Abstract
Two structurally different appendages, thin and thick pili, are found in members of the genus Acinetobacter. The presence of pilus structures correlates with different phenotypes, such as adherence to surfaces, a trait not only observed in pathogenic Acinetobacter species, as well as motility. However, their distinct individual roles were unknown. To characterize the role of different pili in the physiology of Acinetobacter, we isolated the thin pili from the cell surface of Acinetobacter sp. strain BD413 (recently recognized as representative of Acinetobacter baylyi), a soil bacterium that rapidly takes up naked DNA from its environment. Electron microcopy revealed that the pilus has an external diameter of 2 to 3 nm for single filaments. The filaments are packed into right-handed bundles. The major protein constituting the pilus was purified, and the encoding gene, acuA, was cloned. AcuA was found to be weakly related to the structural subunit of F17 pili of Escherichia coli. Analyses of the acuA flanking DNA region led to the identification of three closely associated genes, acuD, acuC, and acuG, whose deduced proteins are similar to chaperone, usher, and adhesin of F17-related pili, respectively. Transcriptional analyses revealed that acuA expression is maximal in the late-stationary-growth phase. Mutation of acuA led to a loss of thin pili and concomitantly loss of adhesion to polystyrene and erythrocytes but not loss of competence. Therefore, thin pili of Acinetobacter sp. strain BD413 are suggested to be assembled by the chaperone/usher pathway and are involved in adherence to biotic and abiotic surfaces.
Pili are 1- to 3-μm-long hair-like bacterial appendages with diameters ranging from 2 to 8 nm and are built by protein subunits called fimbrins or pilins. Pili can be classified on the basis of physical properties, antigenic determinants, adhesion characteristics, characteristics of the major protein subunits, or assembly pathways. To date, at least four distinct assembly pathways for pili are known, and the prototypes for each assembly classification are: P and type I pili (chaperone-usher pathway), curli pili (extracellular nucleation-precipitation pathway), CS1 pili (alternate chaperone pathway), and type IV pili (general secretion pathway) (38, 41).
Pili play an important rule in twitching motility, in adhesion to an inert or living surface, and thus in biofilm formation (26, 27). Biofilms represent a structured community of bacterial cells enveloped in a self-produced matrix. It is estimated that 99.9% of bacteria in nature are attached to a surface in the form of a biofilm (4). Moreover, bacterial biofilm formation has been recognized as important cause of a variety of human infections, including infections of prosthetic devices, endocarditis, dental caries, pneumonia in cystic fibrosis, prostatitis, and others (4, 5, 7, 19, 27, 30, 39).
Members of the genus Acinetobacter are gram-negative, ubiquitously distributed bacteria that are known for their broad metabolic diversity. Other members of the genus Acinetobacter exhibiting a narrow nutritional range have been identified as agents of nosocomial infections often exhibiting multidrug resistance (2, 25, 40). Furthermore, some representatives of the genus Acinetobacter are known for their ability to move on solid surfaces, a trait termed twitching motility, which was first discovered in Acinetobacter calcoaceticus and found to correlate with the presence of polar fimbriae (12). Other representatives, such as Acinetobacter venetianus (formerly A. calcoaceticus RAG-1), are known for their ability to adhere to abiotic and biotic surfaces, which correlates with the presence of thin pili (36).
The encapsulated Acinetobacter strain BD4 and its microencapsulated mutant strain BD413 exhibit growth phase-dependent high competence for natural transformation, a rare trait among the members of the heterogeneous genus Acinetobacter (16, 17). Recently, strain BD413 (also referred to strain ADP1) was recognized as representative of Acinetobacter baylyi, a species including members with unusually high competence for natural transformation (44). Furthermore, strain BD4 and its derivative are also known for the production of two types of pili, bundle-forming thin pili (2 to 3 nm in diameter), and individual thick pili (6 nm in diameter) (12). The thick pili are polar and correlate with twitching motility, whereas the thin pili are peritrichous and do not correlate with observable twitching (12).
Our studies of the natural transformation system of Acinetobacter sp. strain BD413 led to the identification of competence proteins, many of which are related to structural subunits of type IV pili (pilins) or other proteins of type IV pili biogenesis machineries (3, 13, 23, 33, 34). These findings raised the question of whether the BD413 pili are implicated in natural transformation. In order to analyze the structure and function of the BD413 thin pili and to clarify their role in natural transformation, we purified the thin pili, identified the genes, performed genetic and molecular analyses of the thin pilus biogenesis system, monitored their transcriptional regulation, and analyzed the thin-pilus functions. This is the first report on the molecular basis, structure, and function of the Acinetobacter sp. strain BD413 thin pili, which are predicted to be assembled by a chaperone/usher pilus assembly pathway.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The microencapsulated Acinetobacter sp. strain BD413 and a p-hydroxybenzoate hydroxylase (pobA) mutant strain of BD413, designated Acinetobacter sp. strain ADP239 (11), was used in the studies. Escherichia coli strain DH5α (Gibco-BRL) was used as the host for maintenance of plasmids and was grown at 37°C in Luria-Bertani (LB) medium. Acinetobacter wild-type and mutant strains were cultured routinely at 30°C in LB medium or mineral medium (32). Growth conditions were used as described previously (33). The following antibiotics were used: ampicillin (Ap; 100 μg/ml), kanamycin (Km; 20 μg/ml), and tetracycline (Tc; 15 μg/ml).
Molecular techniques.
The molecular and genetic procedures used were standard techniques (37). Transformation studies, conjugation, and complementation studies of Acinetobacter mutants and Southern hybridization experiments were performed as described previously (33). For triparental mating, helper plasmid pRK2013 (8) was used. Recombinant plasmids that were generated and sequenced by using standard primers or primers generated from the genomic sequence information. DNA sequences were analyzed with the BlastN/P programs (National Center for Biotechnology Information database).
Pilus purification.
Pili were purified according to a modified protocol of Kohonen et al. (18). A 3-liter culture of Acinetobacter sp. strain BD413 was grown overnight in mineral medium and harvested by centrifugation at 5,000 × g. The cells were resuspended in 20 ml of TBS (30 mM Tris-HCl, 0.9% NaCl [pH 7.5]). To shear off the pili, the cell suspension was pressed twice through a needle (26 G×1/"Luer-Lock [0.45 by 12 mm]; Braun-Melsungen). The cells were separated from the shear fraction by centrifugation twice for 5 min each at 5,000 × g. The supernatant was dialyzed against high-salt buffer (30 mM Tris-HCl, 150 mM NaCl, 100 mM MgCl2 [pH 7.5]). The pili were sedimented by ultracentifugation (180,000 × g, 1 h, 4°C), and the pellet was resuspended in 0.5 ml of high-salt buffer and loaded on top of a sucrose density gradient (20 to 70%) in high-salt buffer. After centrifugation for 16 h at 96,000 × g at 4°C, the sediment of the gradient was diluted in 10 ml of TBS and then ultracentrifuged again (180,000 × g, 1 h, 4°C). The resulting pellet was resuspended in 0.1 ml of TBS.
SDS-PAGE.
Samples were boiled for 5 to 10 min in a water bath prior to analysis by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) on a 15% polyacrylamide gel as described previously (22).
Cloning of acuA.
Since the N terminus of the 16-kDa subunit was not accessible to sequencing the direct amplification of this gene by PCR was not possible. In the meantime we got access to the recently sequenced genome of Acinetobacter sp. strain BD413 (http://www.genoscope.cns.fr/ [1]). Using the internal sequence of 16 amino acids GITLGGPTNSAQYVAG, we performed homology searches in the genome of Acinetobacter sp. strain BD413 and identified one corresponding open reading frame (ORF). The information of the corresponding ORF was used to generate a pair of primers: FimA_PstI (5′-ATTAAATTGATCTGCAGCTAGTGATG-3′) and FimA_XbaI (5′-ATGCGAGTATCTAGAATGCTG-3′). Using these primers, a 690-bp fragment was amplified, which was digested with PstI and XbaI and ligated to pBluescript II KS, resulting in plasmid pBN01.
Mutant generation and complementation studies.
In order to disrupt acuA by inserting the nptII gene into the EcoRI restriction site present in the insert in pBN01, an additional EcoRI restriction site within the multiple cloning site of vector pBluescript II KS had to be deleted. Therefore, pBN01 was digested with PstI and EcoRV, incubated in the presence of deoxynucleotide triphosphates and Klenow enzyme, and religated, resulting in pBN02. nptII from pUC4K was inserted into the EcoRI restriction site within the insert of pBN02, resulting in pBN03. The plasmid pBN03 was digested to separate the insert from the vector DNA, and the resulting insert carrying the disrupted ORF plus flanking wild-type DNA was introduced into Acinetobacter sp. strain ADP239 by natural transformation. Transformants were selected on LB medium containing kanamycin.
Construction of acuA::lacZ transcriptional fusion and assay for β-galactosidase activities.
To generate the acuA::lacZ transcriptional fusion, a 290-bp BamHI/XbaI fragment carrying a conserved σ70 promoter region (100 bp upstream of the acuA gene) and 190 bp of the 5′-end of acuA gene, was amplified by using purified BD413 DNA and primer Dü_lacZ_BamHI_N1.2 (5′-TTTGAGATCGGATCCATTTTTTTTAAAAAG-3′) and Dü_lacZ_XbaI_C1 (5′-CAAGTACTTGTAATCTCTAGACCATTAATCG-3′). The PCR-product was digested with BamHI and XbaI and then cloned into pUC18 (BamHI/XbaI), resulting in pUN09. Correct amplification was assured by sequencing before pUN09 was digested with EcoRI and XbaI to separate the insert from the vector. The EcoRI/XbaI fragment was subsequently cloned into the broad-host-range plasmid pBK (21), which carries a promoterless β-galactosidase gene resulting in plasmid pBN10. pBN10 was transferred into Acinetobacter sp. strain ADP239 via triparental mating. As a control, Acinetobacter sp. strain ADP239 carrying pBK was used. This control plasmid carrying a promoterless β-galactosidase gene was transferred into Acinetobacter sp. strain ADP239, and the β-galactosidase activities obtained with this strain were subtracted from the experimental data with Acinetobacter sp. strain ADP239 carrying pBN10. The β-galactosidase assays were performed as described by Miller (28), and the activities were expressed as Miller units, which were proportional to the increase in absorbancy of free o-nitrophenol per minute and cell density.
Movement on solid surfaces.
The motility of Acinetobacter strains on solid surfaces was analyzed by streaking an overnight culture across a freshly prepared plate of agar containing 5 g of tryptone, 2.5 g of yeast extract, 5 g of NaCl, and 20 g of agar per liter of distilled water. Since a humid atmosphere facilitates motility on solid surfaces, the plates were incubated for 12 h at 30°C in a gas-tight jar which in addition to the culture plates contained a petri dish filled with water. The cells were analyzed for motility by the appearance of spreading zones along the central streak of growth.
Electron microscopy.
Negative staining with a 3% (wt/vol) aqueous solution of phosphotungstic acid (pH 7.0) or a 4% (wt/vol) aqueous solution of uranyl acetate (pH 4.5) was performed according to the method of Valentine et al. (45). Electron micrographs were taken, at calibrated magnifications, with a Philips EM 301 transmission electron microscope (Philips, Eindhoven, The Netherlands) operated in the conventional bright-field mode. Digital images were acquired by scanning photographs or petri dishes (UMAX Power Look II). Image processing, which was limited to cropping, conversion to grayscale, and adjustment of brightness and/or contrast, was done with Photoshop (Adobe, Inc.) software. Image enhancement of pili was performed by translational analysis (14).
Adhesion to polystyrene.
Overnight cultures were washed twice with mineral medium and suspended in mineral medium to an optical density at 600 nm (OD600) of 1.5. Then, 1 ml of cell suspension was dropped onto a petri dish and incubated at 30°C for 2 h. The petri dish was washed three times with 15 ml of phosphate-buffered saline (PBS; pH 7.5) by shaking it gently for 1 min. Cells attached to the bottom of the petri dish were visualized with phase-contrast microscopy.
Agglutination of erythrocytes.
Separation of erythrocytes of 20 ml of human blood was performed with a LeucoSep kit (Greiner Bio-One, Inc.). Cells of an overnight culture of Acinetobacter were washed and suspended in PBS (10 mM phosphate, 0.9% NaCl [pH 7.5]) to an OD600 of 1.5. Then, 50 μl of purified erythrocytes and 200 μl of bacterial suspension were gently mixed in a 1.5-ml reaction tube, followed by incubation at 25°C for 2 h. In 20-min intervals the settled blood cells were mixed again with the bacterial cells by inverting the tube. After 2 h, the supernatant was aspirated, and the settled blood cells were washed twice with 1.5 ml of PBS (4°C) and suspended in 1.5 ml of PBS at 4°C. The blood cells with attached bacterial cells were visualized with phase-contrast microscopy.
Nucleotide sequence accession number.
The nucleotide sequence of the acu gene cluster has been deposited in the GenBank database under accession no. AY566571.
RESULTS
Purification of thin pilus structures.
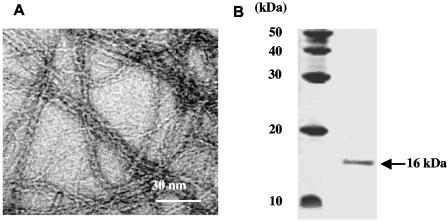
Acinetobacter sp. strain BD413 pili were sheared of the cell body and subjected to sucrose density centrifugation. After centrifugation, a pellet which was visible at the bottom of the gradient was resuspended and inspected by electron microscopy. The pellet fraction contained exclusively bundles of thin filaments with a diameter of ∼3 nm (Fig. 1A). In contrast, the thick pili, which do not form bundles, were not sedimented. SDS-PAGE of the thin pili preparation revealed one major protein of ∼16 kDa (Fig. 1B), demonstrating that the 16-kDa protein represents the major subunit of the thin pilus structures.
FIG. 1.
Electron micrograph of bundles of purified Acinetobacter sp. strain ADP239 thin pili (A) and SDS-PAGE of the structural subunit of purified thin pili (B).
Supramolecular structure of thin pili.
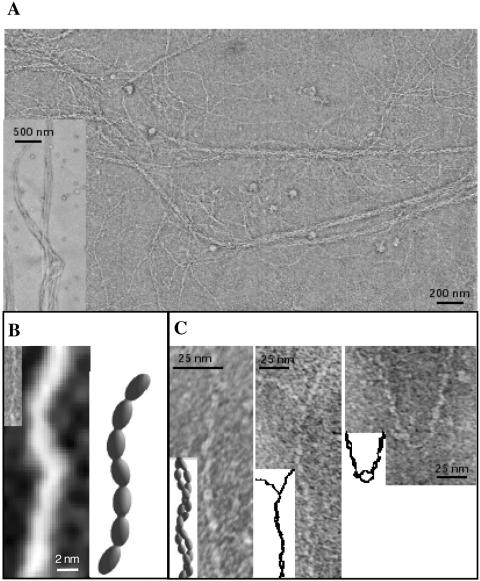
High-resolution electron micrographs of purified thin pili reveal the uniform structure of these filamentous organelles (Fig. 2A). After overall inspection of a specimen grid, five randomly selected areas of 12 μm2 were inspected in detail with respect to the presence of impurities (i.e., other structures than pili) and the homogeneity of pilus preparations. These analyses led to the detection of only one pilus type, other filamentous structures were absent (Fig. 1A and 2A). Occasionally, small lipid vesicles could be observed. Among the representative areas, five 0.8-by-0.6 μm areas were closely inspected to analyze pilus size, structure, and bundle formation. The major portion of pili forms bundles comprising of at least two single pili twisted to right-handed bundles (Fig. 2C). The subunits of a single pilus are arranged in 3-nm-wide, semiflexible, and wire-like filaments. A short portion of a single pilus filament is shown in three-dimensional reconstruction in Fig. 2B. The twisting of thin filaments to right-handed bundles could be a consequence of the hydrophobicity of the pilus itself, enabling multiple hydrophobic interactions between individual filaments.
FIG. 2.
Electron micrographs and three dimensional reconstructions of Acinetobacter sp. strain BD413 thin (Acu) pili. (A) The overview depicts bundles of various thickness (see also the inset). (B) Single filament (inset: original preparation; large image after image enhancement) and respective model of the arrangement of subunits. (C) Pairs of filaments twisted around each other. In the two right images, stretches of unpaired filaments are visible.
Identification and characterization of acuA, encoding the 16-kDa structural subunit of the thin pili.
In order to identify the gene encoding the structural subunit of the thin pili, N-terminal sequencing of the 16-kDa protein was performed. This strategy was not successful, indicating that the N terminus is blocked. Therefore, we generated internal tryptic fragments and the amino acid sequence of one fragment (GITLGGPTNSAQYVAG) was determined. This sequence was sufficient to identify the gene in the genome sequence (http://www.genoscope.cns.fr/).
The ORF encoding the 16-kDa structural subunit of the thin pili consists of 579 bp, is preceded by a well-conserved and well-placed Shine-Dalgarno sequence (AGGT), and encodes a protein of 193 amino acids (19.5 kDa). Further analysis of the deduced N-terminal sequence of the pilus subunit suggests the presence of a sec-dependent signal peptide composed of 19 amino acids (Fig. 3): (i) a positively charged N terminus (Lys 2 and Lys 3); (ii) a hydrophobic core (Ala 7 to Leu 13); and (iii) small, uncharged residues at positions −1 and −3 (Met −1, Gly −3). The mature protein results from cleavage of the signal peptide between the last amino acid of the signal peptide (Met −1) and the first amino acid of the mature protein (Thr +1). The apparent molecular mass of 16 kDa corresponds to the calculated mass for the mature protein. The database searches revealed weak similarities of the 16-kDa protein to structural subunits of pili that are assembled by chaperone/usher pathways (Fig. 3). Highest similarities of 21% were found to the class III structural subunits of F17 pili of pathogenic Escherichia coli strains, which are thin, flexible structures assembled by a chaperone/usher pathway (24, 38). Therefore, the 16-kDa structural subunit of the thin pili was designated AcuA (for Acinetobacter chaperone/usher).
FIG. 3.
Alignment of the Acinetobacter sp. strain BD413 major thin-pilus subunit AcuA with pilins of chaperone/usher pilus biogenesis systems of the F17 pili in E. coli CK210, Proteus mirabilis uroepithelial cell adherence protein (Uca), Xylella fastidiosa fimbrial subunit (AE003862), and a putative fimbrial protein of Yersinia pestis (AE013790). Identical residues are indicated by black boxes. Pilins contain conserved cysteines (arrowheads), a conserved C-terminal motif (underlined), which includes a glycine and a conserved aromatic amino acid, tyrosine. The highly conserved region located near the N terminus is indicated (underlined).
The major structural subunits of pili assembled by chaperone/usher pathways share conserved residues required for proper folding and assembly (38, 41). The alignment of AcuA with the major pilus subunit of the F17 pilus of E. coli, the uroepithelial cell adherence protein (Uca) in Proteus mirabilis, and the fimbrial subunits of Xylella fastidiosa and Yersinia pestis point out conserved residues and domains (Fig. 3). The conserved residues include (i) cysteines that are oxidized by a periplasmic oxidoreductase to form a disulfide bridge, which is necessary for their proper folding and incorporation into the growing pilus; (ii) a C-terminal motif characterized by a common pattern of alternating hydrophobic amino acids flanked by a glycine and the conserved aromatic amino acid tyrosine essential for transient insertion of pilins into the cytoplasmic membrane, the binding to the periplasmic chaperone, and for pilus polymerization by intersubunit quaternary interactions; and (iii) a highly conserved domain located near the N terminus, which is postulated to be a “second site” for chaperone binding. Except for the conserved glycine residue at the C terminus, these motives for the proper folding and assembly are present in AcuA of Acinetobacter sp. strain BD413.
Identification and genetic organization of an acu gene cluster.
Analyses of acuA flanking DNA regions led to the identification of three tightly clustered, tandemly arranged genes immediately downstream of acuA. The deduced proteins of these genes are similar to proteins implicated in the membrane targeting of pili subunits, biogenesis, and adhesion functions of F17-related enteropathogenic pili. Therefore, these genes were designated acuD, acuC, and acuG. The tight clustering and the head-to-tail organization suggests that the acu genes are cotranscribed. Furthermore, the presence of a putative promoter sequence upstream of acuA with −10 signature (TTAAAT) and −35 signature (ATCACA) upstream of acuA, suggest that the cotranscription of the acu genes is mediated by a σ70 promoter. AcuD (26 kDa) is similar to chaperones that form periplasmic chaperone-pilus subunit complexes targeting outer membrane ushers (44% similarity to the chaperone F17a-D of E. coli F17 pili). The third gene of the acu gene cluster encodes AcuC, a 94-kDa protein related to the usher protein F17a-C of E. coli F17 pili (32% similarity). The C terminus (residues 160 to 349) of the deduced protein of the last gene of the acu gene cluster, AcuG (36 kDa), is similar to the C terminus of the adhesive protein F17a-G of E. coli F17 pili. Taken together, these data suggest that AcuA, the structural subunit of the thin pilus from Acinetobacter sp. strain BD413, forms a complex with the periplasmic chaperone AcuD. AcuD then targets AcuA to the outer membrane usher AcuC, which mediates subunit assembly into pili and secretion to the cell surface.
The deduced protein of the gene preceding the acuA gene is similar (70% identity) to the polyphosphate-AMP phosphotransferase from Acinetobacter johnsonii and, therefore, we conclude that acuA represents the first gene of the thin pilus biogenesis gene cluster in strain BD413. The deduced protein of an ORF immediately downstream of acuG, the fourth gene of the acu gene cluster, shows significant similarities to bacterial hydrolases of the isochorismatase family. These similarities, together with the finding that the ORF located immediately downstream of acuG is transcribed in the opposite direction, led us to conclude that acuG is the last gene of the thin pilus biogenesis gene cluster.
Function of the thin pili.
To address the function of the thin pili, we generated a thin pilus mutant by using the pobA mutant Acinetobacter sp. strain ADP239. Therefore, the acuA gene was cloned and an acuA mutant was generated by inserting the nptII marker into the acuA gene. Correct allelic replacement of chromosomal wild-type acuA gene by acuA::nptII was verified by Southern hybridization (data not shown).
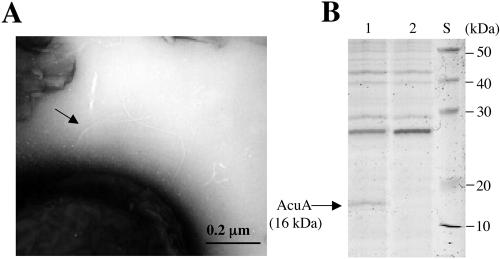
Examination of the acuA mutant strain, N100, revealed that the mutant cells still exhibit single thick pilus structures but are devoid of thin pilus structures (Fig. 4A). Analysis of the shear fraction of the acuA mutant strain N100 by SDS-PAGE revealed that the shear fraction is devoid of AcuA (Fig. 4B). Taken together, these results provide clear evidence that mutant strain N100 carrying a mutation in acuA is devoid of thin pilus structures.
FIG. 4.
Piliation analyses of BD413 wild-type cells and acuA mutant strain N100. (A) Representative electron micrograph of the thin-pilus mutant N100. Arrow, thick pilus. (B) SDS-PAGE of shear fractions of wild-type cells (lane 1) and thin-pilus mutant N100 (lane 2). S, molecular mass standard.
Analyses of the transformation phenotype by complementation of the pobA mutation with wild-type DNA revealed that mutant N100 displays wild-type transformation frequencies. This finding provides clear evidence that the thin pili are not implicated in natural transformation. Furthermore, no difference in movement on agar solid surfaces between wild type and the acuA mutant strain could be detected (data not shown), suggesting that AcuA is not directly involved in twitching motility. However, it cannot be ruled out that the thin pilus is indirectly involved in twitching motility, such as by coordinating cell-to-cell signaling within twitching rafts of cells.
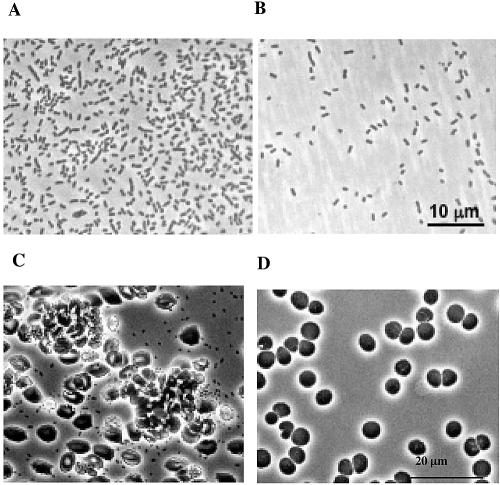
Next, we compared adhesion phenotypes of the wild-type and the acuA mutant strain N100. Phase-contrast microscopy of cells and counting the cells attached to the bottom of a polystyrene petri dish revealed that the wild-type cells formed a dense monolayer covering 35 to 40% of the petri dish surface (Fig. 5A). In contrast, attachment of strain N100 to polystyrene was reduced by 95% (Fig. 5B). Since several Acinetobacter strains are opportunistic pathogens, we sought to determine whether Acinetobacter sp. strain BD413 would adhere to erythrocytes. Indeed, Acinetobacter wild-type cells agglutinated erythrocytes (Fig. 5C), whereas the acuA mutant cells no longer adhered to erythrocytes (Fig. 5D). Taken together, these experiments provide clear evidence that the thin pili of Acinetobacter sp. strain BD413 promote binding to hydrophobic surfaces and to erythrocytes.
FIG. 5.
Phase-contrast microscopic analyses of Acinetobacter sp. strain BD413 cells (A) and acuA mutant cells (B) adhering to polystyrene and BD413 cells (C) and acuA mutant cells (D) adhering to erythrocytes.
Regulation of pilus gene expression.
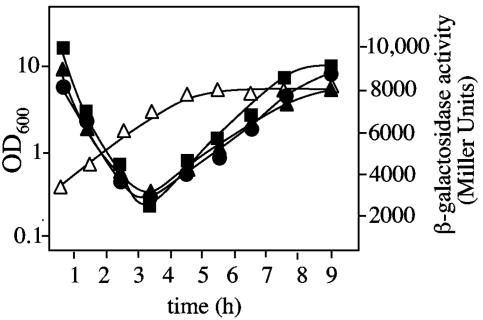
Pilus-mediated biofilm formation is often regulated by environmental stimuli on the transcriptional level by altering expression of pilus subunits. To study the regulation of thin pilus formation in Acinetobacter sp. strain BD413, we fused the potential σ70 promoter region upstream of acuA to a promoter-free lacZ gene, and this fusion was transferred into Acinetobacter sp. strain ADP239. β-Galactosidase activities were monitored during growth at 30°C in mineral medium with succinate as carbon source and parallel during growth in complex (LB) medium at 30 and 37°C, respectively. High transcription of acuA was detected in a freshly inoculated culture (Fig. 6). During prolonged exponential growth, transcription of acuA decreased gradually and minimal acuA transcription levels were detected at the end of exponential growth phase, but levels increased thereafter. Maximal acuA transcription was found in the late stationary growth phase. An analogous growth phase-dependent regulation of acuA expression was found independent of growth temperature (30 or 37°C) and growth medium (Fig. 6). Taken together, these results clearly show that the expression of the major thin pilus subunit is upregulated in the stationary phase and therefore under nutrient limitation independent of carbon source and growth temperature. The upregulation of the major thin pilus subunit under nutrient limitations correlates with the presence of thin pili in later growth stages (12).
FIG. 6.
Growth-phase-dependent expression of acuA in Acinetobacter sp. strain ADP239. acuA induction was monitored by using a acuA::lacZ reporter fusion located on the low-copy plasmid pBK. acuA transcription was parallel monitored during growth in mineral medium with succinate as a carbon source at 30°C (▪) and 37°C (▴), respectively, and during growth in complex (LB) medium at 30°C (•). ▵, OD600. The values are averages of two to three experiments with standard deviations of maximal ± 800 Miller units. The growth curve comprises of averages of the nearly identical optical densities and is applicable to all growth conditions.
DISCUSSION
The presence of thin pili on the surface of the encapsulated Acinetobacter sp. strain BD4, a strain giving rise to the formation of quite broad spreading zones due to sliding on solid medium, has already been discovered by Henrichsen and Blom in 1975 (12). However, the biogenesis machinery and the function of these pili on the surface of strain BD4 has thus far been uncharacterized. We present here for the first time a comprehensive investigation of the supramolecular structure, molecular basis, and transcriptional regulation of the thin pilus of the microencapsulated Acinetobacter sp. strain BD413. The AcuA subunit of Acinetobacter sp. strain BD413 thin pili was found to carry conserved N-terminal and C-terminal motives similar to the C- and N-terminal motives found in pili assembled by chaperone/usher pathways in enteropathogenic E. coli strains (15, 42). Highest similarities were found to the motives of the class III structural subunits of E. coli F17 pili (24). These motives are required for a proper folding and assembly of pilus structural subunits into pili which is mediated by chaperone and usher proteins (38, 41). The presence of these conserved N-terminal and C-terminal residues in AcuA indicates an F17 pilus-subunit analogous folding and assembly of AcuA. Beyond these conserved folding and assembly motives, AcuA shares only little similarities with the structural subunits of F17-related pili, which distinguishes the thin pilus subunits of Acinetobacter sp. strain BD413 from the subunits of F17 pili.
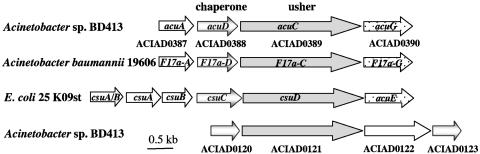
Analyses of the acuA locus led to the identification of three additional tightly clustered genes encoding proteins, designated AcuD, AcuC, and AcuG, which were found to be similar to chaperones, outer membrane ushers, and adhesins implicated in pili biogenesis and pili adhesion. Taken together, the organization of the acu gene cluster in Acinetobacter sp. strain BD413 corresponds to the organization of F17-related pili, which are assembled by a chaperone/usher pathway (Fig. 7). Pili assembled by chaperone/usher pathways exhibit different structures; thick and rigid or thin and flexible or atypical structures are known (15, 20, 31, 38, 41, 42). Our electron microscopic studies revealed that the thin pili of BD413 are flexible, wire-like structures that are 2 to 3 nm wide and twisted into right-handed bundles. This structure is comparable to F17 pili, which are ∼3-nm-wide, flexible, wire-like filaments exposing an adhesin at their tips. Genome sequence analyses of the BD413 genome with the conserved AcuD chaperone and AcuC usher proteins led to the identification of a second potential pilus gene cluster (1). The deduced proteins of this predicted pilus assembly cluster are similar to pilus subunits (ACIAD0122), chaperones (ACIAD0120 and ACIAD0123), and usher proteins (ACIAD0121) of pili assembled by chaperone/usher systems. Although the chaperone and usher genes of this predicted second pilus assembly cluster in Acinetobacter are analogously organized (Fig. 7), the similarities to the Acu proteins are very low. Since an acuA mutant strain was completely devoid of thin pili, the second pilus cluster might be a nonfunctional pilus cluster, or be expressed under different conditions.
FIG. 7.
Alignment of the BD413 acu gene locus and acu-like ORFs present in the genomes of Acinetobacter baumannii and Escherichia coli, and the predicted second pilus assembly system in the genome of Acinetobacter sp. strain BD413. The arrows indicate direction of transcription. Numbers indicate gene labels in the Acinetobacter sp. strain ADP1 (i.e., BD413) chromosome database.
Our studies provide clear evidence that the thin pili are not essential for motility on solid surfaces, but for adhesion to hydrophobic surfaces and to erythrocytes. Pilus-mediated adherence and biofilm formation is thought to begin when bacteria sense environmental conditions and when bacterial cells communicate with each other to coordinate group activities (9, 29). Environmental signals triggering this transition vary among microorganisms (6, 35, 47). One signal having a pronounced effect on the expression of different pilus systems is growth temperature. Growth temperature significantly effects expression of the pap operon, the prototype of chaperone/usher assembly system for thin and rigid pili, and therefore affects bacterial adhesion to human cells (46). Furthermore, studies of the transcriptional regulation of the E. coli P pilus major subunit gene, papA, revealed that the temperature has a significant effect on the transcriptional regulation of the pilus genes (10). These results clearly show the correlation between pilus synthesis of uropathogenic bacteria and growth temperature in the host. In contrast, our results demonstrated that transcription of the thin pilus subunit of Acinetobacter sp. strain BD413 does not vary between 30 and 37°C. This finding together with the fact that Acinetobacter sp. strain BD413 is not a pathogenic strain might reflect that the thin pili of strain BD413 are adhesive organelles not primarily used to encounter host cells but mediating adhesion to abiotic surfaces and carbohydrates. However, it cannot be ruled out that lower temperatures have an effect on pilus expression, which is probably more relevant to Acinetobacter strains in soil.
Thin pilus-mediated biofilm formation of Acinetobacter strain BD413 might support the nutritional versatility of this organism due to the high retention times of bacteria adhering to a substratum. This conclusion corresponds to the finding that Acinetobacter sp. strain BD413 displays a very broad metabolic diversity, such as ca. 20% of the genes of the strain ADP1 are oriented toward the degradation of different organic compounds (1). Extended retention times of Acinetobacter sp. strain BD413 might facilitate degradation of stable compounds such as the many different aromatic substrates serving as a carbon source.
Recently, a novel chaperone/usher pilus system has been identified in the opportunistic pathogen Acinetobacter baumannii (43). The organization of the pilus gene cluster in A. baumannii is analogous to the organization of the acu gene cluster in Acinetobacter sp. strain BD413 except for the presence of three genes encoding proteins with similarities to type I pilus structural subunits (Fig. 7). Comparison of the BD413 chaperone/usher pathway proteins with the A. baumannii pilus system revealed that the major pilus subunit AcuA of BD413 and the A. baumannii pilus subunit do not show any significant similarities, leading to the conclusion that the thin pilus system of Acinetobacter sp. strain BD413 and A. baumannii are significantly different. Analyses of the Acinetobacter sp. strain BD413 genome sequence revealed that strain BD413 lacks genes associated with pathogenesis, such as toxins, invasins, and secretory systems (1, 2). Taken together, the distinctiveness of the Acinetobacter sp. strain BD413 pili could provide opportunities to discriminate between opportunistic pathogens, such as A. baumannii strains and nonpathogenic Acinetobacter strains.
Acknowledgments
This study was supported by the Deutsche Forschungsgemeinschaft (grants Av 9/4-4 and Av 9/4-5). A. Friedrich was supported by the foundation Stipendien Fonds des Verbandes der Chemischen Industrie. We are also grateful to G. Gottschalk, Göttingen, Germany, for generous support.
We thank J. Kellermann (Martinsried, Germany) for N-terminal sequence analyses and K. Fehmeyer (Göttingen, Germany) for preparation of erythrocytes. We acknowledge the Genome Project (Genoscope Cedex, France), which was the source of information on organization of genes in the acuA locus.
REFERENCES
- 1.Barbe, V., D. Vallenet, N. Fonknechten, A. Kreimeyer, S. Oztas, L. Labarre, et al. 2004. Unique features revealed by the genome sequence of Acinetobacter sp. ADP1, a versatile and naturally transformation competent bacterium. Nucleic Acids Res. 32:5766-5779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bergogne-Berenzin, E., and K. J. Towner. 1996. Acinetobacter spp. as nosocomial pathogens: microbiological, clinical, and epidemiological features. Clin. Microbiol. Rev. 9:148-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Busch, S., C. Rosenplänter, and B. Averhoff. 1999. Identification and characterization of ComE and ComF, two novel pilin-like competence factors involved in natural transformation of Acinetobacter sp. strain BD413. Appl. Environ. Microbiol. 65:4568-4574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infection. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 5.Davey, M. E., and G. A. O'Toole. 2000. Microbial biofilms: from ecology to molecular genetics. Microbiol. Mol. Biol. Rev. 64:847-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dewanti, R., and A. C. Wong. 1995. Influence of culture conditions on biofilm formation by Escherichia coli O157:H7. Int. J. Food Microbiol. 26:147-164. [DOI] [PubMed] [Google Scholar]
- 7.Donlan, R. M. 2001. Biofilm formation: a clinically relevant microbiological process. Clin. Infect. Dis. 33:1387-1392. [DOI] [PubMed] [Google Scholar]
- 8.Figurski, D. H., and D. R. Helinski. 1979. Replication of an origin-containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc. Natl. Acad. Sci. USA 76:1648-1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: homoserine lactone signaling. Nat. Rev. Mol. Cell Biol. 3:685-695. [DOI] [PubMed] [Google Scholar]
- 10.Göransson, M., and B.-E. Uhlin. 1984. Environmental temperature regulates transcription of a virulence pili operon in E. coli. EMBO J. 3:2885-2888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hartnett, G. B., B. Averhoff, and L. N. Ornston. 1990. Selection of Acinetobacter calcoaceticus mutants deficient in the p-hydroxybenzoate hydroxylase gene (pobA), a member of a supraoperonic cluster. J. Bacteriol. 172:6160-6161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henrichsen, J., and J. Blom. 1975. Correlation between twitching motility and possession of polar fimbriae in Acinetobacter calcoaceticus. Acta Pathol. Microbiol. Scand. B 83:103-115. [DOI] [PubMed] [Google Scholar]
- 13.Herzberg, C., A. Friedrich, and B. Averhoff. 2000. comB, a novel competence gene required for natural transformation of Acinetobacter sp. BD413: identification, characterization, and analysis of growth-phase-dependent regulation. Arch. Microbiol. 173:220-228. [DOI] [PubMed] [Google Scholar]
- 14.Hoppert, M., and A. Holzenburg. 1999. Electron microscopy in microbiology. Bios. Scientific Publications, Oxford, England.
- 15.Hultgren, S. J., S. Normark, and S. N. Abraham. 1991. Chaperone-assisted assembly and molecular architecture of adhesive pili. Annu. Rev. Microbiol. 45:383-415. [DOI] [PubMed] [Google Scholar]
- 16.Juni, E. 1978. Genetics and physiology of Acinetobacter. Annu. Rev. Microbiol. 32:349-371. [DOI] [PubMed] [Google Scholar]
- 17.Juni, E., and A. Janik. 1969. Transformation of Acinetobacter calcoaceticus (Bacterium anitratum). J. Bacteriol. 98:281-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kohonen, T. K., Nurmiaho, E. L., Ranta, H., and C. S. Eden. 1980. New method for isolation of immunologically pure pili from Escherichia coli. Infect. Immun. 27:569-575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolenbrander, P. E. 2000. Oral microbiological communities: biofilms, interactions, and genetic systems. Annu. Rev. Microbiol. 54:413-437. [DOI] [PubMed] [Google Scholar]
- 20.Kuehn, M. J., J. Heuser, S. Normark, and S. J. Hultgren. 1992. P pili in uropathogenic Escherichia coli are composite fibers with distinct fibrillar adhesive tips. Nature 356:252-255. [DOI] [PubMed] [Google Scholar]
- 21.Kusian, B., R. Bednarski, M. Husemann, and B. Bowien. 1995. Characterization of the duplicate ribulose-1,5-bisphosphate carboxylase genes and cbb promoters of Alcaligenes eutrophus. J. Bacteriol. 177:4442-4450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laemmli, U. 1970. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680-685. [DOI] [PubMed] [Google Scholar]
- 23.Link, C., S. Eickernjäger, D. Porstendörfer, and B. Averhoff. 1998. Identification and characterization of a novel competence gene, comC, required for DNA binding and uptake in Acinetobacter sp. strain BD413. J. Bacteriol. 180:1592-1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lintermans, P., P. Pohl, F. Deboeck, A. Bertels, C. Schlicker, J. Vandekerckhove, J. Van Damme, M. Van Montagu, and H. De Greve. 1988. Isolation and nucleotide sequence of the F17-A gene encoding the structural protein of the F17 fimbriae in bovine enterotoxigenic Escherichia coli. Infect. Immun. 56:1475-1484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Manfredi, R., A. Nanetti, R. Valentini, and F. Chiodo. 2001. Acinetobacter infections in patients with human immunodeficiency virus infection: microbiological and clinical epidemiology. Chemotherapy 47:19-28. [DOI] [PubMed] [Google Scholar]
- 26.Mattick, J. S. 2002. Type IV pili and twitching motility. Annu. Rev. Microbiol. 56:289-314. [DOI] [PubMed] [Google Scholar]
- 27.McBride, J. S. 2001. Bacterial gliding motility: multiple mechanisms for cell movement over surfaces. Annu. Rev. Microbiol. 55:49-75. [DOI] [PubMed] [Google Scholar]
- 28.Miller, J. H. 1972. Assay of β-galactosidase, p. 319-353. In T. Platt, B. Miller-Hill, and J. H. Miller (ed.), Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 29.Miller, M. B., and B. L. Bassler. 2001. Quorum sensing in bacteria. Annu. Rev. Microbiol. 55:165-199. [DOI] [PubMed] [Google Scholar]
- 30.Mulvey, M. A., Y. S. Lopez-Boado, C. L. Wilson, R. Roth, W. C. Parks, J. Heuser, and S. J. Hultgren. 1998. Induction and evasion of host defenses by type 1-piliated uropathogenic Escherichia coli. Science 282:1494-1497. [DOI] [PubMed] [Google Scholar]
- 31.Normark, S., M. Baga, M. Göransson, F. P. Lindberg, B. Lund, M. Norgren, and B.-E. Uhlin. 1986. Genetics and biogenesis of E. coli adhesins, p. 113-143. In D. Mirelman (ed.), Microbial lectins and agglutinins. Wiley Interscience, New York, N.Y.
- 32.Ornston, L. N., and R. Y. Stanier. 1966. The conversion of catechol and protocatechuate to β-ketoadipate by Pseudomonas putida. J. Biol. Chem. 241:3776-3786. [PubMed] [Google Scholar]
- 33.Porstendörfer, D., U. Drotschmann, and B. Averhoff. 1997. A novel competence gene, comP, is essential for natural transformation of Acinetobacter sp. strain BD413. Appl. Environ. Microbiol. 63:4150-4157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Porstendörfer, D., O. Gohl, F. Mayer, and B. Averhoff. 2000. ComP, a pilin-like protein essential for natural competence in Acinetobacter sp. strain BD413: regulation, modification, and cellular localization. J. Bacteriol. 182:3673-3680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pratt, L. A., and R. Kolter. 1998. Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis, and type I pili. Mol. Microbiol. 30:285-293. [DOI] [PubMed] [Google Scholar]
- 36.Rosenberg, M., E. A. Bayer, J. Delarea, and E. Rosenberg. 1982. Role of thin fimbriae in adherence and growth of Acinetobacter calcoaceticus RAG-1. Appl. Environ. Microbiol. 44:929-937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 38.Sauer, F. G., H. Remaut, S. J. Hultgren, and G. Waksman. 2004. Fiber assembly by the chaperone-usher pathway. Biochim. Biophys. Acta 1696:259-267. [DOI] [PubMed] [Google Scholar]
- 39.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 40.Smolyakov, R., A. Borer, K. Riesenberg, F. Schlaeffer, M. Alkan, A. Porath, and D. Rimar. 2003. Nosocomial multidrug resistant Acinetobacter baumannii bloodstream infection: risk factors and outcome with ampicillin-sulbactam treatment. J. Hosp. Infect. 54:32-38. [DOI] [PubMed] [Google Scholar]
- 41.Soto, G. E., and S. J. Hultgren. 1999. Bacterial adhesins: common themes and variations in architecture and assembly. J. Bacteriol. 181:1059-1071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thanassi, D. G., E. T. Saulino, and J. S. Hultgren. 1998. The chaperone/usher pathway: a major terminal branch of the general secretory pathway. Curr. Opin. Microbiol. 1:223-231. [DOI] [PubMed] [Google Scholar]
- 43.Tomaras, A. P., C. W. Dorsey, R. E. Edelmann, and L. A. Actis. 2003. Attachment to and biofilm formation on abiotic surfaces by Acinetobacter baumannii: involvement of a novel chaperone-usher pili assembly system. Microbiology 149:3473-3484. [DOI] [PubMed] [Google Scholar]
- 44.Vaneechoutte, M., D. M. Young, L. N. Ornston, T. D. Baere, A. Nemec, T. V. D. Reijden, and L. Dijkshoorn. 2006. The naturally transformable Acinetobacter strain ADP1 belongs to the newly described species Acinetobacter baylyi. Appl. Environ. Microbiol. 72:932-936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Valentine, R. C., B. M. Shapiro, and E. R. Stadtman. 1968. Regulation of glutamine synthetase. XII. Electron microscopy of the enzyme from Escherichia coli. Biochemistry 7:2143-2152. [DOI] [PubMed] [Google Scholar]
- 46.White-Ziegler, C. A., M. L. Angus Hill, B. A. Braaten, M. W. van der Woude, and D. A. Low. 1998. Thermoregulation of Escherichia coli pap transcription: H-NS is a temperature-dependent DNA methylation blocking factor. Mol. Microbiol. 28:1121-1137. [DOI] [PubMed] [Google Scholar]
- 47.White-Ziegler, C. A., A. Villapakkam, K. Ronaszeki, and S. Young. 2000. H-NS controls pap and daa fimbrial transcription in Escherichia coli in response to multiple environmental cues. J. Bacteriol. 182:6391-6400. [DOI] [PMC free article] [PubMed] [Google Scholar]