Abstract
The impact of environmental perturbation (e.g., nitrogenous fertilizers) on the dynamics of methane fluxes from soils and wetland systems is poorly understood. Results of fertilizer studies are often contradictory, even within similar ecosystems. In the present study the hypothesis of whether these contradictory results may be explained by the composition of the methane-consuming microbial community and hence whether methanotrophic diversity affects methane fluxes was investigated. To this end, rice field and forest soils were incubated in microcosms and supplemented with different nitrogenous fertilizers and methane concentrations. By labeling the methane with 13C, diversity and function could be coupled by analyses of phospholipid-derived fatty acids (PLFA) extracted from the soils at different time points during incubation. In both rice field and forest soils, the activity as well as the growth rate of methane-consuming bacteria was affected differentially. For type I methanotrophs, fertilizer application stimulated the consumption of methane and the subsequent growth, while type II methanotrophs were generally inhibited. Terminal restriction fragment length polymorphism analyses of the pmoA gene supported the PLFA results. Multivariate analyses of stable-isotope-probing PLFA profiles indicated that in forest and rice field soils, Methylocystis (type II) species were affected by fertilization. The type I methanotrophs active in forest soils (Methylomicrobium/Methylosarcina related) differed from the active species in rice field soils (Methylobacter/Methylomonas related). Our results provide a case example showing that microbial community structure indeed matters, especially when assessing and predicting the impact of environmental change on biodiversity loss and ecosystem functioning.
The loss of biodiversity from ecosystems and the potential consequences in terms of ecosystem functioning and stability have been central issues for environmental sciences in the last decade (35). Microbial communities comprise much of Earth's biodiversity and have a critical role in ecosystem functioning (42, 43) by collectively determining the biogeochemical processes that regulate the Earth system. Nevertheless, microorganisms have not been considered in the ongoing debate about global biodiversity loss and global change.
This neglect is probably due to the huge diversity and associated functional redundancy within microbial communities (42), which make it unlikely that losing a single bacterial species will result in altered ecosystem functioning. Changes in microbial community composition in concordance with changes in the functions that the microorganisms catalyze have been observed for specific groups of low diversity catalyzing a very narrow range of biogeochemical functions, such as ammonia-oxidizing bacteria (23, 44). However, in these and other cases separate community members could not be linked to the functions they catalyze, and therefore a direct link between diversity and biogeochemical functioning cannot be made. The latter is possible with bacteria that consume methane.
Methane-consuming microbes play a vital role in global warming issues, as they are the only biological sink for methane. In dry upland soils (e.g., forest and grassland) they account for approximately 6% of the global sink strength of atmospheric methane, and in wetlands they attenuate the source strength by 10 to 30% (26, 33). Methane-oxidizing bacteria (MOB) utilize methane as their sole carbon and energy source and are subdivided into types I and II on the basis of phylogeny, physiology, morphology, and biochemistry (8, 18). There are 12 recognized genera within the Alphaproteobacteria (type II MOB/MOB) and Gammaproteobacteria (type I (8, 22). A very distinct characteristic of these bacteria is the presence of specific phospholipid ester-linked fatty acids (PFLA) which differentiate them from each other (type I, C16:1ω8c and C16:1ω5t; type II, C18:1ω8c) and also from all other organisms (9). Tracing the C of methane (which these organisms use as the sole carbon and energy source) back into these PLFA by using stable isotope (13C) (6, 36) or radioisotope (14C) (3) labeling enables linkage of the biogeochemical function of these bacteria with their phylogeny and community composition. The activity of MOB is regulated mainly by the availability of oxygen and methane and the presence of mineral nitrogen (2, 18). The effect of nitrogen, mainly supplemented in the form of inorganic fertilizer or by atmospheric deposition, has been intensively investigated since nitrogen was found to be inhibitory to methane consumption in forest soils (40) 2 decades ago. Many studies conducted since then have shown conflicting results. Inhibition, stimulation, and no effect were observed in upland soils (e.g., forest and grassland soils) as well as in wetland soils (e.g., rice paddies and freshwater marshes). Several mechanisms of inhibition or stimulation by nitrogen have been proposed (2), but none of them has been proven experimentally.
In this study we tested whether the diverse fertilizer effects may be explained by different compositions of the methane-oxidizing community in different soils and by changes to them effected by nitrogen addition (38). For this purpose, we incubated forest and rice field soils with different nitrogenous fertilizers and at different methane concentrations and assessed the growth and activity of the methane-oxidizing microbiota by analyzing PLFA concentrations and measuring the incorporation of 13CH4 into the various PLFAs.
MATERIALS AND METHODS
Soils.
The rice field soil originated from a rice field in Vercelli, Italy. The soil was collected from a drained field on 18 April 1999 before flooding. This soil had a maximum water-holding capacity corresponding to a gravimetric water content of 47% ± 1% (wt/wt) (n = 5) (19). The estimated C and N contents (percent dry weight) in the rice field soil were 1.8 ± 0.13 and 0.15 ± 0.01 (n = 4), respectively. This soil has repeatedly been used in many microbial ecological investigations (see, e.g., references 10, 21, and 32), and detailed soil characteristics have been described previously (39). The forest soil was collected from a 5- to 20-cm soil depth from a forest near Marburg, Germany (51°00.000′N, 9°50.625′E). The site was located on a slope in a deciduous forest consisting mainly of beech (Fagus sylvatica) and oak (Quercus robur). The soil type was a cambisol with Ah (2 to 6 cm), Bv (6 to 28 cm), and sandstone C horizons. The water pH values of the organic Ah horizon and the mineral subsoil were 3.8 and 4.3, respectively. Detailed characteristics of this forest soil have been described elsewhere (20, 29). Both soils were air dried and stored at room temperature. Soils were homogenized by sieving through 2-mm mesh. The incubations described here were carried out in 2001 and 2002.
Soil incubations.
About 100 g air-dried soil was incubated in 1,000-ml Erlenmeyer flasks that were closed with silicone septa. Soils were moistened and mixed well with sterile water to maintain a water-holding capacity of 40%. These flasks represented the controls. In the case of fertilized soils, the water to moisten the soils was supplemented with either NH4Cl or KNO3, corresponding to 30 μg N per g soil, which is equal to 60 kg N per ha. Half of all soil incubations were supplemented with methane to maintain a low concentration (1,000 ppm by volume [ppmv]), and the other half was supplemented to maintain a high concentration (10,000 ppmv). The 12CH4 was spiked with 13CH4 (99%; Cambridge Isotope laboratory, MA) at a mixing ratio of 97% 12CH4 to 3% 13CH4. Tubes containing 5 ml of 5 M NaOH solution were inserted into the flasks to trap emerging 13CO2 to prevent secondary labeling effects. Incubations were in triplicate for each treatment and were performed at 25°C. Subsamples (5 g) for PLFA and DNA extraction were collected after each complete consumption of the added CH4.
CH4 oxidation measurement.
The decrease of CH4 in the headspace was measured for each sample using an SRI 8610C gas chromatograph (GC) (SRI Instruments, Torrance, CA) equipped with a flame ionization detector (FID) (140°C) and a stainless steel column (6 ft; diameter, 1/8 in.) filled with Porapack Q (80/100 mesh) (oven temperature, 100°C). Activities were calculated using the slope of the steepest part (maximum consumption rate) of the methane depletion curve.
DNA extraction from soil.
DNA was extracted from 0.5-g soil subsamples (stored at −20°C) by using the Bio 101 Fast DNA Spin kit (Qbiogene, Heidelberg, Germany) according to the manufacturer's instructions. To increase the DNA recovery, the final elution of the DNA from a matrix was performed twice with 100 μl of DNase-free water. Additional purification was achieved using polyvinlypolypyrrolidone columns as described previously (20), again with two elutions of the DNA with TE buffer (10 mM Tris base, 1 mM EDTA, pH 8).
PCR amplification.
The pmoA gene fragment was amplified with the primer set A189f/A682R (19). PCRs were performed with 0.5 μM of each primer, 1× Premix F (Epicentre Technologies, Madison, WI), 1 U Taq DNA polymerase (Perkin-Elmer), and 100 ng template DNA. A touchdown PCR program (19) with decreasing annealing temperatures from 62 to 55°C was applied with 30 cycles. PCR was performed on a GeneAmp PCR system 9700 (Perkin-Elmer, Applied Biosystems, Weiterstadt, Germany).
T-RFLP analysis.
In general, the terminal restriction fragment length polymorphism (T-RFLP) analysis was carried out as previously described for PCR-amplified 16S rRNA and pmoA genes (25, 34), using the tetrameric restriction enzyme MspI (recognition site, C/CGG) (Promega, Mannheim, Germany). The fluorescently labeled PCR products (∼2 μg DNA) were purified using the QIA Min Elute PCR purification kit (QIAGEN, Hilden, Germany). Aliquots (1 to 8 μl) of the purified PCR products were digested with MspI under conditions recommended by the manufacturer (Promega). Aliquots (2 to 5 μl) of the digest were mixed with 2.0 μl formamide and 0.5 μl of an internal length standard (GeneScan-1000 ROX; PE Applied Biosystems, Weiterstadt, Germany). The internal length standard consisted of 17 different 6-carboxy-X-rhodamine-labeled fragments ranging in length from 29 to 928 nucleotides. The samples were denatured at 94°C for 5 min and immediately placed on ice until being loaded onto a 24-cm-long denaturing polyacrylamide gel (6%). Electrophoresis was carried out in the GeneScan mode of an automated DNA sequencer (model 373; PE Applied Biosystems) for up to 6 h using the following settings: 2,500 V, 40 mA, and 27 W. After electrophoresis, the size of each terminal restriction fragment (TRF) was determined in comparison to the fluorescently labeled size fragments of the internal length standard by using the GeneScan analysis software provided by the manufacturer (PE Applied Biosystems). The relative abundances of individual TRFs in a given pmoA PCR product were calculated based on the peak areas of the individual TRFs in relation to the total peak area of all TRFs detected in the respective T-RFLP community fingerprint pattern and standardized to uniform DNA quantity as described by Dunbar et al. (13). The GeneScan analysis software automatically quantified peak areas.
Lipid analyses and stable isotope probing of PLFA (SIP-PLFA) of lipids.
Lipids were extracted from 4 g of freeze-dried soil with a Bligh-Dyer extraction procedure as modified and described previously (4, 6). The lipid extract was fractionated on silicic acid into different polarity classes by sequential elution with chloroform, acetone, and methanol. The methanol fraction containing the PLFA was derivatized using mild-alkaline methanolysis to yield fatty acid methylesters (FAME). FAME standards of both C12:0 and C19:0 were used for calculating retention indices and for FAME quantification. Identification of FAME was based on retention time data with known standards. Additional identification was gained by GC-mass spectrometry (GC-MS) using a Thermo Finnagan TRACE GC-MS system. For identification of methanotroph-specific PLFA, extracts of cultures of Methylomonas methanica S1 NCIMB 11130, Methylomicrobium album NCIMB 11123, Methylobacter luteus NCIMB 11914, Methylocystis parvus NCIMB 11129, Methylosinus trichosporium NCIMB 11131, and Methylosinus sporium NCIMB 11126 were used as references. PLFA nomenclature used is as described previously (17). Fatty acids are designated by the number of carbon atoms. The degree of unsaturation is indicated by a number separated from the chain length by a colon and is followed by ωxc, where x indicates the position of the double bond nearest to the aliphatic end (ω) of the molecule and c indicates a cis stereoisomeric position of the double bond on the molecule.
FAME concentrations were determined using a GC-FID system (Thermo Finnagan TRACE GC) equipped with a polar capillary column (SGE, BPX-70; 50 m by 0.32 mm by 0.25 μm), using the following oven conditions: initial temperature of 50°C for 1 min, and then the temperature is programmed to 130°C using a ramp of 40°C min−1 followed by an increase to 230°C with a ramp of 3°C min−1.
Stable carbon isotope ratios for individual FAME were determined using a Varian 3400 GC equipped with an ATAS Optic 2 programmable direct thermal desorption injection system. The GC was coupled via a type II combustion interface to a Finnagan Delta S isotope ratio mass spectrometer. The same polar capillary column was used as for FAME identification and quantification on the GC-FID and GC-MS systems. The oven temperature for the GC-isotope ratio mass spectrometry analyses was as follows: initial temperature of 50°C for 4 min, and then the temperature is programmed to 130°C using a ramp of 30°C min−1, which is immediately followed by an increase to 200°C using a ramp of 6°C min−1, a subsequent increase to 220°C using a ramp of 5°C min−1, and a final increase to 250°C using a ramp of 20°C min−1. The sample is injected into the direct thermal desorption system at 50°C, after which the temperature is programmed to 260°C with a ramp of 10°C s−1. The absolute amount of label incorporated into every separate PLFA was calculated as described previously (5).
Calculation of intrinsic growth rates.
To calculate in situ intrinsic growth rates based on PLFA concentrations, the PLFA were transformed to cell numbers as described previously (41). For type I it was assumed that 33% of the total PLFA content of the cells is C16:1ω8c; a cell contains 100 μmol PLFA · g−1 dry cells, using a conversion factor from dry weight to number of cells of 0.119 × 1012. For type II it was assumed that 49% of the total PLFA content is C18:1ω8c, using a conversion factor from dry weight to cell numbers of 0.075 × 1012. Growth rates were calculated using cell numbers according to the formula (ln X2 − ln X1)/ (t2 − t1).
Statistical analyses. (i) Effects of methane concentration and N amendment.
Effects of methane concentration and nitrogen amendment on intrinsic growth rates and type-specific 13C incorporation in forest soil samples were tested using a factorial two-way analysis of variance (ANOVA). Effects of methane concentration, nitrogen amendment, and incubation time on intrinsic growth rates and type-specific 13C incorporation in rice field soil samples as well as on the relative abundance of restriction fragment 245 in forest and rice field soil samples were tested using a factorial three-way ANOVA. Before all ANOVAs the data were checked for normality (by plots of standard deviations versus means) and for homogeneity of variances (by Levene's test). If necessary, the data were transformed to meet the assumptions of the ANOVAs. All analyses were performed using the STATISTICA software package version 6.1 (Statsoft Inc., Tulsa, OK).
(ii) Multivariate analyses of SIP profiles.
SIP 13C-PLFA profiles of forest and rice field soil samples were compared to profiles of methanotrophic cultures to identify active MOB in these samples, as defined by the most similar profile available from cultivated MOB. A prerequisite for making these comparisons, however, is that only label incorporation in type I- and type II-related PLFAs separately can be determined. Otherwise, a mixed profile of type I and II species would be compared to profiles from separate cultures. Since in soil SIP profiles there are PLFA that occur in both type I and type II MOB, a selection of PLFA that were regarded as occurring predominantly in type II (>0.1%) but not in type I (<0.1%) and vice versa was made. The selected PLFAs for type I MOB were C14:0, i-C15:0, C16:0, C16:1ω9t, C16:1ω5t, C16:1ω8c, C16:1ω7c, C16:1ω6c, C16:1ω5, C17:0 cyc, and C19:0 cyc. The selected PLFAs used for type II MOB were i-C16:0, i-C17:0, i-C17:1ω7c, C17:0, C17:1ω6c, C18:0, C18:1ω9t, C18:1ω8c, C18:1ω5c, and C18:2?. (The question mark in the last PLFA indicates that the precise isomeric structure and double bond position are not known and are still under investigation [P. L. E. Bodelier, unpublished data]). The total 13C taken up in these selected PLFA was used to calculate the relative uptake in all individual PLFA. These relative PLFA concentration profiles of soil samples and known cultures formed the matrix which was used for cluster analyses and nonmetric multidimensional scaling (MDS) analyses. The inputs of cluster analyses as well as MDS were Bray-Curtis similarity matrices which were log(x + 1) transformed to even out the contributions of very rare and very dominant PLFA. Clustering was done using the group average linking routine. The MDS analysis results in a two-dimensional plot where the distance between samples indicates the similarity of these samples to other samples in the plot. The accuracy of the two-dimensional representation is indicated by the “stress” value (Kruskall's stress formula). Stress values of <0.1 indicate a good ordination with no prospect of a misleading interpretation. Stress values of <0.2 still give a good two-dimensional representation, but not too much reliance should be put on the detail. In this case, other methods of representation should be used in parallel, such as clustering analyses. All clustering and MDS analyses were performed using the Primer-E software (Plymouth Marine Laboratory, Plymouth, United Kingdom). All theoretical aspects of the cluster and MDS analyses used have been described previously (11).
RESULTS
Gross methane consumption.
The consumption of methane by both forest and rice field soils was found to be stimulated by the application of both ammonium and nitrate at an elevated methane concentration after addition of N fertilizer (Table 1). Only nitrate inhibited methane consumption in forest soils at a low methane concentration.
TABLE 1.
Effects of methane concentration and N amendment on methane consumption rates in rice and forest soil incubations during the first 12 days of incubation
| CH4 concn (ppmv) | N treatment | Methane consumption (nmol CH4 · g−1 · day−1)a
|
|
|---|---|---|---|
| Forest soil | Rice field soil | ||
| 1,000 | No N | 156.56 ± 7.71 bd | 39.61 ± 1.96 e |
| NH4+ | 148.31 ± 6.45 abc | 138.40 ± 5.02 ab | |
| NO3− | 133.47 ± 5.08 a | 161.03 ± 11.08 d | |
| 10,000 | No N | 858.12 ± 40.07 a | 2,193.75 ± 96.10 c |
| NH4+ | 1,342.55 ± 26.32 b | 2,289.58 ± 145.58 c | |
| NO3− | 1,253.60 ± 13.31 b | 2,856.85 ± 134.40 d | |
Values are means and standard deviations for three replicate incubations. Significant differences between means for the two soils are indicated for each methane concentration separately by different letters (Tukey's test, P < 0.05).
Type-specific biomass and growth rates.
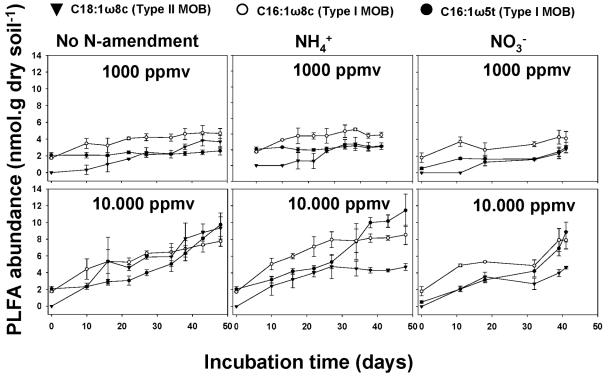
Methanotrophic biomass was assessed by extracting and quantifying the specific PLFA C16:1ω8c and C16:1ω5t (characteristic of type I MOB) and C18:1ω8c (characteristic of type II MOB) (3). At a low methane concentration (1,000 ppmv), the biomass increase of type II MOB (i.e., C18:1ω8c) in forest soil was delayed during the first 20 days of incubation when ammonium or nitrate fertilizer was applied, whereas type I biomass increased from the start of the incubation (Fig. 1). At an elevated methane concentration (10,000 ppmv), biomass accumulation of type II was repressed after 25 days of incubation with ammonium and nitrate, while without N addition type II biomass reached even higher levels than type I PLFA (i.e., C16:1ω8c) (Fig. 1). Since PLFA represent methanotrophic biomass, in situ intrinsic growth rates were calculated from the graphs in Fig. 1. Growth rates of type I were significantly stimulated by methane concentration as well as N amendment (Table 2; see Table SA1 in the supplemental material). In contrast, growth of type II was significantly inhibited by N amendment, with ammonium having the strongest effect.
FIG. 1.
Dynamics of MOB-specific PLFA in forest soil incubations with different methane concentrations and N amendments. Table 1 shows the growth rates calculated from the increase of PLFA over time. Table SA1 in the supplemental material shows the statistical analyses.
TABLE 2.
Effect of methane concentration and N amendment on intrinsic in situ growth rates of type I and II MOB in forest soil incubations
| CH4 concn (ppmv) | N treatment | Growth rate (day−1)a
|
|
|---|---|---|---|
| Type I | Type II | ||
| 1,000 | No N | 0.020 ± 0.004 | 0.029 ± 0.010 |
| NH4+ | 0.017 ± 0.001 | 0.012 ± 0.008 | |
| NO3− | 0.022 ± 0.007 | 0.032 ± 0.020 | |
| 10,000 | No N | 0.031 ± 0.002 | 0.036 ± 0.005 |
| NH4+ | 0.033 ± 0.002 | 0.013 ± 0.010 | |
| NO3− | 0.043 ± 0.008 | 0.034 ± 0.014 | |
Growth rates were calculated using cell numbers calculated from PFLA C16:1ω8c (type I) and C18:1ω8c (type II). For all samples n = 3. Values are means and standard deviations.
In rice field soil incubations amended with ammonium, a distinct effect on growth rates was observed during the first 2 weeks of incubation (data not shown). Growth rates of type II were significantly suppressed in the presence of ammonium (see Table SA2 in the supplemental material), especially during the first 14 days of incubation, whereas the growth rates of type I MOB were not affected by fertilization or incubation time (see Table SA2 in the supplemental material).
Type-specific methane consumption determined by using SIP of PLFA.
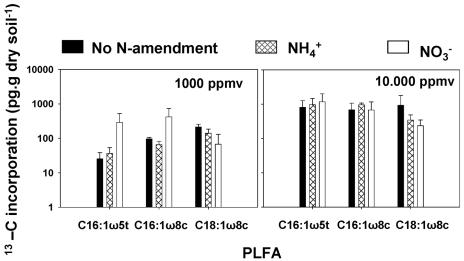
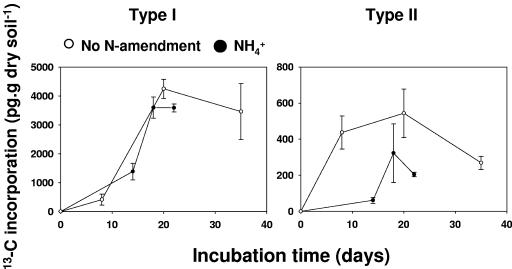
Methane consumption activities specific for type I and II methane-oxidizing bacteria were assessed by quantifying the incorporation of 13C from methane into specific PLFA, i.e., by using stable isotope probing of PLFA (6, 29). Similar to the case for biomass accumulation and growth rate, there was a differential effect on type-specific methane consumption activity (Fig. 2; see Table SA3 in the supplemental material). Type II activity was inhibited in forest soil, while type I was stimulated. In rice field soil the distinct effect observed on the growth rate was also observed for type-specific methane consumption activity (Fig. 3; see Table SA4 in the supplemental material), which was again repressed with type II MOB when ammonium was applied.
FIG. 2.
Total 13C incorporation in MOB-specific PLFA in forest soil after 48 days of incubations. Table SA3 in the supplemental material shows the statistical analyses of the effects of methane concentration and N amendment on 13C incorporation.
FIG. 3.
Total 13C incorporation in MOB-specific PLFA in rice field soil incubations. Incubations were performed using 10,000 ppmv methane and were supplemented with or without ammonium. Table SA4 in the supplemental material shows the statistical analyses.
Linking function and identity by using SIP-PLFA profiles and T-RFLP.
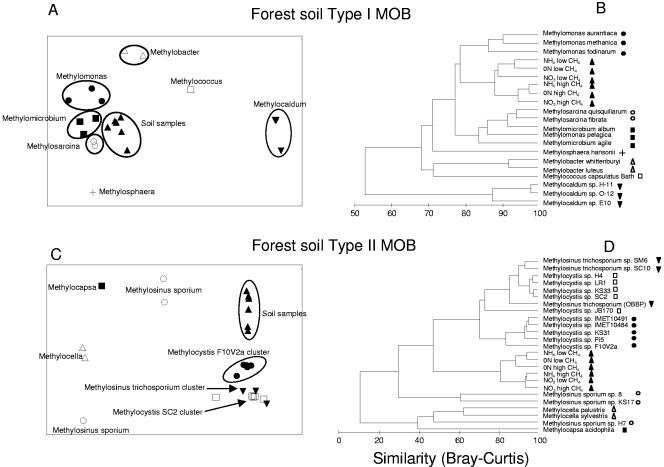
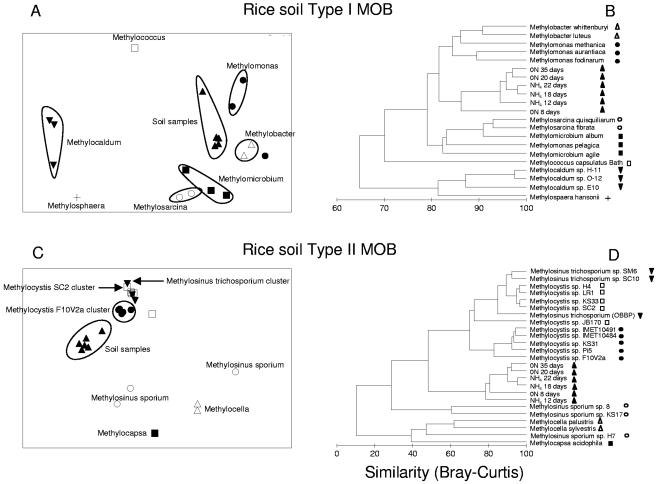
To find out in more detail which type I and II MOB were actually affected by fertilizer application, the SIP-PLFA data were used for multivariate statistical analyses. These analyses revealed that the active type II MOB in both forest and rice field soils were most closely related to a cluster of Methylocystis strains (Fig. 4 and 5) that are well separated from other Methylocystis and Methylosinus species on the basis of their PLFA profiles. The results were confirmed by T-RFLP analysis of the gene encoding the α subunit of the particulate methanemonooxygenase (pmoA) (25). Analysis of the relative abundance of the specific TRF of 245 bp, which is characteristic of Methylocystis species (16), confirmed the PLFA data and demonstrated again a negative effect of fertilizer application in both rice field and forest soils on type II MOB (Fig. 6; see Table SA5 in the supplemental material). For the active type I MOB, the PLFA-SIP data showed that forest soils differed from rice field soils. The active type I methanotrophs in forest soil were most closely related to representatives of the genera Methylomicrobium/Methylosarcina (Fig. 4A and B) while in rice field soil representatives of the genera Methylobacter/Methylomonas (Fig. 5A and B) were the most related to the soil SIP profiles. N amendment obviously stimulated a specific part of the type I community and did so differentially in the two types of soils.
FIG. 4.
Multivariate statistical analyses of SIP 13CH4-PLFA profiles of the MOB community in forest soil samples. The inputs of the MDS (A and C) and the cluster analyses (B and D) were PLFA profiles of MOB cultures (expressed as percentage of total PLFA content) and of SIP profiles of soil samples (expressed as percentage of 13C incorporated in separate PLFAs of the total PLFA 13C uptake). The two-dimensional distances between samples in the MDS graph show the relative similarity between samples. Stress values of the MDS plots were 0.12 (A) and 0.09 (C). For the cluster analyses, the profiles were transformed [log(x + 1)] before the Bray-Curtis similarity matrix was established. The clustering was done using the group average linking method.
FIG. 5.
Multivariate statistical analyses of SIP 13CH4-PLFA profiles of the MOB community in rice field soil samples. The inputs of the MDS (A and C) and the cluster analyses (B and D) were PLFA profiles of MOB cultures (expressed as percentage of total PLFA content) and of SIP profiles of soil samples (expressed as percentage of 13C incorporated in separate PLFAs of the total PLFA 13C uptake). The two-dimensional distances between samples in the MDS graph show the relative similarity between samples. Stress values of the MDS plots were 0.13 (A) and 0.10 (C). For the cluster analyses, the profiles were transformed [log(x + 1)] before the Bray-Curtis similarity matrix was established. The clustering was done using the group average linking method (11).
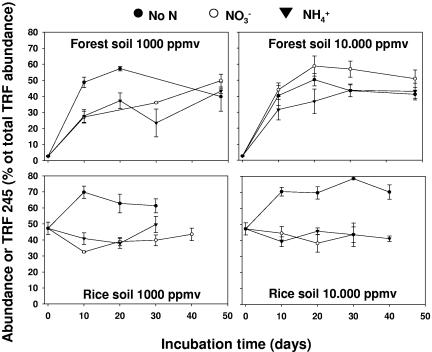
FIG. 6.
Relative abundance of terminal restriction fragment 245, which is representative of type II MOB, as affected by incubation time, methane concentration, and N amendment in forest and rice field soils. Relative abundance is expressed as the percentage of the total fluorescence recorded per sample. All points represent the means and standard errors for three replicate samples. The statistical evaluation of the data is presented in Table SA5 in the supplemental material.
DISCUSSION
The consumption of methane of both forest and rice field soils was stimulated by both ammonium and nitrate fertilizer application. For rice field soil this is consistent with earlier field studies on rice field soils (3), where a strong positive correlation between application of N fertilizer and methane oxidation was observed (32). The immediate stimulating effect in these studies was suggested to be the consequence of an immediate response of the methane-consuming enzyme machinery (2) rather than community growth following the relief of N limitation. A similar response was observed in our study. However, for forest soils this stimulation is rather surprising and has not yet been observed. Usually forest soils have been found to consume atmospheric methane, a process that is particularly sensitive to inhibition by fertilizer application and carried out by an as-yet-uncultivated group of methanotrophs (29). In our study the forest soil was exposed to higher methane concentrations supposedly favoring the “low-affinity” methane oxidation carried out by most methanotrophs available in culture (28). It is striking, however, that even in the forest soils at hand these low-affinity methanotrophs become active immediately upon exposure to elevated methane levels, a situation which can occur in forest soils when the soils get wet as a result of heavy rainfall. However, in other studies methane consumption by low-affinity MOB at high methane concentrations was inhibited (37). As suggested earlier (38), many of these discrepancies with respect to fertilizer effects on methane oxidation may be explained by assuming that not all community members are affected in the same way, making MOB diversity an explanatory variable in methane dynamics. That is actually what we observed in this study.
Growth rates as well as methane incorporation were generally stimulated in type I MOB, whereas these parameters were inhibited in type II MOB, by nitrogenous fertilizers. The growth rates themselves are unique data, since actual growth rates in soils are hardly available for microbes. The rates we measured represent doubling times of 20 to 60 days, which in the case of the growth rates observed at 1,000 ppmv are in the same range as those of MOB cultures growing under methane-limited conditions (27). The rates at 10,000 ppmv methane, however, are orders of magnitude lower than those of laboratory cultures grown under the same conditions (27), indicating some other limitation (e.g., N) or grazing by protozoa.
Surprisingly, the methane concentration did not affect the growth rate of type II MOB. It is generally assumed that type II methane oxidizers, in comparison to type I methanotrophs, thrive especially in environments with high methane and low oxygen concentrations (8, 18), a fact that is certainly not supported by our results with forest soil. Very recently, some Methylocytis spp. were shown to be very oligotrophic with respect to methane, which also indicates that no generalizations can be made about the effect of methane concentration on type II MOB (27).
With respect to the effects of nitrogenous fertilizers on methane consumption, also no generalizations can be made. The general idea until now was that type I MOB profit much more from the presence of mineral nitrogen than type II MOB, possibly because of the ability of type II MOB to fix molecular nitrogen (2, 3, 16). However, the data presented on forest and rice field soil incubation clearly demonstrate that methane-oxidizing microbial species belonging to type I or II were differentially affected by the availability of nitrogen. Type II MOB were repressed by the presence of mineral N, especially ammonium. This could be an effect of competition for N between types I and II in the incubations. However, for rice field soil the effect of ammonium is immediate (Fig. 3), while the T-RFLP analyses demonstrate that in forest soil the inhibition also acts at the beginning of the incubation (Fig. 6).
The SIP-PLFA profiles enable us to pinpoint at the genus level which organisms were actually active in the incubations. The genera identified by our SIP-PLFA approach have already been detected in rice field and forest soils by using molecular approaches such as denaturing gradient gel electrophoresis, cloning, and real-time PCR (7, 15, 19, 21, 29, 30). In forest soil the observed type I genera are most certainly not the dominant members of the methane-consuming community. However, they are present and can become active and grow, as was the case in our incubations, where apparently representatives of the genera Methylobacter/Methylomonas (rice field soil) and Methylomicrobium/Methylosarcina (forest soil) were dominant in methane consumption over other type I genera. Similarly, we know from this study that the type II genus Methylocystis is repressed by N addition. There is as yet no evidence that this also holds true for other MOB genera such as Methylocaldum, Methylococcus, Methylosinus, Methylocella, and Methylocapsa and the uncultured soil clusters USCα and USCγ (29), which can dominate the soil methanotrophic community (31). This makes the picture of the effects of nitrogenous fertilizers on MOB communities far less “black and white” than has been assumed until now.
The nature of the effect (i.e., stimulation or inhibition) obviously depends on the community composition and hence on the biodiversity of the MOB present. We hypothesize that methane consumption in soil or sediment with a predominance of type I MOB will not be affected by fertilizer application, while methane uptake by a soil or sediment with a predominance of type II will be inhibited. Moreover, differentiation can also be expected within type I and II representatives. Clear examples of habitats with a dominance of a specific type of MOB are acid peat, dominated by Methylocella and Methylocystis (12); periodically water-saturated gleyic soils, with a dominance of Methylocystis and USCγ (28); and lake sediments, dominated by Methylomonas (1, 14). A major determinant of the outcome of fertilizer effects will also be the methane concentration in the respective environment, because this determines the active MOB type. In periodically water-saturated upland soils, type I MOB participated in methane consumption only from 500 ppm methane upward (28), while type II MOB consumed at lower methane concentrations. With respect to the latter, we have to keep in mind that in the case of forest soils, the effects in our study were observed at a far higher methane concentration than that to which MOB in dry forest soils are normally exposed. Hence, we cannot say whether this differential effect will also act at atmospheric methane concentrations. However, the bacteria that consume atmospheric methane in forest and other upland soils are still not identified or in culture (29) or phenotypically characterized. Nevertheless, we know from culture studies that Methylocystis and other MOB were present and activated immediately upon exposure to elevated methane, a situation which could emerge when methane availability in forest or other upland soils is elevated due to flooding or increased precipitation, as predicted for future climatic conditions (26).
However, our study demonstrated that the composition of the methanotrophic microbial community apparently affected methane fluxes and hence that microbial diversity has to be taken into account in the global biodiversity debate. Change in land use and global environmental change are both factors that may provoke shifts in methane-consuming microbial communities (24). The latter may have consequences for the flux that the microbes catalyze. However, to confirm this, extensive ecophysiological experiments have to be carried out with pure cultures and with experimental systems differing in MOB diversity. Fertilizer addition studies have to be carried out in habitats with a dominance of either type of MOB, where methane consumption has to be linked with diversity by using, e.g., stable isotope probing of PLFA, DNA, or RNA.
In conclusion, it can be said that biodiversity conservation and land use policy makers should also consider prokaryotic diversity, which is apparently important for certain ecosystem functions, such as methane oxidation in soil.
Supplementary Material
Acknowledgments
S. R. Mohanty and P. L. E. Bodelier contributed equally to this work.
We thank Peter Nichols for providing data on PLFA profiles of methanotrophic cultures and Mary Lidstrom and Sergey Stolyar for the kind provision of Methylomonas strains.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
This is publication no. 3739 of The Netherlands Institute of Ecology.
REFERENCES
- 1.Auman, A. J., and M. E. Lidstrom. 2002. Analysis of sMMO-containing type I methanotrophs in Lake Washington sediment. Environ. Microbiol. 4:517-524. [DOI] [PubMed] [Google Scholar]
- 2.Bodelier, P. L. E., and H. J. Laanbroek. 2004. Nitrogen as a regulatory factor of methane oxidation in soils and sediments. FEMS Microbiol. Ecol. 47:265-277. [DOI] [PubMed] [Google Scholar]
- 3.Bodelier, P. L. E., P. Roslev, T. Henckel, and P. Frenzel. 2000. Stimulation by ammonium-based fertilizers of methane oxidation in soil around rice roots. Nature 403:421-424. [DOI] [PubMed] [Google Scholar]
- 4.Boschker, H. T. S., W. deGraaf, M. Koster, L. A. MeyerReil, and T. E. Cappenberg. 2001. Bacterial populations and processes involved in acetate and propionate consumption in anoxic brackish sediment. FEMS Microbiol. Ecol. 35:97-103. [DOI] [PubMed] [Google Scholar]
- 5.Boschker, H. T. S., and J. J. Middelburg. 2002. Stable isotopes and biomarkers in microbial ecology. FEMS Microbiol. Ecol. 40:85-95. [DOI] [PubMed] [Google Scholar]
- 6.Boschker, H. T. S., S. C. Nold, P. Wellsbury, D. Bos, W. DeGraaf, R. Pel, R. J. Parkes, and T. E. Cappenberg. 1998. Direct linking of microbial populations to specific biogeochemical processes by 13C-labelling of biomarkers. Nature 392:801-805. [Google Scholar]
- 7.Bourne, D. G., I. R. McDonald, and J. C. Murrell. 2001. Comparison of pmoA PCR primer sets as tools for investigating methanotroph diversity in three Danish soils. Appl. Environ. Microbiol. 67:3802-3809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bowman, J. 2000. The methanotrophs—the families Methylococcaceae and Methylocystaceae. In M. Dworkin et al. (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.1. [Online.] Springer, New York, N.Y. http://link.springer-ny.com/link/service/books/10125/.
- 9.Bowman, J. P., L. I. Sly, P. D. Nichols, and A. C. Hayward. 1993. Revised taxonomy of the methanotrophs—description of Methylobacter gen. nov., emendation of Methylococcus, validation of Methylosinus and Methylocystis species, and a proposal that the family Methylococcaceae includes only the group-I methanotrophs. Int. J. Syst. Bacteriol. 43:735-753. [Google Scholar]
- 10.Chin, K. J., T. Lukow, and R. Conrad. 1999. Effect of temperature on structure and function of the methanogenic archaeal community in an anoxic rice field soil. Appl. Environ. Microbiol. 65:2341-2349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clarke, K. R., and R. M. Warwick. 2001. Change in marine communities: an approach to statistical analysis and interpretation, 2nd ed. Primer-E Ltd., Plymouth, United Kingdom.
- 12.Dedysh, S. N., M. Derakshani, and W. Liesack. 2001. Detection and enumeration of methanotrophs in acidic Sphagnum peat by 16S rRNA fluorescence in situ hybridization, including the use of newly developed oligonucleotide probes for Methylocella palustris. Appl. Environ. Microbiol. 67:4850-4857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunbar, J., L. O. Ticknor, and C. R. Kuske. 2001. Phylogenetic specificity and reproducibility and new method for analysis of terminal restriction fragment profiles of 16S rRNA genes from bacterial communities. Appl. Environ. Microbiol. 67:190-197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eller, G., P. Deines, J. Grey, H.-H. Richnow, and M. Kruger. 2005. Methane cycling in lake sediments and its influence on chironomid larval δ13C. FEMS Microbiol. Ecol. 54:339-350. [DOI] [PubMed] [Google Scholar]
- 15.Eller, G., M. Kruger, and P. Frenzel. 2005. Comparing field and microcosm experiments: a case study on methano- and methylo-trophic bacteria in paddy soil. FEMS Microbiol. Ecol. 51:279-291. [DOI] [PubMed] [Google Scholar]
- 16.Graham, D. W., J. A. Chaudhary, R. S. Hanson, and G. A. Ranold. 1993. Factors affecting competition between type 1 and type 2 methanotrophs in two-organism, continuous-flow reactors. Microb. Ecol. 25:1-17. [DOI] [PubMed] [Google Scholar]
- 17.Guckert, J. B., C. P. Antworth, P. D. Nichols, and D. C. White. 1985. Phospholipid, ester-linked fatty-acid profiles as reproducible assays for changes in prokaryotic community structure of estuarine sediments. FEMS Microbiol. Ecol. 31:147-158. [Google Scholar]
- 18.Hanson, R. S., and T. E. Hanson. 1996. Methanotrophic bacteria. Microbiol. Rev. 60:439-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henckel, T., M. Friedrich, and R. Conrad. 1999. Molecular analyses of the methane-oxidizing microbial community in rice field soil by targeting the genes of the 16S rRNA, particulate methane monooxygenase, and methanol dehydrogenase. Appl. Environ. Microbiol. 65:1980-1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henckel, T., U. Jackel, S. Schnell, and R. Conrad. 2000. Molecular analyses of novel methanotrophic communities in forest soil that oxidize atmospheric methane. Appl. Environ. Microbiol. 66:1801-1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henckel, T., P. Roslev, and R. Conrad. 2000. Effects of O2 and CH4 on presence and activity of the indigenous methanotrophic community in rice field soil. Environ. Microbiol. 2:666-679. [DOI] [PubMed] [Google Scholar]
- 22.Heyer, J., U. Berger, M. Hardt, and P. F. Dunfield. 2005. Methylohalobius crimeensis gen. nov., sp. nov., a moderately halophilic, methanotrophic bacterium isolated from hypersaline lakes of Crimea. Int. J. Syst. Evol. Microbiol. 55:1817-1826. [DOI] [PubMed] [Google Scholar]
- 23.Horz, H. P., A. Barbrook, C. B. Field, and B. J. M. Bohannan. 2004. Ammonia-oxidizing bacteria respond to multifactorial global change. Proc. Natl. Acad. Sci. USA 101:15136-15141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horz, H. P., V. Rich, S. Avrahami, and B. J. M. Bohannan. 2005. Methane-oxidizing bacteria in a California upland grassland soil: diversity and response to simulated global change. Appl. Environ. Microbiol. 71:2642-2652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horz, H. P., M. T. Yimga, and W. Liesack. 2001. Detection of methanotroph diversity on roots of submerged rice plants by molecular retrieval of pmoA, mmoX, mxaF, and 16S rRNA and ribosomal DNA, including pmoA-based terminal restriction fragment length polymorphism profiling. Appl. Environ. Microbiol. 67:4177-4185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Intergovernmental Panel on Climate Change. 2001. Climate change 2001: the scientific basis. Contribution of the Working Group I to the third assessment report of the Intergovernmental Panel on Climate Change. Cambridge University Press, Cambridge, United Kingdom.
- 27.Knief, C., and P. F. Dunfield. 2005. Response and adaptation of different methanotrophic bacteria to low methane mixing ratios. Environ. Microbiol. 7:1307-1317. [DOI] [PubMed] [Google Scholar]
- 28.Knief, C., S. Kolb, P. L. E. Bodelier, A. Lipski, and P. Dunfield. 2006. The active methanotrophic community in hydromorphic soils changes in response to changing methane concentration. Environ. Microbiol. 8:321-333. [DOI] [PubMed]
- 29.Knief, C., A. Lipski, and P. F. Dunfield. 2003. Diversity and activity of methanotrophic bacteria in different upland soils. Appl. Environ. Microbiol. 69:6703-6714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kolb, S., C. Knief, S. Stubner, and R. Conrad. 2003. Quantitative detection of methanotrophs in soil by novel pmoA-targeted real-time PCR assays. Appl. Environ. Microbiol. 69:2423-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kolb, S., C. Knief, P. Dunfield, and R. Conrad. 2005. Abundance and activity of uncultured methanotrophic bacteria involved in consumption of atmospheric methane in two forest soils. Environ Microbiol. 7:1150-1162. [DOI] [PubMed] [Google Scholar]
- 32.Krüger, M., and P. Frenzel. 2003. Effects of N-fertilisation on CH4 oxidation and production, and consequences for CH4 emissions from microcosms and rice fields. Global Change Biol. 9:773-784. [Google Scholar]
- 33.LeMer, J., and P. Roger. 2001. Production, oxidation, emission and consumption of methane by soils: a review. Eur. J. Soil Biol. 37:25-50. [Google Scholar]
- 34.Liu, W. T., T. L. Marsh, H. Cheng, and L. J. Forney. 1997. Characterization of microbial diversity by determining terminal restriction fragment length polymorphisms of genes encoding 16S rRNA. Appl. Environ. Microbiol. 63:4516-4522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Loreau, M., S. Naeem, P. Inchausti, J. Bengtsson, J. P. Grime, A. Hector, D. U. Hooper, M. A. Huston, D. Raffaelli, B. Schmid, D. Tilman, and D. A. Wardle. 2001. Ecology—biodiversity and ecosystem functioning: current knowledge and future challenges. Science 294:804-808. [DOI] [PubMed] [Google Scholar]
- 36.Nold, S. C., H. T. S. Boschker, R. Pel, and H. J. Laanbroek. 1999. Ammonium addition inhibits 13C-methane incorporation into methanotroph membrane lipids in a freshwater sediment. FEMS Microbiol. Ecol. 29:81-89. [Google Scholar]
- 37.Reay, D. S., and D. B. Nedwell. 2004. Methane oxidation in temperate soils: effects of inorganic N. Soil Biol. Biochem. 36:2059-2065. [Google Scholar]
- 38.Schimel, J. P., and J. Gulledge. 1998. Microbial community structure and global trace gases. Global Change Biol. 4:745-758. [Google Scholar]
- 39.Schutz, H., A. Holzapfelpschorn, R. Conrad, H. Rennenberg, and W. Seiler. 1989. A 3-year continuous record on the influence of daytime, season, and fertilizer treatment on methane emission rates from an Italian rice paddy. J. Geophys. Res. Atmos. 94:16405-16416. [Google Scholar]
- 40.Steudler, P. A., R. D. Bowdem, J. M. Melillo, and J. D. Aber. 1989. Influence of nitrogen fertilization on methane uptake in temperate forest soils. Nature 341:314-316. [Google Scholar]
- 41.Sundh, I., P. Borga, M. Nilsson, and B. H. Svensson. 1995. Estimation of cell numbers of methanotrophic bacteria in boreal peatlands based on analysis of specific phospholipid fatty acids. FEMS Microbiol. Ecol. 18:103-112. [Google Scholar]
- 42.Torsvik, V., L. Ovreas, and T. F. Thingstad. 2002. Prokaryotic diversity—magnitude, dynamics, and controlling factors. Science 296:1064-1066. [DOI] [PubMed] [Google Scholar]
- 43.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Y. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y. H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 44.Webster, G., T. M. Embley, T. E. Freitag, Z. Smith, and J. I. Prosser. 2005. Links between ammonia oxidizer species composition, functional diversity and nitrification kinetics in grassland soils. Environ. Microbiol. 7:676-684. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.