Abstract
The use of plants for rehabilitation of heavy-metal-contaminated environments is an emerging area of interest because it provides an ecologically sound and safe method for restoration and remediation. Although a number of plant species are capable of hyperaccumulation of heavy metals, the technology is not applicable for remediating sites with multiple contaminants. A clever solution is to combine the advantages of microbe-plant symbiosis within the plant rhizosphere into an effective cleanup technology. We demonstrated that expression of a metal-binding peptide (EC20) in a rhizobacterium, Pseudomonas putida 06909, not only improved cadmium binding but also alleviated the cellular toxicity of cadmium. More importantly, inoculation of sunflower roots with the engineered rhizobacterium resulted in a marked decrease in cadmium phytotoxicity and a 40% increase in cadmium accumulation in the plant root. Owing to the significantly improved growth characteristics of both the rhizobacterium and plant, the use of EC20-expressing P. putida endowed with organic-degrading capabilities may be a promising strategy to remediate mixed organic-metal-contaminated sites.
Heavy metals such as lead, mercury, and cadmium are ranked second, third, and seventh, respectively, on the 2003 Comprehensive Environmental Response, Compensation, and Liability Act (CERCLA, commonly known as Superfund) priority list for hazardous substances because they are toxic widespread pollutants (http://www.atsdr.cdc.gov/clist.html). Soil contamination is a particularly serious environmental concern, as the majority of Superfund sites are highly contaminated with heavy metals (21). Conventional remediation methods such as soil excavation followed by coagulation-filtration or ion exchange are expensive and disruptive to the sites. In situ bioremediation is gaining momentum as a low-cost and effective method for restoration and remediation under many site conditions.
Biosorption and immobilization are major mechanisms utilized by animals and plants to limit the concentrations of internal reactive metal species (12, 13, 16, 18). Naturally occurring metallothioneins (MTs) and phytochelatins (PCs) are examples of peptides that effectively bind a wide range of heavy metals with high affinity. PCs are particularly attractive as they offer higher metal-binding capacity than MTs, due to their repeating Glu-Cys moieties (17). However, the presence of a γ bond between Glu and Cys indicates that these peptides must be synthesized enzymatically. An attractive alternative is to employ synthetic phytochelatins (ECs), which are protein analogs of PCs with similar heavy-metal-binding affinities that can be easily produced from a synthetic DNA template by standard molecular cloning techniques.
Biosorption using microbially produced ECs has been shown to be a promising technique for ameliorating heavy-metal contamination. Bacteria such as Escherichia coli and Moraxella sp. expressing EC20 (with 20 cysteines) on the cell surface or intracellularly have been shown to accumulate up to 25-fold-more cadmium (4, 5) or mercury (6) than the wild-type strain. However, one major obstacle for utilizing these engineered microbes is sustaining the recombinant bacteria population in soil, with various environmental conditions and competition from native bacterial populations.
Symbiosis between plants and microbes in the rhizosphere has long been studied by microbial ecologists (3, 14). The rhizosphere is an area encircling the plant root system, which is characterized by enhanced biomass productivity. Rhizosphere bacteria obtain nutrients excreted from roots, such as organic acids, enzymes, amino acids, and complex carbohydrates (3, 24, 30). In return, the bacteria convert nutrients into available forms of mineral for the plants. For example, maize and lettuce inoculated with Rhizobium leguminosarum were demonstrated to have increased growth through enhanced solubilization of phosphate (9), and sunflowers inoculated with Rhizobium sp. exhibited increased nitrogen uptake (1). Furthermore, the root tips provide a steady-state redox condition and a structural surface for bacterial colonization. The plant root system aerates the soil, distributes bacteria through soil, and penetrates otherwise-impermeable soil layers while drawing soluble forms of the pollutants in the soil water towards the plant and the microbes. Researchers have exploited this symbiotic relationship for rhizoremediation and have documented attenuation of compounds such as volatile organic carbon contaminants, parathion (3), atrazine (2), trichloroethylene (24, 30), and polychlorinated biphenyls (8, 27).
In this study, we aim to utilize the symbiotic plant-microbe relationship to remediate heavy-metal contamination. Pseudomonas putida 06909, an antifungal bacterium isolated from citrus root that is modestly cadmium resistant, was selected as the host strain because it is environmentally robust (15, 29). We demonstrate that expression of EC20 in P. putida 06909 improves both cell growth and cadmium binding in the presence of high levels of cadmium. Furthermore, we detail the process for developing a rhizoremediation system, which results in an engineered symbiosis where the recombinant bacterium significantly reduces the toxic effects of cadmium on the growth of sunflower seedlings while it colonizes the root effectively.
MATERIALS AND METHODS
Bacterial strains, growth conditions, and plasmids.
E. coli strains DH5α and XL1 Blue were used for construction and replication of plasmids. E. coli strains were cultured in Luria-Bertani (LB) medium at 37°C. The rhizobacterium P. putida 06909 was cultured in low-phosphate MJS minimal medium (22) containing 200-μg/ml ampicillin at 30°C for liquid culture cadmium-binding experiments. Expression of maltose-binding protein-EC20 (MBP-EC20) was induced at mid-exponential growth phase with 0.8 mM of isopropyl-β-d-thiogalactopyranoside (IPTG). The optical density of the cultures was measured at an optical density at 600 nm (OD600) with a spectrophotometer (DU640; Beckman).
All molecular cloning techniques were performed according to Sambrook and Russell (22). For the expression of MBP-EC20 in P. putida 06909, the broad-host-range vector pVLT33 (10) was used. A DNA fragment encoding MBP-EC20 was obtained from the plasmid pMC20 (4) by first carrying out digestion with NdeI, blunt ending with Klenow fragment (New England BioLabs, Beverly, MA), and then digestion with HindIII. The 1.3-kb fragment was separated and extracted from DNA agarose gel with the GeneClean kit (QBioGene, Irvine, CA). The vector pVLT33 was digested with EcoRI, and a blunt end was created using Klenow fragment. The vector was then digested with HindIII and purified by phenol chloroform precipitation, and the MBP-EC20 fragment was ligated to the opened plasmid to create pVMC20. The plasmid was electroporated into P. putida at 1.8 kV, 25 μF, and 400 Ω in a 1-mm-gap cuvette; it was grown with 100-μg/ml kanamycin (11).
The plasmid pRCD32, containing a lacZ reporter gene fused to the cadR promoter, was obtained from Lee et al. (15). Expression of β-galactosidase (β-Gal) from the cadR promoter has been demonstrated to be inducible only by Cd2+ but not by copper, zinc, mercury, cobalt, nickel, manganese, or lead. The pRCD32 plasmid was electroporated into P. putida 06909 under the same conditions as above, for determination of root colonization efficiency. Positive clones were selected on an LB plate containing CdCl2, 5-bromo-4-chloro-3-indolyl-beta-d-galactopyranoside (X-Gal), IPTG, and 80-μg/ml tetracycline.
Cadmium binding in growing cultures.
Overnight cultures of P. putida 06909 and P. putida 06909/pVMC20 were inoculated into 10 ml MJS minimal medium containing the appropriate antibiotics in 125-ml flasks and grown in a 30°C shaker incubator. When the culture OD600 reached ∼0.6 (mid-exponential growth phase), 0.8 mM IPTG and 80 μM CdCl2 (final concentrations) were added to each flask. Subsequently, 1-ml samples were taken at 10, 21, and 28 h after inoculation for expression and cadmium-binding analyses. The cell density of the samples was determined at each time point. Whole-cell binding of cadmium was determined by atomic absorption spectrometry. Triplicate samples from independent flasks were taken for each data point.
Cadmium binding in resting cultures.
Overnight cultures were inoculated into MJS medium and induced with IPTG at mid-exponential growth phase. Cells were grown for 17 h, iced for 20 min, and centrifuged at 3,000 rpm at 4°C for 30 seconds. The cell pellet was washed with 50 mM Tris chloride buffer at pH 7.4 and resuspended in the same buffer. CdCl2 (80 μM) was added to the cell suspensions, and samples were taken at 0, 5, 30, 60, and 150 min. Triplicate samples were taken for each time point. Whole-cell binding of cadmium was determined by atomic absorption spectrometry.
Cadmium analysis by atomic absorption spectrometry (AAS).
Samples were centrifuged at maximum speed (13,000 rpm) for 5 min, and the supernatant was transferred to a new tube. The pellets were washed three times with 0.8% sodium chloride in 5 mM HEPES buffer (pH 7.1). The washed pellets were dried in an oven set to 65°C for 24 h and digested with 100 μl of concentrated nitric acid for at least 48 h. The digested cell pellets were reconstituted to 1 ml by the addition of 900 μl of deionized water and diluted to the correct concentration for flame analysis with an atomic absorption spectrometer (AA-6701; Shimadzu, Columbia, MD). The data were normalized to the number of nanomoles of cadmium per milligram of dry cell weight.
Expression of MBP-EC20.
Western blot analysis was used to probe the expression of MBP-EC20. Samples were centrifuged at maximum for 5 min, and the supernatant was discarded. The pellets were stored in a freezer set to −20°C until further processing. Samples were concentrated to an OD600 of 20 and boiled at 95°C for 10 min. The cell lysate was loaded onto 12% (wt/vol) polyacrylamide gel (22), and the proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis. Proteins were then transferred to a nitrocellulose membrane and incubated with rabbit MBP antisera (New England BioLabs, Beverly, MA) preincubated with P. putida wild-type cell lysate. Western blot analysis was performed using an Immun-Blot GAR-AP kit (Bio-Rad, Hercules, CA).
Seed germination and hydroponic plant growth conditions.
Cowpea, corn, wheat, and sunflower seedlings were used for the experiments. Seeds were surface sterilized by being washed and shaken in 95% ethanol for 45 min and in 5.25% bleach for 15 min and then rinsed three times with sterile water for 10 min each wash. Seeds were germinated in paper towels soaked in 200 ml of germination solution containing 200 μM NH4NO3, 1,800 μM Ca(NO3)2 · 4H2O, 1,000 μM KNO3, 500 μM MgSO4 · 7H2O, 1,000 μM MES (morpholineethanesulfonic acid), 500 μM NaOH, 80 μM KH2PO4, 10 μM H3BO3, and 0.1 μM Na2MoO4 · 2H2O in a 600-ml flask. Seeds were 1 in. apart, and each paper towel roll contained 12 seeds. The flasks containing the seeds in paper towels were placed in a growth chamber with a light cycle consisting of 8 h of darkness and 16 h of light and with 65% humidity at 25°C. The seeds germinated after 3 to 7 days. On day 7, seedlings were transferred into hydroponic solutions aerated with tubes bubbling air. The chelator-buffered nutrient solution (20) contained the germinating solution and the following: 8 μM ZnCl2, 0.6 μM MnCl2 · 4H2O, 2 μM CuCl2 · 2H2O, 0.1 μM NiCl2 · 6H2O, 110.7 μM hydroxyethylenediaminetriacetic acid, 53.55 μM HCl, and 75 μM FeCl3 · 6H2O. Each seed was held above the solution with a plastic mesh, while the root grew through the mesh into the solution. White silicone chips held the seedling upright and blocked light away from the root. The root section was also covered by a black cloth. The hydroponic solution was replaced every 7 days, and the pH was maintained at 6.0 with 0.1 M NaOH. Triplicate samples were taken for each data point.
Colonization efficiency for plant selection.
To determine the colonization efficiency of the recombinant bacteria in the rhizosphere of different plants, P. putida harboring the plasmid pRCD32 was used. The purpose of the colonization efficiency experiment is to use the β-Gal activity of the recombinant strains as an indicator of colonizing bacterial density in the rhizosphere of the four plant species to determine the best plant to use for subsequent rhizoremediation experiments. Overnight P. putida 06909/pRDCD32 cultures grown in LB medium were centrifuged and washed with sterile deionized water. Cells were diluted into 100 ml of sterile deionized water. Roots of 5-day-old seedlings were dipped into overnight-grown cultures for 1 min before being returned to the hydroponic solution. Tetracycline (80 μg/ml) and 12.5 μM CdCl2 were added to the nutrient solutions at that time. After 4 days of colonization, all of the roots were harvested and vigorously vortexed for 2 min in 50-ml disposable polypropylene centrifuge tubes with 20 ml of pH 7.0 Z buffer (0.06 M Na2HPO4 · 7H2O, 0.04 M NaH2PO4 · H2O, 0.01 M KCl, 0.001 M MgSO4 · 7H2O, and 0.05 M β-mercaptoethanol) (19). The roots were removed, dried at 105°C for 48 h, and weighed. The bacterial extracts in Z buffer were centrifuged at 3,000 rpm for 15 min, and the supernatants were discarded. The pellets were stored in −20°C until the β-galactosidase assay was performed according to the protocols by Miller (19). β-Galactosidase activities for the different plant species were expressed as Miller units per milligram (dry weight) of roots.
Plant growth in hydroponic solutions and cadmium binding.
Gray-striped sunflower seeds were germinated in paper towels, and each seedling was transferred into 1-liter Erlynmeyer flasks at 7 days old. The seedlings were grown for 21 days in the growth chamber under the same conditions as for germination before the addition of antibiotics and inoculation with the respective bacterial strains. Cultures of P. putida 06909 and P. putida 06909/pVMC20 were grown overnight in 250 ml of LB medium containing respective antibiotics at 30°C. Cells were centrifuged at 3,000 rpm for 10 min and washed with phosphate buffer solution (pH 7.4). The cell pellets were then suspended in the chelator-buffered nutrient solution and added to each flask with antibiotics to obtain the final inoculum density of 5 × 107 CFU/ml. The bacteria were grown in the hydroponic solution with the plants for 4 days to allow colonization. After 4 days of colonization, the hydroponic solutions with antibiotics were replaced, and 80 μM CdCl2 was added at that time. The pH of the hydroponic solution was adjusted to pH 6 with 0.1 M NaOH. The plants were grown for 3 days and harvested. The height and total weight of the plants were recorded. One half of the root was put into aluminum trays, dried at 105°C for 48 h, weighed, and digested with concentrated nitric acid for cadmium analysis by AAS. The other half of the root was suspended with 20 ml of 0.8% sodium chloride in 5 mM HEPES buffer and vortexed vigorously to dislodge the rhizosphere bacteria. The roots were removed, dried at 105°C for 48 h, and weighed. The buffer containing the cells was centrifuged at 3,000 rpm for 10 min, and the supernatant was discarded. A total of 1.1 ml of the HEPES buffer was used to wash and resuspend the pellet, and the OD600 was measured. The pellets were centrifuged at 13,000 rpm, and the supernatant was discarded. The pellets were then stored in −20°C until the Western blot analysis was performed as mentioned above for the detection of MBP-EC20 fusion protein production. Four independent samples were taken for each data point.
Statistical analysis methods.
Data were stored in Microsoft Excel and analyzed using SAS statistical analysis software (SAS Institute, Inc., Cary, NC). Normal distribution of the data was determined by SAS statistical analysis software. All data were statistically tested and found significant at the 95% confidence level by the nonparametric one-sided Wilcoxon rank sum test.
RESULTS
Enhanced cell growth and cadmium binding by EC20 production.
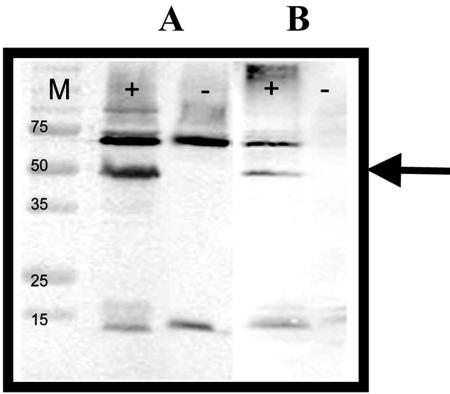
To investigate whether expression of EC20 would enhance cadmium binding to P. putida 06909, our initial work was focused on the intracellular expression of EC20. Plasmid pVMC20, which allows the cytoplasmic expression of EC20 in different gram-negative species as a fusion to MBP, was constructed. Production of EC20 was verified by probing with a MBP antibody, and a strong band at 50 kDa corresponding to the expected size of the MBP-EC20 fusion was detected (Fig. 1).
FIG. 1.
Western blot analysis of EC20 production. EC20 production in liquid culture (A) and rhizosphere (B) from P. putida 06909/pVMC20 (+) and P. putida 06909 (−) was detected with an MBP rabbit antisera (Bio-Rad). Bands of ∼50 kDa were detected in P. putida cells with pVMC20 in liquid culture and rhizosphere experiments.
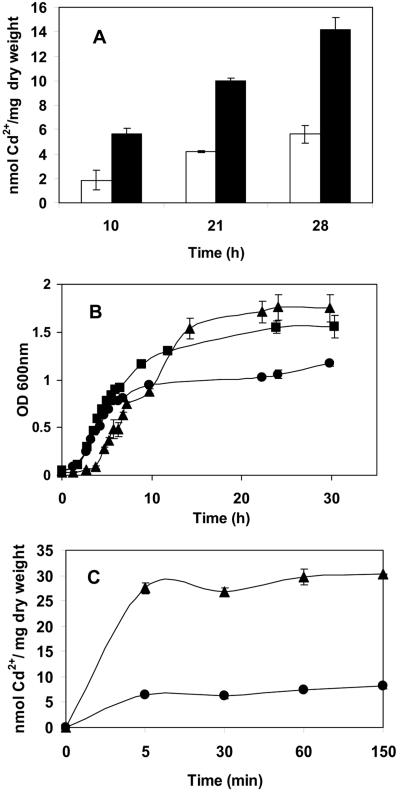
Experiments were performed with growing cultures to investigate the functionality of MBP-EC20 in whole-cell binding of Cd2+. Cultures of P. putida 06909/pVMC20 and the parental control strain were grown in the presence of 80 μM CdCl2, and whole-cell binding of Cd2+ was determined at 10, 21, and 28 h postinoculation. Expression of MBP-EC20 enabled the engineered strain to bind 14.20 ± 0.94 nmol Cd2+/mg (dry weight) of cells, a level 2.5-fold higher than that of the control strain at 28 h (Fig. 2A). More importantly, expression of MBP-EC20 also conferred increased cadmium resistance to the engineered strain. The final cell density for P. putida 06909/pVMC20 was 1.5-fold higher than the control strain grown in the presence of 80 μM CdCl2 (Fig. 2B). The specific growth rates before and after the addition of cadmium were compared (Table 1). Both strains grew at a similar rate until the addition of cadmium, when the growth rate for the wild type was twofold lower than the rate for the EC20-producing strain. This difference in cell growth was a direct result of cadmium toxicity, as the control strain grown in the absence of cadmium reached a final cell density similar to that of the recombinant strain grown in the presence of cadmium. Similar benefits in cell growth and cadmium binding were also observed in the presence of 300 μM CdCl2 (data not shown). These results suggest that presence of the EC20 moiety could effectively sequester intracellular Cd2+ ions and provide a competitive growth advantage to the engineered strain even in highly contaminated environments.
FIG. 2.
Cadmium-binding experiments with growing and resting cells. (A) Cadmium accumulation from growing cultures of P. putida 06909 (white bars) and P. putida 06909/pVMC20 (black bars) in the presence of 80 μM CdCl2. (B) Growth curves of P. putida 06909 in the presence of 80 μM CdCl2 (•), P. putida 06909/pVMC20 in the presence of 80 μM CdCl2 (▴), and P. putida 06909 without addition of cadmium (▪). Cells were grown in MJS medium, and cadmium was added at 5 h after inoculation. (C) Cadmium accumulation from resting cells of P. putida 06909 (•) and P. putida 06909/pVMC20 (▴). Data are presented as means ± the standard error of the mean (SEM) (P = 0.05; n = 3).
TABLE 1.
Specific growth rates of P. putida 06909 and recombinant P. putida 06909/pVMC20 before and after cadmium addition
| Strain | Specific growth rate (h−1)
|
|
|---|---|---|
| Pre-addition of Cd2+ (R2) | Post-addition of Cd2+ (R2) | |
| P. putida 06909 | 0.16 (0.99) | 0.06 (0.91) |
| P. putida 06909/pVMC20 | 0.17 (0.98) | 0.12 (0.98) |
Cadmium binding by resting cells.
To mimic the slow-growing state of P. putida in the rhizosphere, cadmium-binding experiments were also performed with resting cultures. Resting cells of P. putida 06909/pVMC20 bound cadmium rapidly, with 95% of maximum binding occurring with the first 5 min (Fig. 2C). Consistent with the result obtained with growing cultures, the maximum cadmium content of 30.77 ± 0.78 nmol Cd2+/mg (dry weight) of cells for P. putida 06909/pVMC20 was threefold higher than that of the control strain, 9.69 ± 0.27 nmol Cd2+/mg (dry weight) of cells, suggesting the enhanced binding effect by the EC20 moiety was sustained even under slow-growing conditions. The total amount of cadmium bound in the resting cells was larger than the cadmium bound in the growing cells. The difference may have been due to the inactivation of the cadmium export pump, cadA, in P. putida (15) in the resting culture. In the growing culture, the cells continuously exported cadmium; therefore, the resting cells accumulated more cadmium overall.
Selection of plant species using colonization efficiency.
P. putida 06909 is naturally found on citrus roots and is reported to have antipathogenic properties toward the white rot fungus (29). However, citrus trees are not suitable for rhizoremediation, due to their slow growth rate. To search for a faster-growing plant host able to establish a stable population of engineered P. putida 06909 strains, four species of plants—two dicotyledons, sunflower (Helianthus annuus) and cowpea (Vigna unguiculata), and two monocotyledons, wheat (Triticum sativum) and corn (Zea spp.)—were investigated for colonization efficiency. P. putida 06909/pRCD32 expressing a lacZ reporter gene (15) under control of a cadmium-inducible cadR promoter was used to determine the root colonization efficiency. The advantages of using P. putida 06909/pRCD32 were twofold. First, the number of cells that colonized the plant can be directly correlated to the presence of β-galactosidase activity in the plant root, which provided the relative colonization efficiencies among the four plant species. Second, the cadmium-inducible nature of the β-galactosidase activity elucidated whether cadmium in the hydroponic solution was available to the root-colonizing recombinant P. putida.
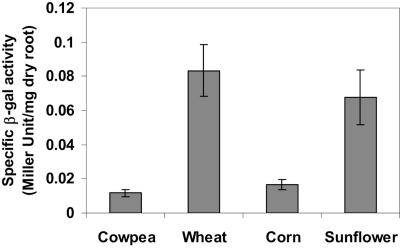
Roots of 5-day-old seedlings were dipped into overnight-grown cultures for 1 min before being returned to the hydroponic solution. After 4 days of colonization, the total bacterial population on the roots was extracted, and the β-galactosidase activity was assessed. By a comparison of the specific β-galactosidase activities (in Miller units per milligram [dry weight] of roots), sunflower and wheat seedlings were found to have the higher rates of colonization (Fig. 3). This result demonstrates that P. putida 06909 is a versatile root colonizer, capable of inhabiting the roots of both dicotyledon and monocotyledon plant species. The versatility of P. putida in adapting to a wide array of plant hosts will be advantageous for future rhizosphere remediation under different site and plant growth conditions. It has been reported that the sunflower seedling root exhibits longer taproots with wider and more extensive lateral branching than the wheat seedlings (28). Therefore, sunflower was chosen for the rhizoremediation studies, due to the large root system area and similarity to citrus, both being dicotyledons.
FIG. 3.
The root-colonizing efficiency of P. putida 06909 in four different plant species. A P. putida 06909 strain harboring pRDCD32 that contains a cadR promoter fusion with a promoterless lacZ gene was used to determine root colonization efficiency. Specific β-galactosidase activity from the root extract was used to indicate the number of colonizing bacteria. Data are presented as means ± SEM (P= 0.0102; n = 3).
Reduction of cadmium phytotoxicity on sunflower growth during hydroponic cultivation.
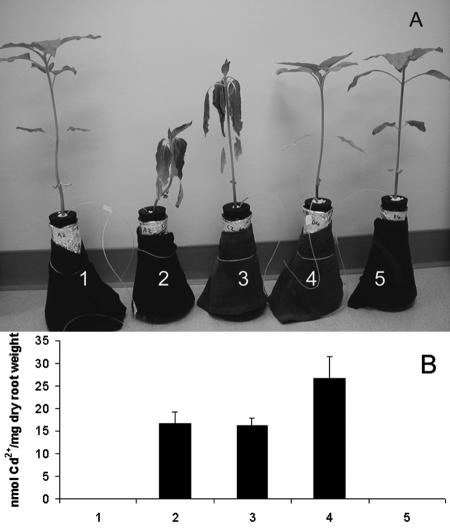
To demonstrate the effectiveness of the engineered P. putida 06909 in cadmium remediation when inoculated onto plant roots, hydroponic plant experiments with sunflower seedlings were performed with 80 μM CdCl2. The seedlings were grown for 14 days before inoculation with P. putida 06909/pVMC20 and the control strain. Sunflower seedlings without inoculation were also used for comparison. Sunflower growth was significantly altered in the presence of Cd2+, as plants without inoculation wilted (Fig. 4A). In contrast, plants inoculated with P. putida 06909/pVMC20, which can effectively sequester Cd2+, showed no signs of phytotoxicity and grew normally, as with plants grown in the absence of Cd2+. Addition of antibiotics to the hydroponic solution has no effect on plant growth, as demonstrated in treatments 1 and 5.
FIG. 4.
Rhizosphere cadmium-binding experiments in hydroponic solution. (A) Plant morphology of 27-day-old sunflower seedlings 3 days after being subjected to cadmium. Treatments 1 and 5 were uncontaminated controls grown with or without antibiotics, respectively, while 80 μM CdCl2 was added to treatments 2 (without inoculation), 3 (incoculated with P. putida 06909), and 4 (inoculated with P. putida 06909/pVMC20). (B) The amount of cadmium bound to the plant root from the same experiments. Data are presented as means ± SEM (P= 0.0286; n = 3).
Colonization of the sunflower roots with P. putida 06909/pVMC20 was verified by removing the bacterial biofilm from the sunflower roots and probing with an anti-MBP serum. A band of 50 kDa was detected from the root extracts inoculated with P. putida 06909/pVMC20 (Fig. 1), indicating the presence of the engineered strain in the sunflower roots, while plants inoculated with the control strain or without inoculation did not produce the expected band.
The amount of cadmium bound to the sunflower roots was determined for the different treatments. Plants without inoculants or inoculated with the control strain bound virtually the same amount of cadmium (Fig. 4B). Plants inoculated with P. putida 06909/pVMC20 bound 1.6-fold-more cadmium than the two controls. These results indicate that the engineered strain provides increased cadmium sequestration at the plant roots, conferring protective effect on sunflower growth in the presence of cadmium.
DISCUSSION
Symbiosis, from the Greek word sumbios, is defined as “living together.” From an ecological perspective, this occurs when several mutually beneficial species coexist in complementary niches. With the aid of biotechnology and genetic engineering, this symbiotic relationship between bacteria and plants has been exploited for in situ bioremediation of a wide range of organic pollutants such as parathion (3), trichloroethylene (24, 30), toluene (7), and PCBs (8, 27) using genetically engineered rhizobacteria or endophytic bacteria. However, very few reports today have attempted to address heavy-metal remediation using this symbiotic relationship. In one report, the Arabidopsis thaliana phytochelatin synthase gene (PCSAT) was expressed in a microsymbiont, Mesorhizobium huakuii subsp. rengei, which resides in the nodules of Astragalus sinicus (25) The symbiont expressing PC synthase was able to increase cadmium accumulation by 1.5 fold in the nodules. However, the quantity of PCs produced depended on the exposure to cadmium, making the amount of cadmium accumulated in the nodules inconsistent and unpredictable. In addition, the production of PCs by the microsymbiont M. huakuii utilizing the bacteroid-specific nifH promoter is restricted to nodules in legume plants. Therefore, the endosymbiont system may not be ideal for remediation under a range of site conditions. To apply the plant-microbe symbiotic system to a variety of environments, a flexible and hardy rhizobacterium-plant system is required.
In this paper, Pseudomonas putida 06909, a robust and versatile antifungal rhizosphere bacterium is engineered to produce MBP-EC20, a metal-binding peptide that has high affinity for cadmium. P. putida 06909 is also modestly cadmium resistant, due to the presence of an efflux pump in the metalloregulatory cad operon (15). Production of MBP-EC20 in P. putida 06909 not only enables enhanced cadmium binding but also protects the engineered strain and the colonized sunflower plants against the toxic effects of cadmium. These results demonstrate that a combination of enhanced microbial biosorption and plant-bacterium symbiosis is a promising strategy for heavy-metal cleanup. The increased resistance to cadmium by the engineered rhizobacteria even at a 300 μM concentration is particularly important, as it provides a competitive advantage to the engineered strain in the contaminated soil environment. This could be crucial for sustaining the growth of the engineered strain in the presence of the native bacterial population.
Unlike the approach reported by Valls et al.(26), which expresses MT in a heavy-metal-resistant soil bacterium, Ralstonia eutropha, resulting in a protective effect on plant growth when inoculated directly into cadmium-contaminated soil, our current strategy utilizes recombinant rhizobacteria that colonize the plant roots to engineer a symbiotic relationship between the rhizobacterium P. putida 06909 and the sunflower seedling. The plant roots sustain a stable bacterial population while drawing contaminated water containing soluble forms of the metals. As a result, the bacterium benefits from colonizing the sunflower roots and exhibits sufficient metabolic activity to produce EC20, which in turn aids the growth of the seedlings in high levels of cadmium by biosorbing and preventing the toxic cadmium from being transported into the plant. Thus, an engineered symbiosis is created between sunflower and the recombinant P. putida 06909. Another advantage of using rhizosphere bacteria is that the cadmium bound on the rhizobacterium could be removed by harvesting the plants. This feature is particularly important because unlike organic pollutants, which are enzymatically degraded, the sequestered heavy metals must be physically removed to remediate the site. This self-sustainable rhizobacterial population is likely to provide both long-term-growth plant protection and removal of cadmium. To study the long-term effect on soil cadmium remediation, chromosomal integration and constitutive production of EC20 in P. putida 06909 are currently in progress to eliminate the need for antibiotic selection and IPTG induction in preparation for rhizoremediation of cadmium in soil.
Another attractive feature of using rhizoremediation is the flexibility of utilizing different engineered rhizobacteria to remediate mixed-waste contaminated soil, as many superfund sites are cocontaminated with a myriad of organics and heavy metals (23). Since most organic-degrading microorganisms (e.g., Pseudomonas sp.) are sensitive to the toxic effects of heavy metals, a successful strategy to address this mixed-waste situation requires the use of microorganisms that will survive and thrive in soil polluted with heavy metals. Introducing the EC20 peptides into different root-colonizing bacteria that are engineered for organic degradation would endow them with both metal resistance and metal remediation capabilities. The rhizosphere bacterial community can be specifically engineered to target various pollutants at cocontaminated sites to provide a customized rhizoremediation system. The versatility of P. putida 06909 in adapting to different plant hosts and the ease in molecular manipulation prove to be invaluable attributes for designing plant-microbe remediation systems. Specific biodegradation genes and plant species can be selected in accordance to the pollutants present and plant growth conditions at the toxic sites. The strategy of simultaneous rhizoremediation of trichloroethylene and cadmium is currently under investigation.
Acknowledgments
This work was supported by a grant from the National Science Foundation (BES-0331416). C.H.W. was supported through a predoctoral traineeship from the University of California Toxic Substances Research and Teaching Program, Lead Campus Program, and a graduate fellowship from the Greater Research Opportunity program of the U.S. EPA (MA-91634801).
We gratefully thank Don Cooksey for supplying the P. putida 06909 strains and David Parker for providing valuable assistance with the hydroponic experiments. We would like to acknowledge U Loi Lao for helping with the statistical analysis.
REFERENCES
- 1.Alami, Y., W. Achouak, C. Marol, and T. Heulin. 2000. Rhizosphere soil aggregation and plant growth promotion of sunflowers by an exopolysaccharide-producing Rhizobium sp. strain isolated from sunflower roots. Appl. Environ. Microbiol. 66:3393-3398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Anderson, T. A., and J. R. Coats. 1995. An overview of microbial degradation in the rhizosphere and its implication for bioremediation, p. 135-143. In H. D. Skipper and R. F. Turco (ed.), Bioremediation: science and applications, vol. 43. Soil Science Society of America, Madison, Wis. [Google Scholar]
- 3.Anderson, T. A., E. A. Guthrie, and B. T. Walton. 1993. Bioremediation in the rhizosphere. Environ. Sci. Technol. 27:2630-2636. [Google Scholar]
- 4.Bae, W., W. Chen, A. Mulchandani, and R. Mehra. 2000. Enhanced bioaccumulation of heavy metals by bacterial cells displaying synthetic phytochelatins. Biotechnol. Bioeng. 70:518-523. [DOI] [PubMed] [Google Scholar]
- 5.Bae, W., R. K. Mehra, A. Mulchandani, and W. Chen. 2001. Genetic engineering of Escherichia coli for enhanced uptake and bioaccumulation of mercury. Appl. Environ. Microbiol. 67:5335-5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bae, W., C. H. Wu, J. Kostal, A. Mulchandani, and W. Chen. 2003. Enhanced mercury biosorption by bacterial cells with surface-displayed MerR. Appl. Environ. Microbiol. 69:3176-3180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barac, T., S. Taghavi, B. Borremans, A. Provoost, L. Oeyen, J. V. Colpaert, J. Vangronsveld, and D. van der Lelie. 2004. Engineered endophytic bacteria improve phytoremediation of water-soluble, volatile organic pollutants. Nat. Biotechnol. 22:583-588. [DOI] [PubMed] [Google Scholar]
- 8.Brazil, G. M., L. Kenefick, M. Callanan, A. Haro, V. de Lorenzo, D. N. Dowling, and F. O'Gara. 1995. Construction of a rhizosphere pseudomonad with potential to degrade polychlorinated biphenyls and detection of bph gene expression in the rhizosphere. Appl. Environ. Microbiol. 61:1946-1952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chabot, R., H. Antoun, and M. P. Cescas. 1996. Growth promotion of maize and lettuce by phosphate-solubilizing Rhizobium leguminosarum biovar phaseoli. Plant Soil 184:311-321. [Google Scholar]
- 10.de Lorenzo, V., L. Eltis, B. Kesslar, and K. N. Timmis. 1993. Analysis of Pseudomonas gene products using lacIq/Ptrp-lac plasmids and transposons that confer conditional phenotypes. Gene 123:17-24. [DOI] [PubMed] [Google Scholar]
- 11.Dennis, J. J., and P. A. Sokol. 1995. Electrotransformation of Pseudomonas, p. 125-133. In J. A. Nickoloff (ed.), Methods in molecular biology, vol. 47. Humana Press, Totowa, N.J. [DOI] [PubMed] [Google Scholar]
- 12.Eccles, H. 1999. Treatment of metal-contaminated wastes: why select a biological process? Trends Biotechnol. 17:462-465. [DOI] [PubMed] [Google Scholar]
- 13.Gadd, G. M. 2000. Bioremedial potential of microbial mechanisms of metal mobilization and immobilization. Curr. Opin. Biotechnol. 11:271-279. [DOI] [PubMed] [Google Scholar]
- 14.Kuiper, I., E. L. Lagendijk, G. V. Bloemberg, and B. J. J. Lugtenberg. 2004. Rhizoremediation: a beneficial plant-microbe interaction. Mol. Plant-Microbe Interact. 17:6-15. [DOI] [PubMed] [Google Scholar]
- 15.Lee, S. W., E. Glickmann, and D. A. Cooksey. 2001. Chromosomal locus for cadmium resistance in Pseudomonas putida consisting of a cadmium-transporting ATPase and a MerR family response regulator. Appl. Environ. Microbiol. 67:1437-1444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malik, A. 2004. Metal bioremediation through growing cells. Environ. Int. 30:261-278. [DOI] [PubMed] [Google Scholar]
- 17.Mehra, R. K., and P. Mulchandani. 1995. Glutathione-mediated transfer of Cu(I) into phytochelatins. Biochem. J. 307:697-705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mejare, M., and L. Bulow. 2001. Metal-binding proteins and peptides in bioremediation and phytoremediation of heavy metals. Trends Biotechnol. 19:67-73. [DOI] [PubMed] [Google Scholar]
- 19.Miller, J. H. 1972. Experiments in molecular genetics. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 20.Pedler, J. F., D. R. Parker, and D. E. Crowley. 2000. Zinc deficiency-induced phytosiderophore release by the Triticaceae is not consistently expressed in solution culture. Planta 211:120-126. [DOI] [PubMed] [Google Scholar]
- 21.Peters, R. W. 1999. Chelant extraction of heavy metals from contaminated soils. J. Hazard. Mater. 66:151-210. [DOI] [PubMed] [Google Scholar]
- 22.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 23.Sandrin, T. R., and R. M. Maier. 2003. Impact of metals on the biodegradation of organic pollutants. Environ. Health Perspect. 111:1093-1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shim, H., S. Chauhan, D. Ryoo, K. Bowers, S. M. Thomas, K. A. Canada, J. G. Burken, and T. K. Wood. 2000. Rhizosphere competitiveness of trichloroethylene-degrading poplar-colonizing recombinant bacteria. Appl. Environ. Microbiol. 66:4673-4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sriprang, R., M. Hayashi, H. Ono, M. Takagi, K. Hirata, and Y. Murooka. 2003. Enhanced accumulation of Cd2+ by a Mesorhizobium sp. transformed with a gene from Arabidopsis thaliana coding for phytochelatin synthase. Appl. Environ. Microbiol. 69:1791-1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Valls, M., S. Atrain, V. de Lorenzo, and L. A. Fernandex. 2001. Engineering a mouse metallothionein on the cell surface of Ralstonia eutropha CH34 for immobilization of heavy metals in soil. Nat. Biotechnol. 18:661-665. [DOI] [PubMed] [Google Scholar]
- 27.Villacieros, M., C. Whelan, M. Mackova, J. Molgaard, M. Sanchez-Contreras, J. Lloret, D. Aguirre de Carcer, R. I. Oruezabal, L. Bolanos, T. Macek, U. Karlson, D. N. Dowling, M. Martin, and R. Rivilla. 2005. Polychlorinated biphenyl rhizoremediation by Pseudomonas fluorescens F113 derivatives, using a Sinorhizobium meliloti nod system to drive bph gene expression. Appl. Environ. Microbiol. 71:2687-2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Weaver, J. E. 1926. Root development of field crops. McGraw-Hill Book Company, Inc., New York, N.Y.
- 29.Yang, C., J. A. Menge, and D. A. Cooksey. 1994. Mutations affecting hyphal colonization and pyoverdine production in pseudomonads antagonistic toward Phytophthora parasitica. Appl. Environ. Microbiol. 60:473-481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yee, D. C., J. A. Maynard, and T. K. Wood. 1998. Rhizoremediation of trichloroethylene by a recombinant, root-colonizing Pseudomonas fluorescens strain expressing toluene ortho-monooxygenase constitutively. Appl. Environ. Microbiol. 64:112-118. [DOI] [PMC free article] [PubMed] [Google Scholar]