Abstract
The relevance of the acyl homoserine lactone (acyl-HSL) quorum signals N-3-oxododecanoyl-homoserine lactone (3OC12HSL) and N-butanoyl-homoserine lactone to the biology and virulence of Pseudomonas aeruginosa is well investigated. Previously, P. aeruginosa was shown to degrade long-chain, but not short-chain, acyl-HSLs as sole carbon and energy sources (J. J. Huang, J.-I. Han, L.-H. Zhang, and J. R. Leadbetter, Appl. Environ. Microbiol. 69:5941-5949, 2003). A gene encoding an enzyme with acyl-HSL acylase activity, pvdQ (PA2385), was identified, but it was not required for acyl-HSL utilization. This indicated that P. aeruginosa encodes another acyl-HSL acylase, which we identify here. A comparison of total cell proteins of cultures grown with long-acyl acyl-HSLs versus other substrates implicated the involvement of a homolog of PvdQ, the product of gene PA1032, for which we propose the name QuiP. Transposon mutants of quiP were defective for growth when P. aeruginosa was cultured in medium containing decanoyl-HSL as a sole carbon and energy source. Complementation with a functional copy of quiP rescued this growth defect. When P. aeruginosa was grown in buffered lysogeny broth, constitutive expression of QuiP in P. aeruginosa led to decreased accumulations of the quorum signal 3OC12HSL, relative to the wild type. Heterologous expression of QuiP was sufficient to confer long-chain acyl-HSL acylase activity upon Escherichia coli. Examination of gene expression patterns during acyl-HSL-dependent growth of P. aeruginosa further supported the involvement of quiP in signal decay and revealed other genes also possibly involved. It is not yet known under which “natural” conditions quiP is expressed or how P. aeruginosa balances the expression of its quorum-sensing systems with the expression of its acyl-HSL acylase activities.
Acyl homoserine lactone (acyl-HSL)-mediated quorum sensing is employed by numerous Proteobacteria in the control of diverse and significant biological activities such as the production of antibiotics (39), the formation of differentiated biofilms (3, 21), cell motility (1, 9), and virulence factor production (7, 25, 30, 33). In recent years, a diverse group of isolated Proteobacteria, Firmicutes, and Actinobacteria (4, 6, 14, 19, 20, 23), as well as naturally occurring microbial communities from the environment (10, 36), have been shown to be capable of degrading acyl-HSL-signaling molecules. A number of mechanisms and genes involved in signal decay have been identified recently (6, 14, 19, 20, 23, 24, 40), and this knowledge has been utilized to exploit or interfere with quorum sensing (QS) (5, 41).
The opportunistic pathogen Pseudomonas aeruginosa causes infections in immunocompromised individuals and forms persistent lung infections in individuals with the genetic disease cystic fibrosis (28). Factors relevant to the success of P. aeruginosa in a diversity of habitats are known to be regulated by its quorum-sensing systems, which consist of at least three interconnected signaling systems (for a review, see reference 17). The acyl-HSL-mediated QS systems of P. aeruginosa are among the best studied of these signaling circuits. These systems employ two distinct acyl-HSL signaling molecules, N-3-oxododecanoyl-homoserine lactone (3OC12HSL) and N-butanoyl-homoserine lactone (C4HSL), which are produced and sensed by the products of the lasI/lasR and rhlI/rhlR gene pairs, respectively (11, 22, 25, 26). The las and rhl QS systems operate in a hierarchy wherein the las system regulates the rhl system at the transcriptional and posttranslational levels (18, 27). 3OC12HSL, the signaling molecule of the las system, is thus believed to be a “gatekeeping” signal for its role in the P. aeruginosa quorum-sensing hierarchy (17).
Previously, we demonstrated that P. aeruginosa PAO1 and a closely related pseudomonad isolated from soil are able to degrade and utilize long-chain (≥8 carbons in the acyl chain) but not short-acyl chain acyl-HSL molecules as sole sources of carbon and energy (14). Quorum signals were degraded via the acylase mechanism (14, 19). Studies of the gene pvdQ from strain PAO1 determined that it was sufficient but not necessary for long-acyl-chain acyl-HSL degradation, as a diversity of pvdQ mutants retained the ability to degrade and utilize acyl-HSLs (14). This established that P. aeruginosa encodes at least one additional acyl-HSL-degrading acylase enzyme. In the current study, we pursued the identification of other genes and enzymes that form the basis for the acyl-HSL degradation and utilization phenotype of P. aeruginosa PAO1.
MATERIALS AND METHODS
Bacterial strains and culture conditions.
The bacterial strains used in this study are listed in Table 1. LB (lysogeny broth) (2), amended with antibiotics when appropriate, was used for growth and maintenance of strains unless otherwise noted. A defined freshwater medium buffered at pH 5.5 with 5 mM 2-(N-morpholino)-ethanesulfonic acid (MES) was used when acyl-HSL signal was provided as the sole carbon source, as previously described (19). Acyl-HSLs used in these studies were as follows: N-3-oxohexanoyl-l-homoserine lactone (3OC6HSL; Sigma), N-butanoyl-dl-homoserine lactone (C4HSL; Fluka), N-hexanoyl-dl-homoserine lactone (C6HSL; Fluka), N-heptanoyl-dl-homoserine lactone (C7HSL; Fluka), N-octanoyl-dl-homoserine lactone (C8HSL; Fluka), N-decanoyl-dl-homoserine lactone (C10HSL; Fluka), N-3-oxododecanoyl-l-homoserine lactone (3OC12HSL; Quorum Sciences, Inc., Iowa City, Iowa); and N-dodecanoyl-dl-homoserine lactone (C12HSL; Fluka). Stock solutions (100 mM) of each acyl-HSL were prepared, aliquoted, and stored as described previously (19). Decanoyl-HSL (C10HSL) was utilized for most experiments as a less-expensive, representative long-chain acyl-HSL. Cells were grown in 5 ml of medium in 18-mm test tubes with shaking at 250 rpm at 37°C unless otherwise noted. Antibiotics were used at the following concentrations: 60 μg/ml tetracycline, 34 μg/ml chloramphenicol, 50 μg/ml spectinomycin, 100 μg/ml ampicillin (Escherichia coli), and 250 μg/ml carbenicillin (P. aeruginosa).
TABLE 1.
Bacterial strains used in this study
| Species and strain | Characteristic(s) | Source |
|---|---|---|
| Pseudomonas aeruginosa PAO1 | ||
| PAO1 | WT | Laboratory stock (originally from B. Iglewski) |
| PAO1UW | WT | 15 |
| PAO1 pPA1032-Nde | This study | |
| PAO1 pPA1893-Nde | This study | |
| PAO1 pPA305-Nde | This study | |
| PAO1 pPA2385-Nde | 14 | |
| PAO1UW mutant ID: 33050 | PA1032::Tn′ phoA/in | 15 |
| PAO1UW mutant ID: 8713 | PA3922::Tn′ lacZ/in | 15 |
| PAO1UW mutant ID: 6619 | PA2385::Tn′ lacZ/in | 15 |
| PAO1UW mutant ID: 7831 | PA1893::Tn′ lacZ/in | 15 |
| PAO1UW mutant ID: 32876 | PA0305::Tn′ phoA/in | 15 |
| PAO1UW PA1032 mutant ID: 33050 with pPA1032-Nde | This study | |
| PAO1UW PA1032 mutant ID: 33050 with empty pucp-Nde | This study | |
| Escherichia coli | ||
| BL21Pro | Clontech | |
| BL21Pro pPA1032 | Shuttle vector | This study |
| BL21PRO PRO-Tet PA1032 | This study | |
| BL21PRO PRO-Tet PA1893 | This study | |
| BL21PRO PRO-Tet PA305 | This study |
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and peptide mass fingerprinting.
Cultures were grown in MES (morpholineethanesulfonic acid; pH 5.5) minimal medium with either 1 mM C10HSL or 1 mM sodium decanoate as a sole carbon and energy source. Cell cultures from the different growth conditions were harvested during the exponential phase at an optical density of 600 nm (OD600) of approximately 0.2, and the volume of culture that would equate to 1 ml of cell suspension at an OD600 of 1.0 was centrifuged at 5,220 × g for 10 min. The cell pellets were resuspended in 150 μl of 1× LDS sample buffer (Invitrogen) with 10% reducing agent (Invitrogen) and heated at 90°C for 10 min. Samples were centrifuged as above for 10 min, and 7 μl of the clarified supernatant was loaded onto a 10% Bis-Tris morpholinepropanesulfonic acid (MOPS)-buffered acrylamide gel (Invitrogen). Prestained SDS-PAGE standards (Bio-Rad) were used as molecular mass markers. Gels were run with MOPS SDS running buffer in an XCell Surelock system (Invitrogen) according to the protocol of the manufacturer, with the exception that the gel was run at 100 V. Gels were stained with Coomassie G-250 and destained with water at 18°C with orbital shaking overnight. Protein bands of interest were carefully excised under a dissecting microscope with sterile razor blades and transferred to sterile microcentrifuge tubes. The in-gel proteins were analyzed at the Peptide Facility at the California Institute of Technology (Caltech) by methods as previously described (12, 16, 42), with several modifications. Proteins were extracted from the gel, trypsin digested, and analyzed by both liquid chromatography-tandem mass spectrometry (LC-MS/MS) with a QSTAR pulsar mass spectrometer (Applied Biosystems) with a C18 reverse-phase 3.5-μm Vydac column (internal diameter, 0.1 mm; length, 60 mm), as well as with matrix-assisted laser desorption ionization-time of flight (MALDI-TOF) MS techniques. Protein identification from mass spectra was performed by peptide mass fingerprinting with Mascot (Matrix Science) software. Results from both LC-MS/MS and MALDI-TOF MS analyses were compared.
Transposon insertion mutant studies and constitutive expression of PA1032 in mutant and wild-type backgrounds.
P. aeruginosa PAO1 mutants carrying transposon insertions into the PA1032 gene and several other loci of interest were obtained from the Manoil laboratory (Table 1) (15). These mutants were chosen on the basis of the results of the protein analysis performed above or because they were obvious homologs of those genes. The transposon location within mutants was confirmed by PCR. Presence of a transposon within each gene of interest was confirmed by PCR with flanking primers, and transposon orientation was determined with transposon internal primers and flanking primers. The mutants were tested for decanoyl-HSL utilization. For starter culture inocula, strains were grown overnight in MES (pH 5.5) medium with tetracycline and 5 mM sodium succinate as the carbon source. Overnight cultures were inoculated (2% [vol/vol]) into medium of the same composition, except that 1 mM C10HSL was the sole carbon source. Cultures were incubated for up to 5 weeks at 37°C in a shaking water bath, and OD600 was measured using a Spectronic 20 spectrophotometer. For insertion mutants exhibiting no growth or strongly defective growth on acyl-HSLs, the impact of complementing the mutation with a functional copy of the gene expressed on a plasmid was examined. The gene PA1032 was amplified from the P. aeruginosa PAO1 genome using the primers 5′-TAGGATACATATGGCCTCGCCAGCCTTCATG-3′ and 5′-AATCTAGATCAGCGAGCGGGAGTGAGCGT-3′ (NdeI and XbaI restriction sites, respectively, are underlined) and ligated into the NdeI-XbaI-digested E. coli-Pseudomonas shuttle expression plasmid pUCP-Nde (2a) with T4 ligase (Invitrogen) according to the manufacturer's protocol. The resulting construct, pPA1032-Nde, was transformed into E. coli and sequenced to ensure that no mutations occurred during PCR amplification. Plasmid pPA1032-Nde was then transformed into wild-type P. aeruginosa PAO1 and P. aeruginosa PA1032 transposon mutants via electroporation using a Bio-Rad Gene Pulser II electroporator.
Wild-type P. aeruginosa PAO1 and transposon insertion mutants transformed with the plasmid pPA1032-Nde were tested for their ability to grow utilizing acyl-HSL signal molecules by inoculating succinate-grown, overnight cultures of these strains into MES (pH 5.5) medium containing 1 mM C10HSL and carbenicillin. Cultures were incubated at 37°C and shaken at 250 rpm. Changes in OD600 were monitored, and culture fluids were analyzed by LC-atmospheric pressure chemical ionization-MS (LC/APCI-MS), essentially as described previously (14). Cells were separated from the culture medium by centrifugation. Clarified culture fluids were diluted 10-fold to decrease the concentration of salts and buffer in the injected samples. Samples contained a final concentration of 50% (vol/vol) methanol and 50% (vol/vol) deionized water acidified with 0.1% (vol/vol) acetic acid. A total of 50 μl of sample was injected and analyzed with a Hewlett-Packard 1100 series LC/APCI-MS outfitted with a 150-mm by 4.6-mm mixed-mode reverse-phase C8 column (Alltech). The mobile phase was 1 ml · min−1 of 75% (vol/vol) methanol and 25% (vol/vol) deionized water acidified with 0.1% (vol/vol) acetic acid. These conditions enabled improved separation of HSL, homoserine (HS), and a variety of chain length acyl-HSLs from culture fluid by the method described previously (14). Concentrations of HSL, HS, and acyl-HSL from samples could be determined by comparison to standard curves of the compounds.
Accumulation of naturally produced 3OC12HSL over time by wild-type P. aeruginosa expressing pPA1032-Nde or the empty control plasmid was examined. Cultures (each, 100 ml) were grown in LB buffered with 50 mM MOPS (pH 7.0) and incubated at 30°C with shaking. Cultures samples (each, 4 ml) were removed over the course of growth and extracted twice with ethyl acetate acidified with 0.01% (vol/vol) acetic acid (8, 26). The extract was evaporated to dryness, and the dried residue was reconstituted in methanol and subsequently mixed with an equal volume of deionized water. Triplicate samples at each time point were analyzed by LC/APCI-MS as described above.
Expression of gene PA1032 and two other P. aeruginosa acylase homologs in E. coli.
The genes PA1032, PA0305, and PA1893, which were predicted to encode N-terminal nucleophile aminohydrolases (Ntn hydrolases) and are homologs of the known acyl-HSL acylase of P. aeruginosa encoded by the gene pvdQ (PA2385), were amplified from strain PAO1 genomic DNA and ligated into pPROTet.E133 (Clontech) using primers 1032F (ATTAGAAGCTTATGGCCTCGCCAGCCTTC), 1032R (ATTACTCTAGATCAGCGAGCGGGAGTG), 305F (ACTACAAGCTTATGAAACGCACCCTGAC), 305R (ATTACTCTAGATCAGCGCTTCGGCTCCAG), 1893F (ACTACAAGCTTATGTCGAAGAACGCACG), and 1893R (ATCACTCTAGATCATGGTCGTGGCTCGC) (HindIII and XbaI restriction sites are underlined). After ligation, the resultant plasmids were electroporated into E. coli BL21Pro (Clontech). E. coli BL21Pro cultures containing the plasmid constructs were induced to express the inserted genes with 10 nM anhydrotetracyline overnight at 16°C upon reaching an OD600 of 0.5. SDS-PAGE analysis of the protein profiles of induced versus noninduced cultures revealed the overexpression of polypeptides of approximately 90 kDa. This mass corresponds to the premodified propolypeptide predicted to be generated by each of these three Ntn hydrolase homolog-encoding genes; however, the presence of the two subunits that would be predicted from the proper processing of the propolypeptide was not readily apparent by SDS-PAGE analysis. To assay for acyl-HSL acylase activity, induced and noninduced cultures were concentrated to an OD600 of 1.2 in 1 ml of MOPS (pH 7) medium amended with each of various-chain-length acyl-HSLs ranging from C4HSL to C14HSL. Reaction mixtures were incubated at 30°C with shaking at 250 rpm. Samples (each, 50 μl) were removed over time, clarified by centrifugation, and examined for acyl-HSL, HSL, acyl-homoserine, and HS content by LC/APCI-MS as described above.
Transcriptome analysis using Affymetrix GeneChip microarrays.
Wild-type P. aeruginosa PAO1 was grown out from freezer stock on LB agar, and single colonies that developed were picked and grown overnight in MES (pH 5.5) medium with 5 mM succinate at 37°C. A total of 1 ml of overnight cell culture was inoculated into flasks containing 15 ml of sterile MES (pH 5.5) medium that contained either 3 mM C10HSL, 8 mM sodium succinate, or 3 mM sodium decanoate as a carbon source. Cultures were grown at 37°C and shaken at 250 rpm. Cell cultures incubated with acyl-HSL signal as the sole carbon source began growing on acyl-HSL signal after a long lag period of approximately 3 weeks, as previously described (14). Once cultures began growing exponentially, they were transferred once into fresh medium of the same composition and allowed to continue growing under the same conditions. Upon reaching exponential phase with an OD600 of 0.4, 10-ml samples of culture from all substrate conditions were processed for RNA. RNA in all culture samples was stabilized immediately with RNA Protect Bacterial Reagent (QIAGEN) and processed according to the manufacturer's protocol. RNA was extracted from cell cultures with the RNeasy kit (QIAGEN) and prepared for hybridization to GeneChips as previously described (29, 31). Hybridization, washing, and scanning of Affymetrix GeneChips were performed at the University of Iowa Genome Center using an Affymetrix Fluidics Station. GeneChip experiments were performed in duplicate, and data were analyzed with Microarray Suite software. We report, as arbitrarily significant, gene activity changes showing >5-fold up- or down-regulation.
RESULTS
Identification of P. aeruginosa proteins putatively involved in acyl-HSL-dependent growth.
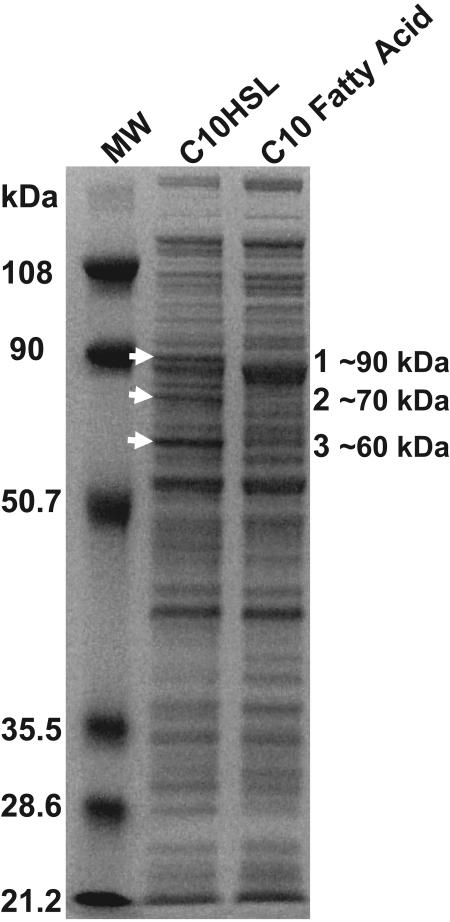
SDS-PAGE analysis and subsequent peptide mass fingerprinting enabled the identification of three proteins of 60 kDa, 70 kDa, and 90 kDa that were readily apparent and consistently present in cultures growing on decanoyl-HSL but not in cultures growing on the corresponding fatty acid, decanoate, as a sole source of carbon and energy (Fig. 1). These three protein bands were not obvious in either (i) cultures grown with decanoate that were inoculated with acyl-HSL-grown cells, (ii) cultures grown with decanoate in the presence of 1 mM C10HSL, or (iii) cultures grown with decanoate in the presence of 10 μM 3OC12HSL, 10 μM C4HSL, or a combination of both of these P. aeruginosa quorum signals (data not shown). Peptide mass fingerprinting identified the proteins with masses of 60 kDa and 70 kDa as the products of P. aeruginosa PAO1 genes PA3922 and PA4022, respectively. Gene PA3922 encodes a hypothetical protein that is highly conserved among several pseudomonads but that is found in few other bacteria. It has no known function and no known obvious structural motifs except for a 25-residue secretion signal sequence (http://www.cbs.dtu.dk/services/SignalP/). Within the P. aeruginosa genome, PA3922 is a close homolog of PA3421 (sharing 69% amino acid identity), and the flanking genes in their respective gene clusters are quite similar to each other as well. The second of the three, the product of gene PA4022, is predicted to be an alcohol dehydrogenase, based on homology. The PA4022 protein is most likely localized to the periplasm, as indicated by the expression of alkaline phosphatase from translational fusions at this locus, described in a previous study (15). The identification of the third, 90-kDa protein became especially intriguing. With LC-MS/MS analyses, 17 peptides generated from this excised 90-kDa protein band were strongly matched, with 21% total coverage, to the predicted protein product of gene PA1032. PA1032 was identified from independent MALDI-TOF MS and LC-MS/MS analysis of the in-gel protein overexpressed by P. aeruginosa-degrading decanoyl-HSL. This locus had previously been annotated as a “putative penicillin amidase” (34). To our knowledge, PA1032 has not been previously studied by others. PA1032 is the final gene of a putative gene cluster that consists of genes PA1033, which is a distant homolog of glutathione S-transferases, and PA1034 and PA1035, which both encode “hypothetical proteins of unknown function” (www.pseudomonas.com). The product of PA1032 was particularly intriguing, as it is one of four Ntn hydrolase homologs encoded by the P. aeruginosa genome, one of which is PvdQ (PA2385), a previously identified acyl-HSL acylase (14). PA1032 shares 21% amino acid identity with PvdQ and 23% amino acid identity with AiiD, the acyl-HSL acylase from a strain belonging to the genus Ralstonia (20), with the conserved regions predominantly in the signal sequence and α and linker regions but less in the β subunit (GeneDoc; http://www.psc.edu/biomed/genedoc/). PA1032 is predicted to be secreted into the periplasm based on its signal sequence (http://www.psort.org/psortb/) and for its similarity to the Ntn hydrolase family of proteins. It is predicted to be cleaved into four peptides: a signal sequence, two peptides corresponding to the α and β subunits of the natural enzyme, and a spacer peptide. In our experience with the overexpression of this gene, much unprocessed propolypeptide remains. Its expression during the acyl-HSL-dependent growth of strain PAO1, coupled with its homology to previously identified, functional acyl-HSL acylases, led us to pursue the study of PA1032, with the hypothesis that this gene encodes the second acyl-HSL acylase of P. aeruginosa that we had previously postulated to exist (14).
FIG. 1.
Protein profiles of cells growing on decanoyl-HSL (C10HSL) compared to cells growing on the corresponding fatty acid. Arrows indicate three readily apparent proteins specific for growth on C10HSL. Proteins 1 to 3 were identified by peptide mass fingerprinting and LC-MS/MS as the products of genes PA1032, PA4022, and PA3922, respectively.
Phenotypic analysis and complementation of PA1032 transposon insertion mutants.
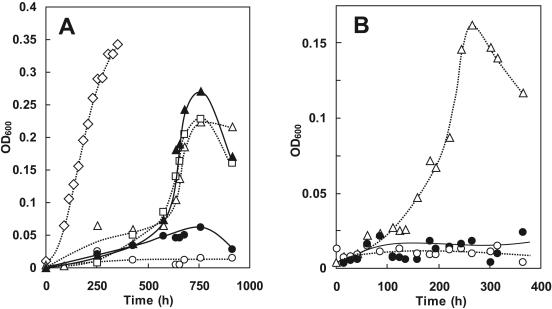
Strains of P. aeruginosa PAO1 carrying transposon insertions into gene PA1032 and into three other Ntn hydrolase-encoding genes, pvdQ/PA2385, PA1893, and PA0305, as well as into gene PA3922, whose protein product was present during acyl-HSL degradation (see above), were examined for their ability to grow while utilizing decanoyl-HSL as a growth substrate. The growth phenotypes of strains carrying transposon insertions into gene PA4022, which encodes another peptide of interest (identified above), were not analyzed as we were unable to confirm insertion positions as we had with the others examined, i.e., after obtaining the relevant strains from the University of Washington—Seattle. Strains carrying transposon insertions into either gene PA1032 or PA3922 exhibited defective growth on decanoyl-HSL in comparison to the wild type, while strains carrying insertions in the other genes exhibited no detectable phenotypic variation (Fig. 2A). The PA1032 mutant did not appear to grow at any rate or to any yield during the 5-week incubation time. The mutant PA3922 strain grew more slowly and reached, at most, one-fourth of the yield of the wild type. These results implicate genes PA1032 and PA3922 in the acyl-HSL-dependent growth phenotype of P. aeruginosa. Moreover, it appears that gene PA1032, and not the other genes encoding the other three Ntn hydrolase homologs, likely encodes the acyl-HSL acylase activity observed during the growth utilization of acyl-HSLs.
FIG. 2.
Acyl-HSL growth characteristics of several transposon insertion mutants of interest. (A) Comparison of growth utilization of acyl-HSLs by the University of Washington—Seattle wild-type strain PAO1 and five insertion mutants derived from it (15). ⋄, wild type; □, PA0305 insertion mutant; ▵, PA1893 insertion mutant; ▴, pvdQ (PA2385) insertion mutant; •, PA3922 insertion mutant; and ○, quiP (PA1032) insertion mutant. (B) Complementation of the quiP (PA1032) transposon insertion mutant with a functional copy of the gene expressed from pUCP-Nde (▵) versus two controls: the uncomplemented mutant (○) and the same mutant containing pUCP-Nde without the gene insert (•). Each growth curve is one representative of the results of at least three replicate experiments.
A plasmid that constitutively expressed a functional, wild-type copy of PA1032 was introduced into the PA1032 transposon insertion mutant. The acyl-HSL-degrading capability of the mutant was complemented by the plasmid expressing PA1032 but not by the plasmid that contained no insert (Fig. 2B). Interestingly, complemented mutants that constitutively expressed the product of PA1032 from a plasmid degraded acyl-HSLs without lag. In this and our previous study (14), naïve P. aeruginosa cells (those not started with an acyl-HSL-grown cell inoculum) were observed to exhibit a lag of many days to several weeks before exponential growth and acyl-HSL utilization commenced. The introduction of the PA1032 expression construct into naïve P. aeruginosa, the PA1032 transposon mutant, or the PA3922 transposon mutant conferred the ability for these strains to grow and utilize long-acyl-chain acyl-HSLs without any significant lag. This result suggests that the control and regulation of gene PA1032 underlie the long lag periods previously observed. Based on its involvement in the inactivation and utilization of acyl-HSLs and contribution to the catabolic and anabolic needs of the cell, we give gene PA1032 the name quiP, for the quorum signal utilization and inactivation protein that it encodes.
Heterologous expression of quiP confers upon E. coli long-chain acyl-HSL acylase activity.
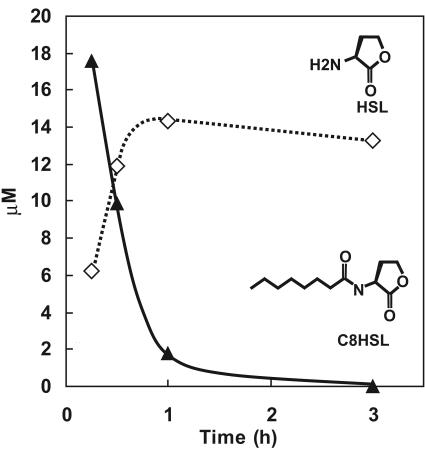
Expression of quiP from the inducible plasmid pPROTetE133 (Clontech) in E. coli resulted in the production of a 90-kDa protein corresponding to the expected mass of the unprocessed propolypeptide, as determined by SDS-PAGE (data not shown). The smaller α and β subunits that would be derived from the properly processed propolypeptide (13) were not visible by SDS-PAGE analysis. Nevertheless, some fraction of the QuiP propolypeptide was apparently processed to maturity, as the recombinant E. coli cells gained the ability to catalyze the degradation of 20 μM C8HSL, generating nearly stoichiometric amounts of HSL as a product of the reaction (Fig. 3). Such cells degraded all acyl-HSLs tested having side chains of seven or more carbons in length (C7HSL, C8HSL, C10HSL, C12HSL, 3OC12HSL, and C14HSL), also releasing HSL as a product (data not shown). The results confirm that quiP encodes a protein with acyl-HSL acylase activity. The same recombinant E. coli cell preparations did not degrade C4HSL, C6HSL, or 3OC6HSL, indicating that the enzyme has specificity for long but not short acyl-HSLs.
FIG. 3.
E. coli cells expressing recombinant quiP (PA1032) degrade long-chain acyl-HSLs and generate stoichiometric amounts of HSL. The graph shows degradation of a representative long-chain acyl-HSL, C8HSL. C8HSL substrate disappearance and HSL product accumulation were determined by LC/APCI-MS. Recombinant E. coli cells expressing PA1032 degraded C7HSL, C8HSL, C10HSL, C12HSL, 3OC12HSL, and C14HSL but did not degrade C4HSL, C6HSL, or 3OC6HSL (results not shown).
Constitutive expression of quiP in wild-type P. aeruginosa PAO1 alters the accumulation of its long-chain acyl-HSL quorum signal, 3OC12HSL, when grown in rich medium.
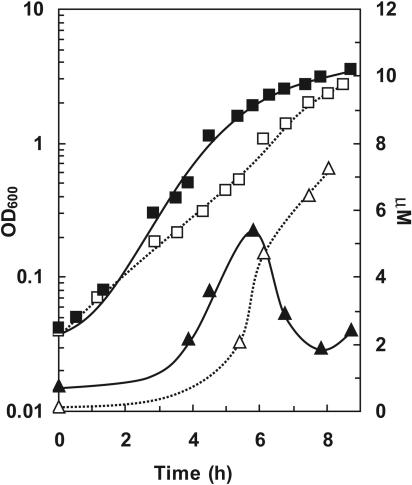
As noted above and previously (14), signal decay by P. aeruginosa appears to be tightly controlled and nontrivial to up-regulate experimentally. To examine the influence of the constitutive expression of quiP on quorum signal accumulations, we grew P. aeruginosa cells that contained a QuiP expression plasmid and a variant of strain PAO1 that contained the insertless control plasmid in the undefined standard laboratory medium LB, buffered at pH 7. Over time, we quantified the accumulation of endogenously produced 3OC12HSL by LC-MS/MS (Fig. 4). For reasons that are unclear, the growth rate of the control culture containing the empty plasmid was reproducibly inferior to that of the QuiP-recombinant strain. Both cultures accumulated similar levels of 3OC12HSL during the initial 7 h of growth. However, upon reaching an OD600 of 0.2, the standing pool size of 3OC12HSL in cultures of QuiP recombinant strains decreased dramatically (Fig. 4). Meanwhile, the pool size in the control strain continued to increase. The initial accumulation followed by the decrease of 3OC12HSL observed in QuiP recombinant cultures was reproducible, but the mechanism of regulation remained unexplained. Whether the initial accumulation phase related to issues of inadequate expression, delays in propolypeptide maturation, or other unknowns (such as a cue-mediated posttranslational activation of the protein) remains to be determined.
FIG. 4.
Impact of the constitutive expression of quiP (PA1032) on 3OC12HSL quorum signal accumulation by recombinant P. aeruginosa cells grown in rich medium. P. aeruginosa cells constitutively expressing quiP accumulate less endogenously produced 3OC12HSL than cells that contain the plasmid alone. P. aeruginosa strains were grown in LB buffered with 50 mM MOPS at pH 7. OD600 values (▪) and acyl-HSL accumulation (▴) by wild-type strain PAO1 expressing the acylase from plasmid pPA1032 are shown. Growth (□) and signal accumulations (▵) by wild-type strain PAO1 containing a control plasmid without insert are also shown.
Examination of gene expression profiles during utilization of acyl-HSLs by P. aeruginosa supports the role of quiP and PA3922 in signal decay.
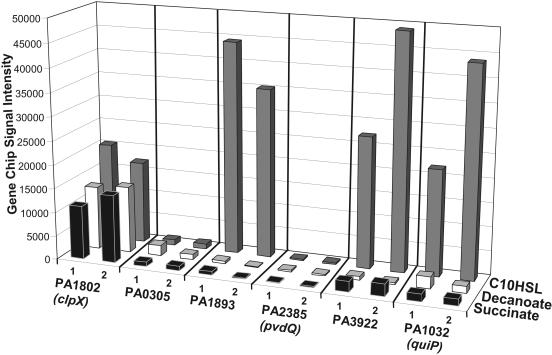
We employed a microarray approach to compare the transcriptional profiles of P. aeruginosa strain PAO1 cultures growing on decanoyl-HSLs versus cultures growing on decanoate or succinate. This was done to further determine whether loci we had identified by SDS-PAGE and mutant analysis and other genes are involved in the utilization of long-chain acyl-HSLs by P. aeruginosa. A listing of all genes significantly up- or down-regulated under these conditions is provided (see the supplemental material). The results were in good agreement with those obtained from comparison of protein profiles under similar growth conditions (see above and Fig. 1). mRNA of genes quiP and PA3922 was strongly overexpressed in cultures actively utilizing decanoyl-HSL (Fig. 5). Curiously, gene PA4022, identified as a locus of interest during the protein analysis (above), is not represented on the commercial P. aeruginosa microarrays. Thus, the expression of this gene was not examined. In agreement with previous results that the acyl-HSL acylase PvdQ was not necessary for acyl-HSL utilization, there was no evidence for the expression of pvdQ mRNA during the growth of P. aeruginosa under these same conditions. Of the two remaining acylase homologs encoded by P. aeruginosa, PA0305 and PA1893, only the latter was expressed in great amount during decanoyl-HSL utilization relative to decanoate- or succinate-dependent growth. PA1893, however, was up-regulated in cultures grown in succinate-defined medium amended with 3 mM decanoyl-HSL (data not presented) and has previously been shown to be quorum regulated (31, 35, 38). In agreement with protein profile comparisons (see above) and previous transcriptome studies (31, 35), quiP was not expressed greatly in succinate-defined medium amended with acyl-HSLs, suggesting that quiP is not up-regulated simply as a function of the acyl-HSL status of the cell.
FIG. 5.
Examination of P. aeruginosa mRNA expression levels of four acylase-encoding genes and other genes of interest in cultures grown utilizing different carbon sources. GeneChip microarrays were employed to examine gene expression in cultures grown utilizing either decanonyl-HSL (gray bars); its corresponding fatty acid, decanoate (white bars); or succinate (black bars) as the sole carbon source. The signal intensity values are quantitative measures of the relative amount of mRNA expressed for each gene under the given growth conditions. The clpX protease gene was used as a control, as it is known to be expressed at similar levels under most growth conditions. A comprehensive list of genes that were up- or down-regulated by at least fivefold during decanoyl-HSL utilization versus growth on decanoate is provided (see Table S1 in the supplemental material).
DISCUSSION
P. aeruginosa has the ability to degrade acyl-HSL of eight or more carbons, including its native long-chain acyl-HSL (14). The gene pvdQ of P. aeruginosa has been shown to be sufficient but not necessary for acyl-HSL degradation (14). In this study, we identify QuiP, the product of the P. aeruginosa gene PA1032, as a second acyl-HSL acylase capable of the inactivation of quorum signals in P. aeruginosa. quiP mRNA and its protein product are expressed during strain PAO1 growth on long-chain acyl-HSL (Fig. 1 and 5), and QuiP is sufficient to catalyze the degradation of long-chain but not short-chain acyl-HSLs in E. coli (Fig. 3). Constitutive expression of QuiP in P. aeruginosa resulted in decreased accumulation of 3OC12HSL signal by strain PAO1, indicating that the enzyme has activity against physiologically relevant concentrations of acyl-HSL produced by P. aeruginosa (Fig. 4). QuiP appears to be the acyl-HSL acylase that underlies the ability of P. aeruginosa to degrade and utilize certain acyl-HSLs for growth, including one of its own quorum signals, 3OC12HSL.
QuiP has a specificity for the degradation of long-acyl-chain acyl-HSL that is similar to that of PvdQ (14) and another recently described AHL-degrading acylase encoded by a Streptomyces species (23). Proteins sharing 61 to 68% amino acid identity with QuiP are encoded by other members of the family Pseudomonadaceae: Azotobacter vinelandii, Pseudomonas fluorescens, Pseudomonas syringae, and Pseudomonas putida. QuiP also shared 29 to 32% amino acid identity with proteins encoded by Ralstonia metallidurans and Ralstonia eutropha, as well as several oxygenic or anoxygenic phototrophs (Gloeobacter violaceus, Rubrivivax gelatinosus, and Nostoc punctiforme). Whether these microbes also share an ability to degrade acyl-HSLs has, to our knowledge, not yet been examined.
These studies also suggest that the protein encoded by PA3922 plays an undetermined yet possibly critical role in the acyl-HSL degradation and utilization process. It was shown to be strongly up-regulated and its protein product to be present in large amounts during acyl-HSL utilization, as revealed by microarray analyses and protein profiling, respectively (Fig. 5 and 1). A mutant strain carrying a transposon insertion into this locus exhibited a marked growth defect during the utilization of long-chain acyl-HSLs (Fig. 2B). However, the nature of its role in the AHL degradation process remains elusive, and its homology with other proteins in the databases provides no obvious clues. Curiously, PA3922 was previously reported to be down-regulated in P. aeruginosa biofilms exposed to the antibiotic tobramycin (37), but the role of this gene in the organism's response to that perturbation is not understood either. The distribution of homologs of gene PA3922 is virtually restricted to the pseudomonads and a few other bacteria; no obvious homologs of this gene are found in E. coli.
Under natural circumstances, when would these enzymes that degrade acyl-HSLs be expressed, and what role could they play in the biology of P. aeruginosa? The expression of QuiP, the acyl-HSL acylase described in this study, is tightly controlled. To our knowledge, up-regulation of quiP had not previously been reported in microarray or other studies. The expression of this trait does not appear to be controlled as a function of the catabolic or anabolic needs of the cell (14). Thus, although the recycling of the carbon and energy through the degradation of quorum signals would no doubt have benefits for the cell, it would appear that the primary function(s) of QuiP is something other than the growth physiology of this bacterium.
Clearly, P. aeruginosa populations must carefully balance and control signal production and reception during quorum sensing with the expression of proteins exhibiting signal decay activities by either ensuring that the latter systems do not interfere with cell-cell communications or (at times) ensuring that they do. Since P. aeruginosa populations maintain different ratios of their long- and short-chain acyl-HSL signals when grown in biofilm versus planktonic states (32), we speculate that the expression of genes such as quiP could potentially occur in spatially distinct subpopulations of P. aeruginosa growing in the biofilm state.
Supplementary Material
Acknowledgments
This research was supported by the W. M. Keck Foundation Fund for Discovery in Basic Medical Research at the California Institute of Technology, by the Powell Foundation, by EPA STAR Fellowship no. 91620301 (to J.J.H.), and by the Oklahoma Center for the Advancement of Science and Technology (to M.W.).
We thank Gary Hathaway and the Peptide Facility at Caltech for peptide mass fingerprinting, Nathan Dalleska for technical discussions and help with performing LC/APCI-MS analyses, and our laboratory colleagues for their helpful comments.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Atkinson, S., J. P. Throup, G. Stewart, and P. Williams. 1999. A hierarchical quorum-sensing system in Yersinia pseudotuberculosis is involved in the regulation of motility and clumping. Mol. Microbiol. 33:1267-1277. [DOI] [PubMed] [Google Scholar]
- 2.Bertani, G. 2004. Lysogeny at mid-twentieth century: P1, P2, and other experimental systems. J. Bacteriol. 186:595-600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2a.Cronin, C. N., and W. S. McIntire. 1999. pUCP-Nco and pUCP-Nde: Escherichia-Pseudomonas shuttle vectors for recombinant protein expression in Pseudomonas. Anal. Biochem. 272:112-115. [DOI] [PubMed] [Google Scholar]
- 3.Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. [DOI] [PubMed] [Google Scholar]
- 4.Dong, Y.-H., A. R. Gusti, Q. Zhang, J.-L. Xu, and L.-H. Zhang. 2002. Identification of quorum-quenching N-acyl homoserine lactonases from Bacillus species. Appl. Environ. Microbiol. 68:1754-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dong, Y.-H., L.-H. Wang, J.-L. Xu, H.-B. Zhang, X.-F. Zhang, and L.-H. Zhang. 2001. Quenching quorum-sensing-dependent bacterial infection by an N-acyl homoserine lactonase. Nature 411:813-817. [DOI] [PubMed] [Google Scholar]
- 6.Dong, Y. H., J. L. Xu, X. Z. Li, and L. H. Zhang. 2000. AiiA, an enzyme that inactivates the acylhomoserine lactone quorum-sensing signal and attenuates the virulence of Erwinia carotovora. Proc. Natl. Acad. Sci. USA 97:3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dunphy, G., C. Miyamoto, and E. Meighen. 1997. A homoserine lactone autoinducer regulates virulence of an insect-pathogenic bacterium, Xenorhabdus nematophilus (Enterobacteriaceae). J. Bacteriol. 179:5288-5291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eberhard, A., A. L. Burlingame, C. Eberhard, G. L. Kenyon, K. H. Nealson, and N. J. Oppenheimer. 1981. Structural identification of autoinducer of Photobacterium fischeri luciferase. Biochemistry 20:2444-2449. [DOI] [PubMed] [Google Scholar]
- 9.Eberl, L., M. K. Winson, C. Sternberg, G. Stewart, G. Christiansen, S. R. Chhabra, B. Bycroft, P. Williams, S. Molin, and M. Givskov. 1996. Involvement of N-acyl-l-homoserine lactone autoinducers in controlling the multicellular behaviour of Serratia liquefaciens. Mol. Microbiol. 20:127-136. [DOI] [PubMed] [Google Scholar]
- 10.Flagan, S., W.-K. Ching, and J. R. Leadbetter. 2003. Arthrobacter strain VAI-A utilizes acyl-homoserine lactone inactivation products and stimulates quorum signal biodegradation by Variovorax paradoxus. Appl. Environ. Microbiol. 69:909-916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gambello, M. J., and B. H. Iglewski. 1991. Cloning and characterization of the Pseudomonas aeruginosa lasR gene, a transcriptional activator of elastase expression. J. Bacteriol. 173:3000-3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hellman, U., C. Wernstedt, J. Gonez, and C. H. Heldin. 1995. Improvement of an in-gel digestion procedure for the micropreparation of internal protein-fragments for amino-acid sequencing. Anal. Biochem. 224:451-455. [DOI] [PubMed] [Google Scholar]
- 13.Hewitt, L., V. Kasche, K. Lummer, R. J. Lewis, G. N. Murshudov, C. S. Verma, G. G. Dodson, and K. S. Wilson. 2000. Structure of a slow processing precursor penicillin acylase from Escherichia coli reveals the linker peptide blocking the active-site cleft. J. Mol. Biol. 302:887-898. [DOI] [PubMed] [Google Scholar]
- 14.Huang, J. J., J.-I. Han, L.-H. Zhang, and J. R. Leadbetter. 2003. Utilization of acyl-homoserine lactone quorum signals for growth by a soil pseudomonad and Pseudomonas aeruginosa PAO1. Appl. Environ. Microbiol. 69:5941-5949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobs, M. A., A. Alwood, I. Thaipisuttikul, D. Spencer, E. Haugen, S. Ernst, O. Will, R. Kaul, C. Raymond, R. Levy, C. R. Liu, D. Guenthner, D. Bovee, M. V. Olson, and C. Manoil. 2003. Comprehensive transposon mutant library of Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 100:14339-14344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeno, P., T. Mini, S. Moes, E. Hintermann, and M. Horst. 1995. Internal sequences from proteins digested in polyacrylamide gels. Anal. Biochem. 224:75-82. [DOI] [PubMed] [Google Scholar]
- 17.Juhas, M., L. Eberl, and B. Tummler. 2005. Quorum sensing: the power of cooperation in the world of Pseudomonas. Environ. Microbiol. 7:459-471. [DOI] [PubMed] [Google Scholar]
- 18.Latifi, A., M. Foglino, K. Tanaka, P. Williams, and A. Lazdunski. 1996. A hierarchical quorum-sensing cascade in Pseudomonas aeruginosa links the transcriptional activators LasR and RhIR (VsmR) to expression of the stationary-phase sigma factor RpoS. Mol. Microbiol. 21:1137-1146. [DOI] [PubMed] [Google Scholar]
- 19.Leadbetter, J. R., and E. P. Greenberg. 2000. Metabolism of acyl-homoserine lactone quorum-sensing signals by Variovorax paradoxus. J. Bacteriol. 182:6921-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lin, Y.-H., J.-L. Xu, J. Y. Hu, L.-H. Wang, S. L. Ong, J. R. Leadbetter, and L.-H. Zhang. 2003. Acyl-homoserine lactone acylase from Ralstonia strain XJ12B represents a novel and potent class of quorum-quenching enzymes. Mol. Microbiol. 47:849-860. [DOI] [PubMed] [Google Scholar]
- 21.Lynch, M. J., S. Swift, D. F. Kirke, C. W. Keevil, C. E. R. Dodd, and P. Williams. 2002. The regulation of biofilm development by quorum sensing in Aeromonas hydrophila. Environ. Microbiol. 4:18-28. [DOI] [PubMed] [Google Scholar]
- 22.Ochsner, U. A., A. Koch, A. Fiechter, and J. Reiser. 1994. Isolation and characterization of a regulatory gene affecting rhamnolipid biosurfactant synthesis in Pseudomonas aeruginosa. J. Bacteriol. 176:2044-2054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park, S.-Y., H.-O. Kang, H.-S. Jang, J.-K. Lee, B.-T. Koo, and D.-Y. Yum. 2005. Identification of extracellular N-acylhomoserine lactone acylase from a Streptomyces sp. and its application to quorum quenching. Appl. Environ. Microbiol. 71:2632-2641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park, S.-Y., S. J. Lee, T.-K. Oh, J.-W. Oh, B.-T. Koo, D.-Y. Yum, and J.-K. Lee. 2003. AhlD, an N-acylhomoserine lactonase in Arthrobacter sp., and predicted homologues in other bacteria. Microbiology 149:1541-1550. [DOI] [PubMed] [Google Scholar]
- 25.Passador, L., J. M. Cook, M. J. Gambello, L. Rust, and B. H. Iglewski. 1993. Expression of Pseudomonas aeruginosa virulence genes requires cell-to-cell communication. Science 260:1127-1130. [DOI] [PubMed] [Google Scholar]
- 26.Pearson, J. P., K. M. Gray, L. Passador, K. D. Tucker, A. Eberhard, B. H. Iglewski, and E. P. Greenberg. 1994. Structure of the autoinducer required for expression of Pseudomonas aeruginosa virulence genes. Proc. Natl. Acad. Sci. USA 91:197-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pesci, E. C., J. P. Pearson, P. C. Seed, and B. H. Iglewski. 1997. Regulation of las and rhl quorum sensing in Pseudomonas aeruginosa. J. Bacteriol. 179:3127-3132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ramsey, D. M., and D. J. Wozniak. 2005. Understanding the control of Pseudomonas aeruginosa alginate synthesis and the prospects for management of chronic infections in cystic fibrosis. Mol. Microbiol. 56:309-322. [DOI] [PubMed] [Google Scholar]
- 29.Ramsey, M. M., and M. Whiteley. 2004. Pseudomonas aeruginosa attachment and biofilm development in dynamic environments. Mol. Microbiol. 53:1075-1087. [DOI] [PubMed] [Google Scholar]
- 30.Rumbaugh, K. P., J. A. Griswold, and A. N. Hamood. 2000. The role of quorum sensing in the in vivo virulence of Pseudomonas aeruginosa. Microbes Infect. 2:1721-1731. [DOI] [PubMed] [Google Scholar]
- 31.Schuster, M., C. P. Lostroh, T. Ogi, and E. P. Greenberg. 2003. Identification, timing, and signal specificity of Pseudomonas aeruginosa quorum-controlled genes: a transcriptome analysis. J. Bacteriol. 185:2066-2079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 33.Smith, R. S., and B. H. Iglewski. 2003. P. aeruginosa quorum-sensing systems and virulence. Curr. Opin. Microbiol. 6:56-60. [DOI] [PubMed] [Google Scholar]
- 34.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. L. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. S. Wong, Z. Wu, I. T. Paulsen, J. Reizer, M. H. Saier, R. E. W. Hancock, S. Lory, and M. V. Olson. 2000. Complete genome sequence of Pseudomonas aeruginosa PAO1, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 35.Wagner, V. E., D. Bushnell, L. Passador, A. I. Brooks, and B. H. Iglewski. 2003. Microarray analysis of Pseudomonas aeruginosa quorum-sensing regulons: effects of growth phase and environment. J. Bacteriol. 185:2080-2095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang, Y.-J., and J. R. Leadbetter. 2005. Rapid acyl-homoserine lactone quorum signal biodegradation in diverse soils. Appl. Environ. Microbiol. 71:1291-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Whiteley, M., M. G. Bangera, R. E. Bumgarner, M. R. Parsek, G. M. Teitzel, S. Lory, and E. P. Greenberg. 2001. Gene expression in Pseudomonas aeruginosa biofilms. Nature 413:860-864. [DOI] [PubMed] [Google Scholar]
- 38.Whiteley, M., K. M. Lee, and E. P. Greenberg. 1999. Identification of genes controlled by quorum sensing in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 96:13904-13909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Williams, P., N. J. Bainton, S. Swift, S. R. Chhabra, M. K. Winson, G. Stewart, G. P. C. Salmond, and B. W. Bycroft. 1992. Small molecule-mediated density-dependent control of gene-expression in prokaryotes: bioluminescence and the biosynthesis of carbapenem antibiotics. FEMS Microbiol. Lett. 100:161-167. [DOI] [PubMed] [Google Scholar]
- 40.Zhang, H. B., L. H. Wang, and L. H. Zhang. 2002. Genetic control of quorum-sensing signal turnover in Agrobacterium tumefaciens. Proc. Natl. Acad. Sci. USA 99:4638-4643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang, L. H., and Y. H. Dong. 2004. Quorum sensing and signal interference: diverse implications. Mol. Microbiol. 53:1563-1571. [DOI] [PubMed] [Google Scholar]
- 42.Zhou, J., F. Rusnak, T. Colonius, and G. M. Hathaway. 2000. Quasi-linear gradients for capillary liquid chromatography with mass and tandem mass spectrometry. Rapid Commun. Mass Spectrom. 14:432-438. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.