DOI] [PubMed] [Google Scholar]
- 22.Mayorga, M. E., and W. E. Timberlake. 1992. The developmentally regulated Aspergillus nidulans wA gene encodes a polyketide homologous to polyketide and fatty acid synthases. Mol. Gen. Genet. 235:205-212. [DOI] [PubMed] [Google Scholar]
- 23.Nelson, P. E., T. A. Toussoun, and W. F. O. Marasas. 1983. Fusarium species—an illustrated manual for identification. The Pennsylvania State University Press, University Park, Penn.
- 24.O'Donnell, K., H. C. Kistler, B. K. Tacke, and H. H. Casper. 2000. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl. Acad. Sci. USA 97:7905-7910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peplow, A. W., A. G. Tag, G. F. Garifullina, and M. N. Beremand. 2003. Identification of new genes positively regulated by TRI101 and a regulatory network for trichothecene mycotoxin production. Appl. Environ. Microbiol. 69:2731-2736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 27.Seo, J.-A., R. H. Proctor, and R. D. Plattner. 2001. Characterization of four clustered and coregulated genes associated with fumonisin biosynthesis in Fusarium verticillioides. Fungal Genet. Biol. 34:155-165. [DOI] [PubMed] [Google Scholar]
- 28.Tanaka, H., and T. Tamura. 1962. The chemical constitution of rubrofusarin, a pigment from Fusarium graminearum. Part I. The zinc dust distillation of rubrofusarin and methylxanthones. Agric. Biol. Chem. 26:767-770. [Google Scholar]
- 29.Todd, R. B., and A. Andrianopoulos. 1997. Evolution of a fungal regulatory gene family: the Zn(II)2Cys6 binuclear cluster DNA binding motif. Fungal Genet. Biol. 21:388-405. [DOI] [PubMed] [Google Scholar]
- 30.Tsai, H.-F., M. H. Wheeler, Y. C. Chang, and K. J. Kwon-Chung. 1999. A developmentally regulated gene cluster involved in conidial pigment biosynthesis in Aspergillus fumigatus. J. Bacteriol. 181:6469-6477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tudzynski, B., and K. Holter. 1998. Gibberellin biosynthetic pathway in Gibberella fujikuroi: evidence for a gene cluster. Fungal Genet. Biol. 25:157-170. [DOI] [PubMed] [Google Scholar]
- 32.Woloshuk, C. P., K. R. Foutz, J. F. Brewer, D. Bhatnagar, T. E. Cleveland, and G. A. Payne. 1994. Molecular characterization of aflR, a regulatory locus for aflatoxin biosynthesis. Appl. Environ. Microbiol. 60:2408-2414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yu, J.-H., Z. Hamari, K.-H. Han, J.-A. Seo, Y. Reyes-Dominguez, and C. Scazzocchio. 2004. Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41:973-981. [DOI] [PubMed] [Google Scholar]
- 34.Yun, S.-H. 1998. Molecular genetics and manipulation of pathogenicity and mating determinants in Mycosphaerella zeae-maydis and Cochliobolus heterostrophus. Ph.D. thesis. Cornell University, Ithaca, N.Y.