Abstract
We isolated a cyanophage (Ma-LMM01) that specifically infects a toxic strain of the bloom-forming cyanobacterium Microcystis aeruginosa. Transmission electron microscopy showed that the virion is composed of anisometric head and a tail complex consisting of a central tube and a contractile sheath with helical symmetry. The morphological features and the host specificity suggest that Ma-LMM01 is a member of the cyanomyovirus group. Using semi-one-step growth experiments, the latent period and burst size were estimated to be 6 to 12 h and 50 to 120 infectious units per cell, respectively. The size of the phage genome was estimated to be ca. 160 kbp using pulse-field gel electrophoresis; the nucleic acid was sensitive to DNase I, Bal31, and all 14 restriction enzymes tested, suggesting that it is a linear double-stranded DNA having a low level of methylation. Phylogenetic analyses based on the deduced amino acid sequences of two open reading frames coding for ribonucleotide reductase alpha- and beta-subunits showed that Ma-LMM01 forms a sister group with marine and freshwater cyanobacteria and is apparently distinct from T4-like phages. Phylogenetic analysis of the deduced amino acid sequence of the putative sheath protein showed that Ma-LMM01 does not form a monophyletic group with either the T4-like phages or prophages, suggesting that Ma-LMM01 is distinct from other T4-like phages that have been described despite morphological similarity. The host-phage system which we studied is expected to contribute to our understanding of the ecology of Microcystis blooms and the genetics of cyanophages, and our results suggest the phages could be used to control toxic cyanobacterial blooms.
Microcystis aeruginosa is one of the highly noxious cyanobacteria that frequently form dense blooms in eutrophic freshwaters throughout the world (9). M. aeruginosa produces potent hepatotoxins (microcystins) that specifically inhibit eukaryotic protein phosphatase types 1 and 2A and cause hepatocellular carcinoma (21, 26, 48). Hence, M. aeruginosa blooms are often responsible for the death of livestock and wildlife and cause serious problems in water management (9).
Despite studies of the effects of various environmental factors on the growth of Microcystis species, the mechanisms that determine bloom dynamics and termination have not been studied sufficiently (27). Recent observations have shown that in addition to physical factors such as temperature and irradiation, chemical factors such as nutrients, and biological factors (predators), mortality induced by virus may be one of the important factors that control these algal blooms (8, 23, 44).
There are a great number of viruses in natural waters, in both marine and freshwater environments (5), and it is suspected that a large proportion of these viruses are infectious for bacteria or cyanobacteria (30). The first isolation of freshwater cyanophages was reported about 40 years ago, and during the following two decades numerous cyanophage strains were isolated (2, 3, 14, 31-34). In the marine environment, phages are thought to be responsible for controlling the dynamics of the most abundant marine primary producers, Synechococcus spp. and Prochlorococcus spp. (39, 40, 42, 46). Recently, a number of cyanophage strains that infect these two marine cyanobacterial groups were isolated and intensively studied (10, 16, 38-40, 46).
Previously, a few phage strains, including SM-1 (34), SM-2 (14), and MA 1 (29), were reported to be lytic for M. aeruginosa; however, the M. aeruginosa NRC-1 strain reported to be sensitive to SM-1 and SM-2 was later found to be a Synechococcus strain, so SM-1 and SM-2 are phages that infect Synechococcus sp. This confusion in phage identification was caused by misidentification (classification) of the host cyanobacterium (40). Phlips et al. (29) reported isolation of a lytic agent that formed plaques on lawns of an M. aeruginosa strain; however, the agent was not identified. Thus, the cyanophages that infect M. aeruginosa have not been characterized or cultured previously. The ecological impact of phages on Microcystis populations is not clear; however, reports have suggested that phage may play an important role in regulating bloom dynamics. Manage et al. (22) observed that an increase in cyanophage titers (the numbers of particles forming plaques on an M. aeruginosa lawn) was accompanied by a large decrease in the abundance of M. aeruginosa in a natural freshwater environment; Tucker and Pollard (45) recently identified two types of podovirus-like particles that inhibited growth of M. aeruginosa in natural lake samples collected during an M. aeruginosa bloom.
Here we describe the first isolation and characterization of a cyanophage that specifically infects a toxic strain of M. aeruginosa. Analysis of this host-phage system is expected to increase our understanding of the ecology and physiology of toxic cyantobacterial blooms.
MATERIALS AND METHODS
Host strains and growth conditions.
The cyanobacterial strains used are listed in Table 1. They were maintained in CB medium (19) by using a cycle consisting of 12 h of darkness and 12 h of light (ca. 40 μmol photons m−2 s−1) provided by cool white fluorescent illumination (FL40SS;, Toshiba Co., Ltd.) at 30°C.
TABLE 1.
Cyanobacterial strains used in this study and their susceptibilities to cyanophage Ma-LMM01
| Species | Straina | Origin | Susceptibilityb |
|---|---|---|---|
| Microcystis aeruginosa | NIES44 | Lake Kasumigaura (Japan) | − |
| NIES87c | Lake Kasumigaura (Japan) | − | |
| NIES90d | Lake Kawaguchi (Japan) | − | |
| NIES98 | Lake Kawaguchi (Japan) | − | |
| NIES102c,d | Lake Kasumigaura (Japan) | − | |
| NIES104 | Chiyoda-ku (Japan) | − | |
| NIES111 | Lake Kasumigaura (Japan) | − | |
| NIES112c | Lake Suwa (Japan) | − | |
| NIES298c,d | Lake Kasumigaura (Japan) | + | |
| NIES604 | Lake Kasumigaura (Japan) | − | |
| MMY 52 | Lake Mikata (Japan) | − | |
| Aphanizomenon flos-aquae | NIES81 | Ukiginuno Pond (Japan) | − |
| Nostoc sp. | PCC7120 | Iowa | − |
| Planktothrix sp. | Pla 3-2 | Lake Mikata (Japan) | − |
| Pla 7-4 | Lake Mikata (Japan) | − | |
| Synechocystis sp. | PCC6803 | California | − |
NIES strains were purchased from the National Institute for Environmental Studies, Environmental Agency, Japan.
+, cell lysis; −, no effect.
Strains used in the screening experiment for cyanophages.
Microcystis strains that produce microcystins (19).
Cyanophage isolation.
A surface water sample was collected from Lake Mikata in Fukui Prefecture, Japan, on 25 August 2003 during an M. aeruginosa bloom. After the water sample was successively filtered through 0.8-μm and 0.2-μm polycarbonate membranes (Advantec Co., Ltd), 100 μl of the filtrate was inoculated into 900-μl exponentially growing cultures of four M. aeruginosa strains (Table 1). The cultures were incubated for 1 week. Growth inhibition was observed only in the M. aeruginosa NIES298 culture inoculated with the filtrate. A clonal infectious agent was isolated from the lysed culture using three cycles of an extinction dilution procedure (25, 41).
Transmission electron microscopy (TEM).
An aliquot of the clonal phage suspension was absorbed onto carbon-coated copper grids, stained with 2% uranium acetate, and observed at 80 kV using a JEOL JEM-1010 transmission electron microscope. The virion size was estimated from the negatively stained images. Exponentially growing cells of M. aeruginosa NIES298 (1 liter) that were collected on a 0.8-μm polycarbonate membrane filter were suspended in 10 ml of fresh CB medium and then inoculated with 2.5 ml of the clonal phage suspension. M. aeruginosa cultures without inoculation served as a control. At 0 and 52 h postinoculation, 250-μl samples were removed, mixed with 50 μl CB medium containing 6% Agarose-LM (Nacalai Tesque Co., Ltd.), and solidified at 4°C. The agarose blocks with M. aeruginosa cells were cut into 1- to 2-mm cubes and fixed with 1% glutaraldehyde for 7 days. After the cubes were washed with phosphate-buffered saline (13 mM NaH2PO4 · 2H2O, 86.8 mM Na2HPO4 · 12H2O, 85.6 mM NaCl; pH 7.4), they were fixed with 1% osmic acid at 4°C for 3 h. After three washes with phosphate-buffered saline, the cubes were dehydrated in a graded ethanol series (50 to 100%) and embedded in Quetol 653 resin (NISSHIN EM Co., Ltd.). Ultrathin sections were stained with 2% uranium acetate and 3% lead citrate and observed at 80 kV using a JEOL JEM-1010 transmission electron microscope.
Host range test.
Forty microliters of a fresh clonal phage suspension was added to 800-μl exponentially growing cultures of the hosts shown in Table 1. The cultures were incubated as described above and monitored daily for host cell lysis using optical microscopy. Cultures that were not lysed after 14 days were considered unsuitable hosts for the infecting agent.
Growth experiments.
We used a semi-one-step growth procedure as described by Sandaa et al. (37). Exponentially growing cultures of M. aeruginosa (300 ml) were inoculated with the pathogen at a multiplicity of infection (MOI) of 0.72 to 0.90. An M. aeruginosa culture inoculated with an autoclaved pathogen suspension served as the control. After inoculation, an aliquot of the cell suspension was collected from each culture every 2 h for 16 h, and the numbers of host cells and pathogens were determined by optical microscopy and the extinction dilution method (25, 41), respectively.
In addition, exponentially growing cultures of M. aeruginosa (300 ml) were inoculated with the virus at MOIs of 0 (autoclaved phage culture), 10−1, 10−3, 10−5, 10−7, and 10−9 and incubated, and the changes in host cell numbers were monitored over time using optical microscopy.
Storage.
An exponentially growing culture of M. aeruginosa was inoculated with the phage and incubated for 4 days. The resulting lysate was sequentially passed through 0.8-μm and 0.2-μm filters to remove cellular debris. The titer of the fresh lysate was determined using the extinction dilution method (25, 41). Aliquots of the lysate were stored at 4, −20, −80, and −196°C (liquid nitrogen) in the dark without cryoprotectants. After 14 days, the samples were titrated to measure the stability of the phage at each temperature (24).
Phage purification.
One liter of lysate was mixed with 20 ml chloroform and 40 g NaCl, mixed for 30 min at 30°C, and then filtered through a 1-μm polytetrafluoroethylene membrane filter (Advantec Co., Ltd). The filtrate was mixed with 10% (wt/vol) polyethylene glycol 6000 (Nacalai Tesque Co., Ltd.), incubated overnight at 4°C, and centrifuged (7,000 × g, 4°C, 40 min). The precipitate was resuspended in 5 ml SM buffer (50 mM Tris-HCl, 100 mM NaCl, 10 mM MgSO4 ·7H2O, 0.01% gelatin), and 5 ml chloroform was added. After vigorous vortexing, the suspension was centrifuged (7,500 × g, 4°C, 20 min), and the aqueous layer was layered on a CsCl step gradient (1.45, 1.50, and 1.70 g ml−1) in an SW40Ti ultracentrifugation tube (Beckman, Inc.) by using the method described previously (36). The gradient was centrifuged using an SW40Ti rotor (Beckman, Inc.) at 111,000 × g and 15°C for 1 h. The concentrated phage band was collected using a 26-gauge needle. The resultant phage suspension was dialyzed in 500 ml of SM buffer at 4°C for 3 h.
Genome extraction.
Twenty microliters of a proteinase K solution (1 mg ml−1; Wako Pure Chemical Industries, Ltd.), 12.5 μl of a sodium dodecyl sulfate (SDS) solution (20%), and 20 μl of 0.5 M EDTA were added to a 500-μl dialyzed phage suspension and incubated at 55°C for 1.5 h. After incubation, an equal volume of phenol-chloroform-isoamyl alcohol (25:24:1) was added to the phage suspension, and the suspension was gently vortexed and centrifuged (17,860 × g, 4°C, 10 min); then the aqueous layer was removed. This procedure was performed twice. An equal volume of chloroform was added to the resultant aqueous layer. After vigorous vortexing, the preparation was centrifuged at 17,860 × g and 4°C for 10 min. Finally, the aqueous layer was removed and dialyzed against 500 ml of TE buffer (10 mM Tris-HCl, 1 mM EDTA; pH 8.0) at 4°C for 3 h. This dialyzed DNA suspension was stored at 4°C.
PFGE and enzyme treatment.
The phage genome size was estimated by pulsed-field gel electrophoresis (PFGE). An aliquot of a phage suspension was filtered through a 0.2-μm polycarbonate membrane (Advantec Co., Ltd). The filtrate was resuspended in 800 μl of 1% Agarose-LM (Nacalai Tesque Co., Ltd.), dispensed into plug molds, and solidified. The plugs were punched out of the molds into a small volume of digestion buffer containing 250 mM EDTA, 1% SDS, and 1 mg ml−1 proteinase K (Wako Pure Chemical Industries, Ltd.) and then incubated at 50°C overnight. The digestion buffer was decanted, and the plugs were washed eight times for 1 h in TE buffer (stored at 4°C). The plugs were placed in wells of 1.2% SeaKem Gold agarose (FMC Bioproducts) in 0.5× TBE gel buffer (90 mM Tris-borate, 1 mM EDTA; pH 8.0) and overlaid with molten 0.5% Agarose-LM. Agarose plugs containing lambda phage concatemers (Promega Co., Ltd.) were used as the molecular weight standards. The samples were electrophoresed using a Gene Navigator system (Amersham Biosciences) at 120 V with pulse ramps from 10 s to 40 s at 14°C for 39 h in 0.5× TBE tank buffer (45 mM Tris-borate, 1 mM EDTA; pH 8.0). Following electrophoresis nucleic acids were visualized by staining for 1 h with SYBR Gold (Molecular Probes Inc., Eugene, OR).
Using the manufacturers' recommendations, we tested the sensitivity of the phage nucleic acid to RNase A (0.01 ng μl−1; 37°C for 1 h; Nippon Gene Co., Ltd.), DNase I (0.02 ng μl−1; 37°C for 1 h; Promega Co., Ltd.), and the following 14 restriction enzymes, which were incubated for 16 h: SpeI (0.5 U μl−1; 37°C; TOYOBO Co., Ltd.), XhoI (0.45 U μl−1; 37°C; TOYOBO Co., Ltd.), XbaI (0.5 U μl−1; 37°C; Roche Molecular Biochemicals), BamHI (0.6 U μl−1; 37°C; TOYOBO Co., Ltd.), EcoRI (0.5 U μl−1; 37°C; TOYOBO Co., Ltd.), EcoRV (0.6 U μl−1; 37°C; TOYOBO Co., Ltd.), HincII (0.5 U μl−1; 37°C; TOYOBO Co., Ltd.), HindIII (0.5 U μl−1; 37°C; Nippon Gene Co., Ltd.), NotI (0.5 U μl−1; 37°C; New England Biolabs Inc.), PstI (0.5 U μl−1; 37°C; TOYOBO Co., Ltd.), SacI (0.5 U μl−1; 37°C; TOYOBO Co., Ltd.), SalI (0.5 U μl−1; 37°C; TOYOBO Co., Ltd.), ScaI (0.5 U μl−1; 37°C; TOYOBO Co., Ltd.), and SmaI (0.6 U μl−1; 30°C; TOYOBO Co., Ltd.). To investigate whether the genome is linear or circular, the phage nucleic acid was heat treated (100°C for 5 min) or treated with Bal31 nuclease (0.1 U μl−1; 30°C for 15 min; TOYOBO Co., Ltd.). The samples were electrophoresed in 1% (wt/vol) Agarose S (Nippon Gene Co., Ltd.). Following electrophoresis, nucleic acids were visualized as described above.
Analysis of phage proteins.
A phage suspension was mixed with 2 volumes of Laemmli sample buffer (62.5 mM Tris-HCl [pH 6.8], 5% 2-mercaptoethanol, 2% SDS, 25% glycerol, 0.01% bromphenol blue) and boiled for 5 min, and the proteins were separated by SDS-polyacrylamide gel electrophoresis (80 by 100 by 1.0 mm; 15% polyacrylamide gel; 200 V) using a Mini-PROTEAN 3 cell system (Bio-Rad Co., Ltd.). Proteins were visualized using Coomassie brilliant blue or the silver stain method. Protein molecular mass standards (Bio-Rad Co., Ltd.) with molecular masses ranging from 10 to 250 kDa were used.
Genome sequencing and phylogenic analysis.
Phage DNA extracted from CsCl-purified virions was digested with HincII or physically sheared by using a Hydroshear (Genomic Solutions, Ltd. Cambridgeshire, United Kingdom). The DNA fragments were ligated into the pUC118/HincII vector, and the plasmids were transformed into ElectroMAX DH10B competent cells (Invitrogen Corp., Carlsbad, CA). Sequencing was performed using the dideoxy method with a 3730xl DNA analyzer (Applied Biosystems). Genome sequences were assembled into contiguous sections using phred/phrap systems (12, 13) (finishing and annotation procedures are under way). Open reading frames (ORFs) were identified using the NCBI ORF finder (http://www.ncbi.nlm.nih.gov/gorf/gorf.html). The protein-encoding genes were predicted using GeneMark (6). Similarity analyses of the ORF sequences were performed using BLASTX (4).
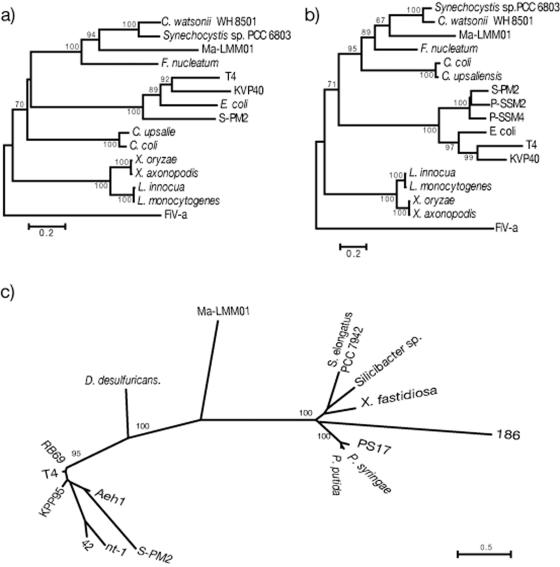
We found amino acid sequences in the conserved regions of three ORFs that probably code for the essential phage proteins, and these sequences were phylogenetically analyzed. Phylogenetic trees were constructed using MEGA 3 with the Jones-Taylor-Thornton matrix (JTT model) (17) and the neighbor-joining method (35). The DNA sequences used to construct the phylogenetic trees are indicated below (see Fig. 6).
FIG. 6.
Phylogenetic neighbor-joining trees calculated from confidently aligned regions of the amino acid sequences of ribonucleotide reductase alpha-subunit (a) and beta-subunit (b) and putative sheath protein (c) of Ma-LMM01. In panels a and b, the corresponding amino acid sequences of Feldmannia irregularis virus a (FiV-a) were used as outgroups. The phylogenetic tree for the putative sheath protein (c) is unrooted due to the lack of suitable outgroup sequences. Nodes with bootstrap values less than 70% were collapsed. Amino acid sequences of the following organisms were used in the phylogenetic analyses (accession numbers are NCBI accession numbers unless indicated otherwise): for the ribonucleotide reductase alpha-subunit, Synechocystis sp. strain PCC6803 (accession number NP_441654), Crocosphaera watsonii WH8501 (ZP_00179687), Fusobacterium nucleatum subsp. nucleatum ATCC 25586 (ZP_00144798), Listeria monocytogenes EGD-e (NP_465679), Listeria innocua Clip11262 (NP_471591), Xanthomonas oryzae pv. oryzae KACC10331 (YP_199113), Xanthomonas axonopodis pv. citri strain 306 (AAM38910), Campylobacter upsaliensis RM3195 (ZP_00371289), Campylobacter coli RM2228 (ZP_00368144), Escherichia coli (5R1R_C), enterobacterial phage T4 (AAD42621), bacteriophage S-PM2 (CAF34215), bacteriophage KVP40 (AAQ64346), and Feldmannia irregularis virus a (AAR26844); for the ribonucleotide reductase beta-subunit, Synechocystis sp. strain PCC 6803 (NP_443040), Crocosphaera watsonii WH8501 (ZP_00178432), Fusobacterium nucleatum subsp. nucleatum ATCC 25586 (NP_603013), Listeria monocytogenes EGD-e (NP_465678), Listeria innocua Clip11262 (NP_471590), Xanthomonas oryzae pv. oryzae KACC10331(YP_199114), Xanthomonas axonopodis pv. citri strain 306 (AAM38909), Campylobacter upsaliensis RM3195 (ZP_00371674), Campylobacter coli RM2228 (ZP_00367491), Escherichia coli (1MXR_B), enterobacterial phage T4 (AAD42624), bacteriophage S-PM2 (CAF34216), cyanophage P-SSM2 (AAW48101), cyanophage P-SSM4 (AAW50173), bacteriophage KVP40 (AAQ64347), and Feldmannia irregularis virus a (AAR2684); and for sheath protein and sheath-like protein, Xylella fastidiosa Dixon (ZP_00039823), Pseudomonas syringae pv. tomato strain DC3000 (NP_793179), Pseudomonas putida KT2440 (NP_745203), Desulfovibrio desulfuricans G20 (ZP_00130650), Synechococcus elongatus PCC 7942 (ZP_00163909), Silicibacter sp. strain TM1040 (ZP_00339531), bacteriophage PS17 (BAA05467), enterobacterial phage RB69 (AAP76079), bacteriophage S-PM2 (CAF34166), bacteriophage Aeh1 (AAQ17879), enterobacterial phage T4 (GKBPT4), Vibrio natriegens bacteriophage nt-1 (AAG02027), Burkholderia cepacia bacteriophage 42 (42) (AAG02028), enterobacterial phage 186 (P13332), and bacteriophage KPP95 (AAS46616). The amino acid sequences of Ma-LMM01 ORFs encoding the ribonucleotide reductase alpha- and beta-subunits and putative sheath protein are available from the DDBJ (accession numbers AB242259, AB242260, and AB242261, respectively).
RESULTS AND DISCUSSION
Physical properties of the phage.
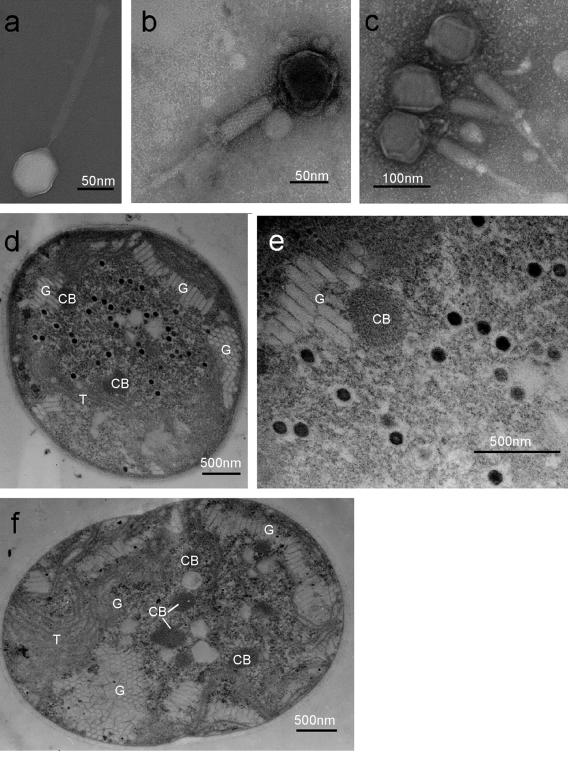
TEM images of the negatively stained phage showed that two morphotypes of phage-like particles coexisted: particles with a polyhedral head and an elongated tail (Fig. 1a) and particles with a polyhedral head and a contracted tail (Fig. 1b and c). The head was 86 nm in diameter, and the tail complex consisted of a central tube (width, 9 nm) and a contractile sheath (width, 24 nm; length, 209 nm) that contracted to a length of 90 nm. A multilayer collar and a base plate were occasionally observed (data not shown). All known cyanophages isolated so far belong to one of three tailed bacteriophage families, the Myoviridae, the Siphoviridae, or the Podoviridae. Myoviruses have a contractile tail connected to an icosahedral head by a neck; siphoviruses have a long noncontractile tail; and podoviruses have a short noncontractile tail (10, 16, 38, 40). Of the three tailed bacteriophage families, the Myoviridae includes the majority of cyanophages isolated from marine waters (16, 40). The phage described here lyses M. aeruginosa and has an icosahedral head and a contractile tail, which is the distinctive morphological feature of the Myoviridae (Fig. 1a and b). Based on the morphological features, this pathogen was considered a member of the family Myoviridae and was apparently distinct from the phage-like particles that inhibit the growth of M. aeruginosa described by Tucker and Pollard (45). The head size and tail length of the new pathogen are similar to the head size and tail length of T4-like phage S-PM2 that infects the marine cyanobacterium Synechococcus sp., which has an isometric head (85 nm) and a long tail (180 nm) (16). In contrast, T4 phage, the most intensively studied myovirus that infects Escherichia coli, has a prolate icosahedral head (78 by 111 nm) and a significantly shorter tail (113 nm) (43). The other noticeable morphological features of the T4-like phages are tail fibers and whisker fibers; many T4-like phages have six long tail fibers carried by a base plate at the end of the contractile tail held in a folded configuration by whisker fibers extending from the collar (43). Cyanophage strains P-SSM2 and P-SSM4, which infect the typical cyanobacterium Prochlorococcus strains, also have tail fibers (16, 38). In contrast, no fiber-like structure was observed here using TEM, which distinguished our strain from T4-like phages or other myoviruses infecting cyanophages (cyanomyoviruses).
FIG. 1.
Transmission electron micrographs of cyanophage Ma-LMM01 and its host, M. aeruginosa NIES298. (a) Negatively stained virion of Ma-LMM01 with an extended tail; (b) negatively stained virion of Ma-LMM01 with a contracted tail; (c) negatively stained virions of Ma-LMM01 with a contracted tail that were purified using CsCl step gradient ultracentrifugation; (d) thin section of an M. aeruginosa cell 52 h after inoculation with Ma-LMM01; (e) higher magnification of the Ma-LMM01 particles in panel d; (f) thin section of a healthy cell of M. aeruginosa. CB, carboxysome; G, gas vesicle; T, thylakoid.
Thin sections of M. aeruginosa cells obtained 52 h after inoculation with the putative pathogen revealed that there had been propagation of intracellular phage-like particles (Fig. 1d and e). In contrast, no virus was observed in healthy cells (Fig. 1f). Serial transfer of the lysate into M. aeruginosa cultures resulted in cell lysis (data not shown), and lytic activity was eliminated by heat treatment at 100°C for 20 min (data not shown). These results fulfilled Koch's postulates; hence, we concluded that the phage-like particles found in the inoculated cells and in the cyanobacterial lysates were particles of a cyanophage lytic for M. aeruginosa. This cyanophage was designated M. aeruginosa Lake Mikata Myoviridae 01 (Ma-LMM01) according to the nomenclature for cyanophages proposed by Suttle (40).
Host specificity.
Ma-LMM01 lysed only M. aeruginosa NIES298 and not an additional 15 cyanobacterial strains tested, including nine strains of M. aeruginosa (Table 1). Based on these results, we concluded that Ma-LMM01 is strain specific rather than species specific. Sullivan et al. (39) reported that myoviruses tend to exhibit broader host ranges with Prochlorococcus and Synechococcus strains than podoviruses or siphoviruses exhibit; in contrast, Ma-LMM01, which morphologically belongs to the family Myoviridae, has a narrow host range.
It was shown previously that the average sequence diversity of the 16S-to-23S internal transcribed spacer region in Microcystis is much higher (∼7%) than the average sequence diversity of 16S rRNA genes (<1%) (28, 47). Internal transcribed spacer analysis also showed that the toxic strain M. aeruginosa NIES298 was most closely related to the nontoxic strain M. aeruginosa NIES87 among the Microcystis strains tested in this study (data not shown). Yoshida et al. (47) found blooms of genetically diverse types of Microcystis populations, and the predominant genotype changed during a bloom. From this we concluded that there are probably a number of distinct host-phage systems in M. aeruginosa blooms and that they may be responsible for the genetic diversity of M. aeruginosa. Here we describe one of the various combinations of M. aeruginosa strains and phage strains; hence, to discuss the relationship between toxicity and phage sensitivity, further analysis using a larger number of host and phage strains is required.
Growth characteristics.
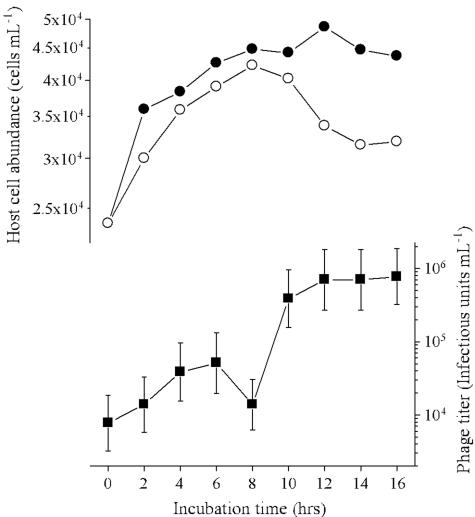
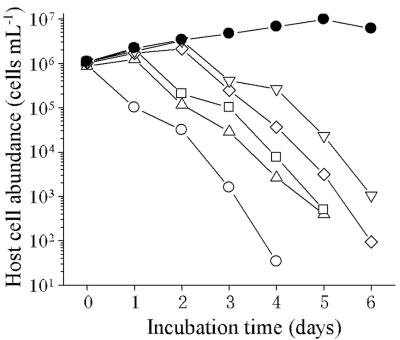
To perform the one-step growth experiments for Ma-LMM01, an MOI greater than 2 was used initially; however, we found that this resulted in a small decrease in the number of host cells (data not shown). Therefore, semi-one-step growth experiments were carried out with MOIs less than 1. The results obtained with an initial MOI of 0.72 are shown in Fig. 2, and in these conditions the latent period was estimated to be 8 to 12 h with 82 infectious units cell−1. In repetitive semi-one-step growth experiments with MOIs ranging from 0.72 to 0.90, decreases in the numbers of host cells were observed 6 to 12 h postinoculation, and the burst size was estimated to be 50 to 120 infectious units cell−1 based on the decreases in the number of host cells and increases in the viral titer. Cell lysis was not complete (Fig. 2). Because the initial MOI was less than 1, the burst size may have been underestimated even though when the MOI was less than 0.1 almost complete lysis was observed (Fig. 3). Further study is required to explain the mechanism that caused these differences during host cell lysis.
FIG. 2.
Changes in the number of M. aeruginosa cells (○) and the titer of Ma-LMM01 (▪) in a semi-one-step growth experiment. An exponentially growing culture of M. aeruginosa was inoculated with Ma-LMM01 at an MOI of 0.72. As a control, an M. aeruginosa culture was inoculated with an autoclaved phage suspension (•).
FIG. 3.
Changes in the cell densities of M. aeruginosa cultures infected with Ma-LMM01 at various MOIs. Exponentially growing cultures of M. aeruginosa were inoculated with Ma-LMM01 at MOIs of 10−1 (○), 10−3 (▵), 10−5 (□), 10−7 (◊), and 10−9 (▿). As a control, an M. aeruginosa culture was inoculated with an autoclaved phage suspension (•).
Storage.
In the storage test, no significant loss of infectivity was detected after 14 days of storage at −80 and −196°C without addition of cryoprotectants; however, storage at −20°C was lethal to Ma-LMM01. Cyanophage Ma-LMM01 is the first phage strain that infects the toxic cyanobacterium M. aeruginosa that was stored for future use at the National Institute of Technology and Evaluation (accession number FERM P-ABP-71).
Genome.
After CsCl density centrifugation, Ma-LMM01 was purified as a single band at 1.45 g ml−1. TEM showed that most of the concentrated virions had a contracted sheath (Fig. 1c). The nucleic acid that was extracted was sensitive to DNase I (data not shown), Bal31 nuclease (data not shown), and all 14 of the restriction enzymes tested (Fig. 4) but not to RNase A and heat treatment (data not shown). The size was estimated to be ∼160 kbp using PFGE (data not shown). Based on these results, we concluded that the Ma-LMM01 genome consists of linear double-stranded DNA that is ∼160 kbp long; thus, in terms of length it is similar to other myovirus genomes, such as those of S-PM2 (194 kbp) (16), P-SSM2 (252 kbp) (38), P-SSM4 (178 kbp) (38), and T4 (169 kbp) (1), but is much larger than those of Mu (36.7kbp) (20), P1 (94.8 kbp) (20), and P2 (33.5 kbp) (11).
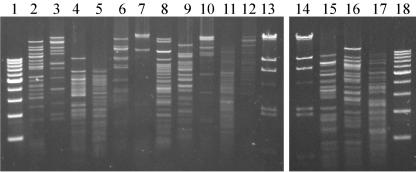
FIG. 4.
Ma-LMM01 genomic DNA digested with various restriction enzymes. Lanes 1 and 18, 1-kb ladder marker; lane 2, BamHI; lane 3, EcoRI; lane 4, EcoRV; lane 5, HincII; lane 6, HindIII; lane 7, NotI; lane 8, PstI; lane 9, SacI; lane 10, SalI; lane 11, ScaI; lane 12, SmaI; lanes 13 and 14, lambda/HindIII marker; lane 15, SpeI; lane 16, XhoI; lane 17, XbaI.
T4 phage often have a genome that contains methylated bases, and methylated DNA is highly resistant to a host's restriction enzymes (7). The genomes of cyanomyoviruses (S-BM1, S-PM1, and S-WHM1) that infect marine Synechococcus strains are resistant to various restriction enzymes (7). In contrast, the Ma-LMM01 genome was sensitive to various restriction enzymes, suggesting that it has a low level of methylation. Similar to T7 phage and T3 phage (7, 15), Ma-LMM01 may synthesize inhibitors of restriction-modification systems to protect its genomic DNA.
Proteins.
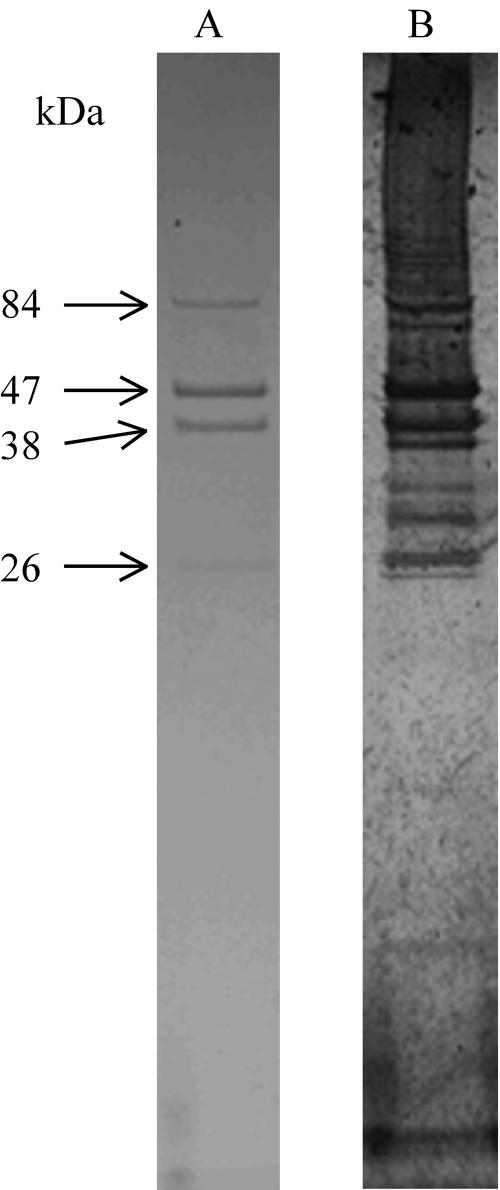
Using SDS-polyacrylamide gel electrophoresis, we found that Ma-LMM01 contains four major polypeptides (84, 47, 38, and 26 kDa) and seven minor polypeptides that were visualized using the more sensitive silver staining method (Fig. 5).
FIG. 5.
Major structural proteins of Ma-LMM01 stained by Coomassie brilliant blue G (lane A) or using the silver staining method (lane B).
Phylogenetic analysis.
We found three ORFs that may be genes coding for essential phage proteins. Two of these ORFs code for the alpha- and beta-subunits of ribonucleotide reductase and exhibit significant similarity to ORFs of Synechocystis sp. strain PCC 6803 (E-values, 0.0 and 3.00E-92, respectively). Ribonucleotide reductase is an essential enzyme that generates precursors of DNA by converting ribonucleotides into deoxyribonucleotides (18). Ribonucleotide reductase genes are commonly found in lytic phages but not in lysogenic phages; hence, this DNA metabolic process is considered essential for rapid replication of lytic phages (10). Ma-LMM01 formed clear plaques on host lawns, and no turbid plaques were observed (data not shown), showing that the phage is lytic, although integrase and transposase genes were also found in the genome (data not shown). We concluded that Ma-LMM01 is a lytic phage similar to P-SSP7 that has an integrase that may be responsible for lysogeny (38). Further analysis of transcription and translation of the Ma-LMM01 genome is necessary to assess the possibility of lysogenic activity. Phylogenetic trees were constructed from the deduced amino acid sequences of the alpha- and beta-subunits of the ribonucleotide reductase, and the cyanophage described here formed a sister group with cyanobacteria, which was supported by bootstrap values of 93 and 71%, respectively (Fig. 6a and b). In contrast to Ma-LMM01, cyanophages S-PM2, P-SSM2, and P-SSM4 formed a monophyletic group with the T4-like phages based on the ribonucleotide reductase alpha- and beta-subunits (Fig. 6a and b) (38). Therefore, the origins of this key enzyme are different for different cyanomyoviruses.
The other ORF identified in this study may code for a sheath protein specific to myoviruses. In T4, gene 18 codes for the tail sheath protein, and the sheath structure consists of 144 molecules of the gp18 product (43). The deduced amino acid sequence of the Ma-LMM01 sheath protein exhibited similarity to the Xylella fastidiosa DIXON prophage sheath-like protein sequence (E-value, 6.00E-08) but did not form a monophyletic group with the sequences of any other T4-like phages, temperate phages, or prophage-like sequences (Fig. 6c). This suggests that this cyanophage is distinct from the other myoviruses that have been described or that the sheath-like protein of Ma-LMM01 has additional functions in the replication process.
Conclusion.
Although recently viruses have been considered an important component of the aquatic ecosystem (5, 8, 23, 40, 42, 44), the viral impact on Microcystis blooms is unknown. Here we isolated a myovirus that infects a toxin-producing strain of M. aeruginosa and characterized it. We believe that information about this host-phage system will contribute to our understanding of the ecology of Microcystis blooms and the genetics of cyanophages and that it is possible that this system could used as a control for toxic cyanobacterial blooms.
Ma-LMM01 was isolated from a natural freshwater source in which M. aeruginosa is the dominate organism. It is therefore probable there is a close ecological interaction between the phage and natural M. aeruginosa blooms. M. aeruginosa often forms colonies in mucilage in the natural environment, whereas in this study its ability to form colonies was lost by serial transfer to fresh media during subcultivation (28). Therefore, the in vitro culture of M. aeruginosa NIES298 used in this study does not form colonies and hence does not necessarily reflect the interaction between the natural host and the phage. Thus, we need to understand how susceptible M. aeruginosa colonial cells are to phage infection.
The genome of this phage is not completely understood; shotgun sequencing is now under way, and we have not identified all of the genes yet. However, in this study we phylogenetically analyzed three genes that show the distinctive features of this phage, and in the future we will fully sequence and perform translation experiments. Since several genes coding for putative integrases or transposases were found (data not shown), analysis of the lysogenic activity of Ma-LMM01 will be a focus of future study.
To improve management of water sources, this phage could be used as a biological control agent to control toxic blooms of cyanobacteria (40). However, this will require studies of the community composition of Microcystis, host resistance mechanisms, the host ranges of cyanophages, the ecological impact of cyanophages in each stage of Microcystis blooms, and the annual life cycle of cyanophages. These studies will be essential to assess the practical application of phages in water management. Expansion of the collection of phages that infect various strains of Microcystis will also be required.
Acknowledgments
This study was supported in part by the Industrial Technology Research Grant Program of the New Energy and Industrial Technology Development Organization of Japan (NEDO).
REFERENCES
- 1.Ackermann, H.-W., and M. S. DuBow. 2000. Family Myoviridae, p. 69-84. In M. H. V. V. Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carsten, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy, classification and nomenclature of viruses, seventh report. Academic Press, San Diego, Calif.
- 2.Adolph, K. W., and R. Haselkorn. 1973. Isolation and characterization of a virus infecting a blue-green alga of the genus Synechococcus. Virology 54:230-236. [DOI] [PubMed] [Google Scholar]
- 3.Adolph, K. W., and R. Haselkorn. 1971. Isolation and characterization of a virus infecting the blue-green alga Nostoc muscorum. Virology 46:200-208. [DOI] [PubMed] [Google Scholar]
- 4.Altschul, S. F., W. Gish, W. Miller, E. W. Myers, and D. J. Lipman. 1990. Basic local alignment search tool. J. Mol. Biol. 215:403-410. [DOI] [PubMed] [Google Scholar]
- 5.Bergh, Ø., K. Y. Børsheim, G. Bratbak, and M. Heldal. 1989. High abundance of viruses found in aquatic environments. Nature 340:467-468. [DOI] [PubMed] [Google Scholar]
- 6.Besemer, J., A. Lomsadze, and M. Borodovsky. 2001. GeneMarkS: a self-training method for prediction of gene starts in microbial genomes. Implications for finding sequence motifs in regulatory regions. Nucleic Acids Res. 29:2607-2618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bickle, T. A., and D. H. Kruger. 1993. Biology of DNA restriction. Microbiol. Rev. 57:434-450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brussaard, C. P. 2004. Viral control of phytoplankton populations—a review. J. Eukaryot. Microbiol. 51:125-138. [DOI] [PubMed] [Google Scholar]
- 9.Carmichael, W. W. 1995. Toxic Microcystis and the environment, p. 1-11. InM. F. Watanabe, K. Harada, W. W. Carmichael, and H. Fujiki (ed.), Toxic Microcystis. CRC Press, Boca Raton, Fla.
- 10.Chen, F., and J. Lu. 2002. Genomic sequence and evolution of marine cyanophage P60: a new insight on lytic and lysogenic phages. Appl. Environ. Microbiol. 68:2589-2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Christie, G. E., L. M. Temple, B. A. Bartlett, and T. S. Goodwin. 2002. Programmed translational frameshift in the bacteriophage P2 FETUD tail gene operon. J. Bacteriol. 184:6522-6531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ewing, B., and P. Green. 1998. Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res. 8:186-194. [PubMed] [Google Scholar]
- 13.Ewing, B., L. Hillier, M. C. Wendl, and P. Green. 1998. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 8:175-185. [DOI] [PubMed] [Google Scholar]
- 14.Fox, J. A., S. J. Booth, and E. L. Martin. 1976. Cyanophage SM-2: a new blue-green algal virus. Virology 73:557-560. [DOI] [PubMed] [Google Scholar]
- 15.Garcia, L. R., and I. J. Molineux. 1999. Translocation and specific cleavage of bacteriophage T7 DNA in vivo by EcoKI. Proc. Natl. Acad. Sci. USA 96:12430-12435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hambly, E., F. Tetart, C. Desplats, W. H. Wilson, H. M. Krisch, and N. H. Mann. 2001. A conserved genetic module that encodes the major virion components in both the coliphage T4 and the marine cyanophage S-PM2. Proc. Natl. Acad. Sci. USA 98:11411-11416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jones, D. T., W. R. Taylor, and J. M. Thornton. 1992. The rapid generation of mutation data matrices from protein sequences. Comput. Appl. Biosci. 8:275-282. [DOI] [PubMed] [Google Scholar]
- 18.Jordan, A., and P. Reichard. 1998. Ribonucleotide reductases. Annu. Rev. Biochem. 67:71-98. [DOI] [PubMed] [Google Scholar]
- 19.Kasai, F., M. Kawachi, M. Erata, and M. M. Watanabe. 2004. NIES-collection list of strains, 7th ed. The National Institution for Environmental Studies, Tsukuba, Japan.
- 20.Lobocka, M. B., D. J. Rose, G. Plunkett III, M. Rusin, A. Samojedny, H. Lehnherr, M. B. Yarmolinsky, and F. R. Blattner. 2004. Genome of bacteriophage P1. J. Bacteriol. 186:7032-7068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.MacKintosh, C., K. A. Beattie, S. Klumpp, P. Cohen, and G. A. Codd. 1990. Cyanobacterial microcystin-LR is potent and specific inhibitor of protein phosphatase 1 and 2A from both mammals and higher plants. FEBS Lett. 264:187-192. [DOI] [PubMed] [Google Scholar]
- 22.Manage, P., Z. Kawabata, and S. Nakano. 1999. Seasonal changes in densities of cyanophage infectious to Microcystis aeruginosa in a hypereutrophic pond. Hydrobiologia 411:211-216. [Google Scholar]
- 23.Nagasaki, K., Y. Tomaru, K. Nakanishi, N. Hata, N. Katanozaka, and M. Yamaguchi. 2004. Dynamics of Heterocapsa circularisquama (Dinophyceae) and its viruses in Ago Bay, Japan. Aquat. Microb. Ecol. 34:219-226. [Google Scholar]
- 24.Nagasaki, K., and M. Yamaguchi. 1999. Cryopreservation of a virus (HaV) infecting a harmful bloom causing microalga, Heterosigma akashiwo (Raphidophyceae). Fish Sci. 65:319-320. [Google Scholar]
- 25.Nagasaki, K., and M. Yamaguchi. 1997. Isolation of a virus infectious to the harmful bloom causing microalga Heterosigma akashiwo (Raphidophyceae). Aquat. Microb. Ecol. 13:135-140. [Google Scholar]
- 26.Nishiwaki-Matsushima, R., T. Ohta, S. Nishiwaki, M. Suganuma, K. Kohyama, T. Ishikawa, W. W. Carmichael, and H. Fujiki. 1992. Liver tumor promotion by the cyanobacterial cyclic peptide toxin microcystin-LR. J. Cancer Res. Clin. Oncol. 118:420-424. [DOI] [PubMed] [Google Scholar]
- 27.Oliver, R. L., and G. G. Ganf. 2000. Fresh water blooms, p. 149-194. In B. A. Whitton and M. Potts (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 28.Otsuka, S., S. Suda, S. Shibata, H. Oyaizu, S. Matsumoto, and M. M. Watanabe. 2001. A proposal for the unification of five species of the cyanobacterial genus Microcystis Kutzing ex Lemmermann 1907 under the rules of the Bacteriological Code. Int. J. Syst. Evol. Microbiol. 51:873-879. [DOI] [PubMed] [Google Scholar]
- 29.Phlips, E. J., R. L. Monegue, and F. J. Aldridge. 1990. Cyanophages which impact bloom-forming cyanobacteria. J. Aquat. Plant Manag. 28:92-97. [Google Scholar]
- 30.Proctor, L. M., and J. A. Fuhrman. 1990. Viral mortality of marine cyanobacteria and bacteria. Nature 343:60-62. [Google Scholar]
- 31.Safferman, R. S., T. O. Diener, P. R. Desjardins, and M. E. Morris. 1972. Isolation and characterization of AS-1, a phycovirus infecting the blue-green algae, Anacystis nidulans and Synechococcus cedrorum. Virology 47:105-113. [DOI] [PubMed] [Google Scholar]
- 32.Safferman, R. S., and M. E. Morris. 1963. Algal virus: isolation. Science 140:679-680. [DOI] [PubMed] [Google Scholar]
- 33.Safferman, R. S., and M. E. Morris. 1964. Growth characteristics of the blue-green algal virus LPP-1. J. Bacteriol. 88:771-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Safferman, R. S., I. R. Schneider, R. L. Steere, M. E. Morris, and T. O. Diene. 1969. Phycovirus SM-1: a virus infecting unicellular blue-green algae. Virology 37:386-395. [DOI] [PubMed] [Google Scholar]
- 35.Saitou, N., and M. Nei. 1987. The neighbor joining method. A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory, Cold Spring Harbor, N.Y.
- 37.Sandaa, R. A., M. Heldal, T. Castberg, R. Thyrhaug, and G. Bratbak. 2001. Isolation and characterization of two viruses with large genome size infecting Chrysochromulina ericina (Prymnesiophyceae) and Pyramimonas orientalis (Prasinophyceae). Virology 290:272-280. [DOI] [PubMed] [Google Scholar]
- 38.Sullivan, M. B., M. L. Coleman, P. Weigele, F. Rohwer, and S. W. Chisholm. 2005. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. 3:e144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sullivan, M. B., J. B. Waterbury, and S. W. Chisholm. 2003. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature 424:1047-1051. [DOI] [PubMed] [Google Scholar]
- 40.Suttle, C. A. 2000. Cyanophages and their role in the ecology of cyanobacteria, p. 563-589. In B. A. Whitton and M. Potts (ed.), The ecology of cyanobacteria. Kluwer Academic Publishers, Dordrecht, The Netherlands.
- 41.Suttle, C. A. 1993. Enumeration and isolation of viruses, p. 121-137. In P. F. Kemp, B. Sherr, E. Sherr, and J. J. Cole (ed.), Handbook of methods in aquatic microbial ecology. Lewis Publishers, Boca Raton, Fla.
- 42.Suttle, C. A., and A. M. Chan. 1993. Marine cyanophages infecting oceanic and coastal strains of Synechococcus: abundance, morphology, cross-infectivity and growth characteristics. Mar. Ecol. Prog. Ser. 92:99-109. [Google Scholar]
- 43.Tetart, F., C. Desplats, M. Kutateladze, C. Monod, H.-W. Ackermann, and H. M. Krisch. 2001. Phylogeny of the major head and tail genes of the wide-ranging T4-typebacteriophages. J. Bacteriol. 183:358-366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tomaru, Y., K. Tarutani, M. Yamaguchi, and K. Nagasaki. 2004. Quantitative and qualitative impacts of viral infection on a Heterosigma akashiwo (Raphidophyceae) bloom in Hiroshima Bay, Japan. Aquat. Microb. Ecol. 34:227-238. [Google Scholar]
- 45.Tucker, S., and P. Pollard. 2005. Identification of cyanophage Ma-LBP and infection of the cyanobacterium Microcystis aeruginosa from an Australian subtropical lake by the virus. Appl. Environ. Microbiol. 71:629-635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wilson, W., I. Joint, N. Carr, and N. Mann. 1993. Isolation and molecular characterization of five marine cyanophages propagated on Synechococcus sp. strain WH7803. Appl. Environ. Microbiol. 59:3736-3743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yoshida, M., T. Yoshida, Y. Takashima, R. Kondo, and S. Hiroishi. 2005. Genetic diversity of the toxic cyanobacterium Microcystis in Lake Mikata. Environ. Toxicol. 20:229-234. [DOI] [PubMed] [Google Scholar]
- 48.Yoshizawa, S., R. Matsushima, M. F. Watanabe, K. Harada, A. Ichihara, W. W. Carmichael, and H. Fujiki. 1990. Inhibition of protein phosphatases by microcystins and nodularin associated with hepatotoxicity. J. Cancer Res. Clin. Oncol. 116:609-614. [DOI] [PubMed] [Google Scholar]