Abstract
We report the isolation and establishment of Rickettsia felis in the C6/36 cell line. Rickettsial growth was intense, always with 90 to 100% of cells being infected after few weeks. The rickettsial isolate was confirmed by testing infected cells by PCR and sequencing fragments of three major Rickettsia genes (gltA, ompB, and the 17-kDa protein gene).
Rickettsia felis is a bacterium belonging to the spotted fever group of the genus Rickettsia. It was first observed by Adams et al. in 1990 (1) within midgut cells of cat fleas (Ctenocephalides felis felis) in the United Sates and was later described as a new Rickettsia species (2, 10). The organism has been detected, by molecular assays, infecting the flea C. felis felis in different countries of the world, suggesting a cosmopolitan distribution (5, 7, 8, 13). There are only a few reports of R. felis infecting other flea species, including Ctenocephalides canis, Pulex irritans, and Anomiopsyllus nudata (15).
R. felis has been reported as the causative agent of the flea-borne spotted fever, an emerging human rickettsiosis that has been diagnosed in Mexico, the United States, Brazil, France, Germany, and Thailand (12, 13, 14, 17). Diagnosis of human infection by R. felis has been performed by molecular assays or serological testing. The latter has been limited by the difficulties of producing R. felis antigens, since only one laboratory has been able to establish R. felis in tissue culture, using the XTC-2 cell line (10, 13). Here we report the isolation and establishment of R. felis in tissue culture, using C6/36 cells, a mosquito cell line derived from Aedes albopictus (6).
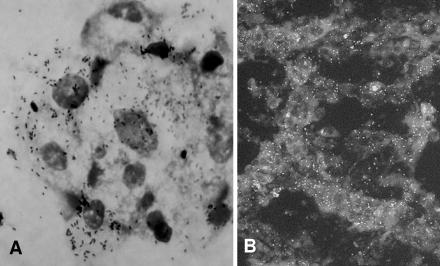
In this experiment, uninfected and infected cells were incubated at 28 and 25°C, respectively, and the level of infected cells was determined visually by Gimenez staining (3). Fleas (C. felis felis) were collected from naturally infested dogs in a farm at Pedreira, São Paulo State, Brazil, where a previous study detected up to 80% of the fleas infected with R. felis (5). In the laboratory, four pools of five live fleas were formed. Each pool was disinfected for 10 min in iodine alcohol; washed twice in sterile water; triturated in 600 μl of Leibovitz-15 medium with amphotericin B (Cultilab, Brazil), 2.5% bovine calf serum (HyClone), and antibiotics (penicillin [100 U/ml] and streptomycin [0.5 mg/ml]), and inoculated into four shell vials (≈150 μl per shell vial) containing a monolayer of C6/36 cells. After inoculation, the shell vials were centrifuged for 1 h at 700 × g and 22°C (12). Thereafter, the monolayer was washed and 1 ml of the same medium was added to each shell vial, which was incubated at 25°C. On the third day, the medium was changed to antibiotic-free medium. On the fifth day, a new change of medium was made, but the aspirated medium was centrifuged and the pellet was examined by Gimenez staining. Typical Rickettsia-like organisms (Fig. 1A) were observed within cells from the four inoculated shell vials. The monolayers of these infected shell vials were harvested and inoculated into a 25-cm2 flask containing an uninfected monolayer of C6/36 cells. When the level of infected cells reached >90%, cells were harvested and inoculated into two new 75-cm2 flasks containing uninfected C6/36 cells. When the level of infected cells reached ≈100%, cells were harvested and a portion of them was inoculated into new uninfected monolayers. The other portion was divided into aliquots to be frozen at −80°C or for the production of R. felis antigen slides to be used in further immunofluorescence assay tests, as previously described (4). Although the first three passages took 30 to 70 days for 90 to 100% of the cells to become infected, the isolate has been established in the C6/36 cell line with several subsequent passages, always reaching 90 to 100% infected cells within 15 to 30 days of incubation at 25°C.
FIG. 1.
Rickettsia felis in C6/36 cells demonstrated by Gimenez staining (A) and by immunofluorescence assay using opossum anti-Rickettsia serum and anti-opossum immunoglobulin G secondary antibody (B).
One aliquot of infected cells from the 10th passage was used for DNA extraction and PCR targeting the rickettsial gltA, ompB, and 17-kDa protein genes, as previously described (9). Fragments of 1,080, 766, and 499 bp of the gltA, ompB, and 17-kDa protein genes were sequenced. These fragments were 99.5 to 100% similar to the corresponding sequences of different strains of R. felis in GenBank (Table 1), confirming that our isolate belongs to the species R. felis. This isolate was named strain Pedreira and has been deposited as a reference strain in the Laboratory of Parasitic Diseases, Department of Preventive Veterinary Medicine, Faculty of Veterinary Medicine, University of São Paulo, São Paulo, Brazil.
TABLE 1.
Percent sequence similarities of gltA, ompB, and 17-kDa protein gene fragments of a Brazilian rickettsia strain cultured in the C6/36 cell line with other available sequences in GenBank
| Rickettsial gene | Fragment length (nucleotides) | Highest percent sequence similarities (no. of identical nucleotides/total) with available sequences in GenBank (accession no.) |
|---|---|---|
| gltA | 1,080 | 100 (1,080/1,080) with Rickettsia felis URRWXCal2 (CP000053), 100 (1,080/1,080) with Rickettsia felis California 2 (AF210692) |
| ompB | 766 | 100 (766/766) with Rickettsia felis URRWXCal2 (CP000053), 100 (748/748) with Rickettsia felis California-2 (AF210695), 99.9 (765/766) with Rickettsia felis (AF182279), 99.5 (762/766) with Rickettsia felis (AY394854) |
| 17-kDa protein gene | 499 | 100 (499/499) with Rickettsia felis URRWXCal2 (CP000053), 100 (499/499) with Rickettsia felis (AF195118), 100 (394/394) with Rickettsia felis California 2 (AF210693) |
Immunofluorescence assay slides made with strain Pedreira were tested with two opossum (Didelphis aurita) sera that had previously shown high titers (8,192 and 2,048) against R. rickettsii antigen (M. C. Horta, A. Pinter, C. E. Souza, A. M. Joppert, L. E. O. Yai, M. B. Labruna, and T. T. S. Schumaker, Abstr. 4th Int. Conf. Rickettsiae Rickettsial Dis., abstr. P.166, 2005). Only the former serum reacted with the R. felis antigen (Fig. 1B), showing a further end point titer of 256.
The present study reports the isolation and establishment of R. felis in a mosquito cell line (C6/36). Previous studies (10, 13) reported the isolation and establishment of R. felis in an amphibian cell line (XTC-2), in which cells became 90% infected after 6 days of incubation at 28°C. In the present study, we also report an efficient cell model for cultivation of R. felis, in which cells became 90 to 100% infected after as few as 15 days of incubation at 25°C. We expect that this alternative cell line for the in vitro growth of rickettsiae will facilitate the study of R. felis through the world.
Spotted fever group rickettsiae are usually in vitro cultured at temperatures above 30°C (16). Thus, in vitro growth restricted to lower temperatures seems to be just another particular feature of R. felis, since its recent genome sequencing showed several specific features, such as the presence of a plasmid and conjugative pili, reported for the first time for the genus Rickettsia (11).
Acknowledgments
This work was supported by FAPESP, São Paulo, Brazil (grants 02/10759-0 to M.C.H., 03/13872-4 to M.B.L., and 03/04728-7 to T.T.S.S), and by CNPq, Brazil (scholarship to M.B.L.).
REFERENCES
- 1.Adams, J. R., E. T. Schmidtmann, and A. F. Azad. 1990. Infection of colonized cat fleas, Ctenocephalides felis (Bouché), with a rickettsia-like microorganism. Am. J. Trop. Med. Hyg. 43:400-409. [DOI] [PubMed] [Google Scholar]
- 2.Bouyer, D. H., J. Stenos, P. C. Valdes, C. G. Moron, V. L. Popov, J. E. Zavala-Velazquez, L. D. Foil, D. R. Stothard, A. F. Azad, and D. H. Walker. 2001. Rickettsia felis: molecular characterization of a new member of the spotted fever group. Int. J. Syst. Evol. Microbiol. 51:339-347. [DOI] [PubMed] [Google Scholar]
- 3.Gimémez, D. F. 1964. Staining rickettsiae in yolk-sac cultures. Stain Technol. 39:135-140. [DOI] [PubMed] [Google Scholar]
- 4.Horta, M. C., M. B. Labruna, L. A. Sangioni, M. C. B. Vianna, S. M. Gennari, M. A. M. Galvão, C. L. Mafra, O. Vidotto, T. T. S. Schumaker, and D. H. Walker. 2004. Prevalence of antibodies to spotted fever group rickettsiae in humans and domestic animals in a Brazilian spotted fever-endemic area in the state of São Paulo, Brazil: serologic evidence for infection by Rickettsia rickettsii and another spotted fever group rickettsia. Am. J. Trop. Med. Hyg. 71:93-97. [PubMed] [Google Scholar]
- 5.Horta, M. C., A. Pinter, A. Cortez, R. M. Soares, S. M. Gennari, T. T. S. Schumaker, and M. B. Labruna. 2005. Rickettsia felis (Rickettsiales: Rickettsiaceae) in Ctenocephalides felis felis (Siphonaptera: Pulicidae) in the state of São Paulo, Brazil. Arq. Bras. Med. Vet. Zootec. 57:321-325. [Google Scholar]
- 6.Igarashi, A. 1978. Isolation of a Singh's Aedes albopictus cell clone sensitive to dengue and chikungunya viruses. J. Gen. Virol. 40:531-544. [DOI] [PubMed] [Google Scholar]
- 7.Kelly, P. J., N. Meads, A. Theobald, P. E. Fournier, and D. Raoult. 2004. Rickettsia felis, Bartonella henselae, and B. clarridgeiae, New Zealand. Emerg. Infect. Dis. 10:967-968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kenny, M. J., R. J. Birtles, M. J. Day, and S. E. Shaw. 2003. Rickettsia felis in the United Kingdon. Emerg. Infect. Dis. 9:1023-1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Labruna, M. B., J. W. McBride, D. H. Bouyer, L. M. A. Camargo, E. P. Camargo, and D. H. Walker. 2004. Molecular evidence for a spotted fever group Rickettsia species in the tick Amblyomma longirostre in Brazil. J. Med. Entomol. 41:533-537. [DOI] [PubMed] [Google Scholar]
- 10.La Scola, B., S. Meconi, F. Fenollar, J. M. Rolain, V. Roux, and D. Raoult. 2002. Emended description of Rickettsia felis (Bouyer et al. 2001), a temperature-dependent cultured bacterium. Int. J. Syst. Evol. Microbiol. 52:2035-2041. [DOI] [PubMed] [Google Scholar]
- 11.Ogata, H., P. Renesto, S. Audic, C. Robert, G. Blanc, P. E. Fournier, H. Parinello, J. M. Claverie, and D. Raoult. 2005. The genome sequence of Rickettsia felis identifies the first putative conjugative plasmid in an obligate intracellular parasite. PLOS Biol. 3:1391-1402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Parola, P., R. S. Miller, P. McDaniel, S. R. Telforf, J. M. Rolain, C. Wongsrichanalai, and D. Raoult. 2003. Emerging rickettsioses of the Thai-Myanmar border. Emerg. Infect. Dis. 9:592-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raoult, D., B. La Scola, M. Enea, P. E. Fournier, V. Roux, F. Fenollar, M. A. M. Galvão, and X. Lamballerie. 2001. A flea-associated Rickettsia pathogenic for humans. Emerg. Infect. Dis. 7:73-81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Richter, J., P. E. Fournier, J. Petridou, D. Hãussinger, and D. Raoult. 2002. Rickettsia felis infection acquired in Europe and documented by polymerase chain reaction. Emerg. Infect. Dis. 8:207-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stevenson, H. L., M. B. Labruna, J. A. Montenieri, M. Y. Kosoy, K. L. Gage, and D. H. Walker. 2005. Detection of Rickettsia felis in a New World flea species, Anomiopsyllus nudata (Siphonaptera: Ctenophthalmidae). J. Med. Entomol. 42:163-167. [DOI] [PubMed] [Google Scholar]
- 16.Weiss, E., and J. W. Moulder. 1984. The rickettsias and chlamydias, p. 687-739. In N. R. Kreig and J. G. Holt (ed.), Bergey's manual of systematic bacteriology, vol. 1. Williams & Wilkins, Baltimore, Md. [Google Scholar]
- 17.Zavala-Velázquez, J. E., J. A. Ruiz-Sosa, R. A. Sánchez-Elias, G. Becerra-Carmona, and D. H. Walker. 2000. Ricketsia felis rickettsiosis in Yucatán. Lancet 356:1079-1080. [DOI] [PubMed] [Google Scholar]