Abstract
The hyperthermophilic bacterium Thermotoga maritima has shared many genes with archaea through horizontal gene transfer. Several of these encode putative oligopeptide ATP binding cassette (ABC) transporters. We sought to test the hypothesis that these transporters actually transport sugars by measuring the substrate affinities of their encoded substrate-binding proteins (SBPs). This information will increase our understanding of the selective pressures that allowed this organism to retain these archaeal homologs. By measuring changes in intrinsic fluorescence of these SBPs in response to exposure to various sugars, we found that five of the eight proteins examined bind to sugars. We could not identify the ligands of the SBPs TM0460, TM1150, and TM1199. The ligands for the archaeal SBPs are TM0031 (BglE), the β-glucosides cellobiose and laminaribiose; TM0071 (XloE), xylobiose and xylotriose; TM0300 (GloE), large glucose oligosaccharides represented by xyloglucans; TM1223 (ManE), β-1,4-mannobiose; and TM1226 (ManD), β-1,4-mannobiose, β-1,4-mannotriose, β-1,4-mannotetraose, β-1,4-galactosyl mannobiose, and cellobiose. For comparison, seven bacterial putative sugar-binding proteins were examined and ligands for three (TM0595, TM0810, and TM1855) were not identified. The ligands for these bacterial SBPs are TM0114 (XylE), xylose; TM0418 (InoE), myo-inositol; TM0432 (AguE), α-1,4-digalactouronic acid; and TM0958 (RbsB), ribose. We found that T. maritima does not grow on several complex polypeptide mixtures as sole sources of carbon and nitrogen, so it is unlikely that these archaeal ABC transporters are used primarily for oligopeptide transport. Since these SBPs bind oligosaccharides with micromolar to nanomolar affinities, we propose that they are used primarily for oligosaccharide transport.
The release of the complete genome sequence of the hyperthermophilic bacterium Thermotoga maritima highlighted the possibility that extensive horizontal gene transfer (HGT) had occurred between T. maritima and archaea (34). This proposal met with considerable discussion in the literature (15, 26, 27). Subsequent analyses demonstrated that several archaeal genes have been inherited by T. maritima and other members of the order Thermotogales and that these genes encode proteins involved in a variety of physiological processes (5, 36, 37, 56).
Among the assigned T. maritima genes that most closely match archaeal genes, 37% are thought to encode transporters (34). ATP binding cassette (ABC) transporters are the largest class of transporters in T. maritima (43). Six putative oligopeptide ABC transporter operons may have been shared with archaea (34). Although no rigorous phylogenetic analysis describing the evolution of these transporters has been published, we will refer to these transporter proteins as archaeal since their sequences are most similar to those from members of the Archaea. These clusters of genes provide excellent subjects to investigate how these genes, perhaps acquired from archaea, came to function in a bacterium.
Archaea transcribe their genes by using promoters unlike those found in bacteria (19). Interdomain HGT events, such as those that involve ABC transporters, raise questions about how genes derived from a foreign transcriptional system are expressed in a new host. Archaeal genes that now function in Thermotogales may have recombined behind existing bacterial promoters to drive their expression. The selective pressures that allowed their expression and stable maintenance at some sites while cleansing genomes of newly recombined genes at other sites are unknown. We are attempting to understand the evolutionary constraints on interdomain HGT events in Thermotogales and to determine how these ABC transporter genes have integrated themselves into the transcriptional and metabolic processes of their new hosts.
To better understand the functional amelioration of these transporters into the physiology of their bacterial hosts, we must first determine their current roles. Their original assignment as oligopeptide transporters probably does not accurately describe their functions. Similarly assigned genes in Sulfolobus acidocaldarius and Pyrococcus furiosus were later found to encode sugar transports (17, 25). These operons in T. maritima lie among genes encoding sugar hydrolases and so likely encode sugar transporters too. We set out to test this hypothesis by determining the binding properties of the substrate-binding proteins (SBPs) encoded in these operons. The T. maritima genome carries several other genes that may also encode sugar ABC transporters, and most of these are apparently of bacterial origin. To better appreciate the physiological environment in which these archaeal transporters operate, it is of interest to know if they have the same substrate affinities as the bacterial sugar transporters. Therefore, we also determined the binding properties of several bacterial SBPs. Finally, to corroborate the proposed functions of all of these ABC transporters, we correlated the results of our binding studies with the results of published studies of expression of the transporter genes and related sugar hydrolase genes in response to different growth conditions.
MATERIALS AND METHODS
Chemicals.
The carbohydrates used in these studies are listed in Table S1 in the supplemental material and were purchased from Sigma Chemical Company (St. Louis, MO), Seikagaku of America (Associates of Cape Cod, Inc., East Falmouth, MA), ICN Biomedicals, Inc. (Irvine, CA), and Megazyme International Ireland, Ltd. (County Wicklow, Ireland). Unless otherwise noted, all carbohydrates were 99% minimum purity.
Growth of T. maritima on peptides.
Cells of T. maritima were grown anaerobically in a defined medium lacking ammonium chloride as a source of fixed nitrogen. The medium contained (per liter) 0.05 g CaCl2, 0.05 g K2HPO4, 1 g KCl, 0.5 g MgSO4 · 7H2O, 20 g NaCl, 4.8 g HEPES, 0.5 g cysteine-HCl, 0.007 g FeSO4 · 7H2O, 0.0033 g NaWO4 · 2H2O, 2.5 g Na2O3S2, and 0.001 g resazurin. Vitamins were added at the concentrations described in a previously published formulation (11). The medium was adjusted to pH 7.5 with sodium hydroxide and dispensed into anaerobic tubes under a nitrogen atmosphere. Stock solutions of peptide and amino acid sources (Bacto peptone [Difco], tryptone [Fisher Biotech], Phytone [BBL], and polypeptone [BBL]) were sterilized by autoclave, except Phytone, which was sterilized by filtration. Aliquots of these stock solutions were added to the basal medium to a final concentration of 0.5% (vol/vol). A 1% inoculum was used, and cultures were incubated at 77°C. Growth was monitored at 600 nm for 48 h. Negative-control cultures had no additions from these stock solutions, and positive controls contained 0.5% (vol/vol) maltose and 0.05 g/liter NH4Cl.
Cloning, expression, and purification of substrate-binding proteins.
The SBP-encoding genes were amplified from T. maritima genomic DNA by use of the synthetic oligonucleotide primers shown in Table S2 in the supplemental material and PCR with Pfu or Platinum Pfx (Invitrogen) DNA polymerase. The PCR primers were designed so that the nucleotide sequences encoding presumed signal peptides were not incorporated into the PCR products (40).
The PCR products were ligated into the pGEMeasy vector (Promega Corp.) and then digested with the appropriate restriction endonucleases (see Table S2 in the supplemental material). The digested PCR products were subcloned into either pET15b (Novagen) or pQE-30 (QIAGEN) expression vectors, which were previously digested with the corresponding restriction endonucleases. PCR products of most open reading frames (ORFs) were ligated into pET15b. ORFs TM0958 and TM0801 were ligated into pQE-30 (QIAGEN). Constructs in pET15b and pQE-30 were transformed into competent cells of Escherichia coli strain BL21(DE3) by electroporation. The PCR products of ORFs TM0460, TM1199, and TM1226 were ligated into pQE-30Xa (QIAGEN) and transformed into strain M15 pRep4 (QIAGEN).
For each construct, a single colony of the transformed cells was inoculated into 10 ml LB medium with 50 μg/ml ampicillin (and 25 μg/ml kanamycin for strain M15 pRep4) and grown at 37°C to an optical density at 600 nm of 0.3. The cells were harvested by centrifugation, and the cell pellets were inoculated into fresh medium (100 ml LB with 50 μg/ml ampicillin) and incubated with shaking at 37°C. When the culture reached an optical density at 600 nm of 0.5, expression of the cloned gene was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) to 1 mM and the cells were incubated for another 4 h at 37°C with shaking. The cells were then harvested by centrifugation (5,500 × g) for 20 min. Cell pellets were stored at −20°C. Some SBPs did not bind to any substrates initially, so cells expressing those genes were grown in M9 minimal medium with 0.4% (wt/vol) glucose to prevent binding of sugars in the LB medium to the expressed SBPs. The soluble recombinant protein was purified by nickel chelation chromatography using Ni-nitrilotriacetic acid resin (Novagen) according to the manufacturer's instructions. The pure protein was dialyzed against 15 liters of 50 mM sodium phosphate buffer (pH 7.5) at 4°C to remove imidazole. The dialyzed samples were stored at 4°C.
Protein concentration determinations.
The molar extinction coefficients of the SBPs were determined from their predicted amino acid sequences (20). Protein concentrations were determined by measuring the absorbance of solutions at 280 nm and using the molar extinction coefficients calculated from their amino acid compositions.
Partial hydrolyzation of polysaccharides.
The polysaccharides listed in Table S1 in the supplemental material were partially hydrolyzed by suspending 0.5 g of the sugar in 5 ml 50 mM H2SO4 and heating the suspension for 12 h at 100°C in a sealed glass tube. The mixture was cooled, and suspended solids were removed by centrifugation at 13,000 × g. The clarified supernatant was neutralized with a measured amount of 1 M sodium phosphate, pH 7.5, and stored at 4°C.
Fluorescence spectroscopy.
All fluorescence spectroscopic measurements were performed using an SLM Aminco-Bowman 2 spectrofluorometer. During each experiment, the temperature of the cuvette was held constant at 20°C. Fluorescence emission spectra were measured at an excitation wavelength of 280 nm. Emission intensities were measured at 300 to 360 nm in the presence (up to a final concentration of 1 mM) and absence of the sugar ligand. The excitation and emission slit widths were 1 nm and 8 nm, respectively. Dissociation constants were measured in a solution containing 0.3 μM binding protein, 50 mM sodium phosphate (pH 7.5), and 300 mM NaCl at 20°C (16). Relative fluorescence (F/F0) was plotted against carbohydrate concentration (L0), and the dissociation constant (Kd) was determined by nonlinear fitting of the data to the equation F = F0 + ΔF/2P0{(Kd + P0 + L0) − [(Kd + P0 + L0)2 − 4L0P0]1/2}, where F is the measured fluorescence of the protein in the presence of the ligand, F0 is the fluorescence of the ligand-free protein, ΔF is the change in fluorescence at saturation of the protein with the ligand, P0 and L0 are the concentrations of the protein and ligand, respectively, and Kd is the dissociation constant. The data were fitted using the Marquardt-Levenberg algorithm.
Determination of ribose binding.
The affinity of the putative ribose-binding protein TM0958 for radiolabeled ribose was measured by precipitating the protein-ligand complex with ammonium sulfate and binding this complex to cellulose nitrate filters for scintillation counting (47). Glass tubes (12 by 100 mm) containing 50 μl (150 μg) of TM0958 (3 mg/ml) and 10 μl water or a solution of unlabeled sugar were preheated in a heating block at 55°C for 30 s. This temperature was used because previous binding studies showed that rapid evaporation of water in the tubes at higher temperatures did not allow reproducible sampling (32). A 10-μl aliquot of 7 μM α-d-[3H]ribose (50 μCi μmol−1; ICN Chemicals) was rapidly added to the samples with a micropipette. For binding-constant determinations, solutions containing different concentrations of radiolabeled ribose were added to separate tubes. After an additional 30 s at 55°C, 2 ml ice-cold, saturated ammonium sulfate in 50 mM Tris-HCl (pH 7.5) was added to the samples and the tubes were transferred to an ice bath for at least 10 min. The contents of the tubes were then filtered through Whatman cellulose nitrate membrane filters (25 mm diameter, 0.45 μm porosity) and washed with an additional 2 ml ice-cold, saturated ammonium sulfate in 50 mM Tris-HCl (pH 7.5). The filters were then placed in scintillation fluid (Optifluor; Packard) to be counted in a Beckman LS3801 liquid scintillation counter. For ligand competition experiments, a 100-fold excess of unlabeled ribose was added prior to the addition of radiolabeled ribose.
RESULTS
Oligopeptides alone do not support growth of T. maritima.
The archaeal ORFs listed in Table 1 were annotated as oligopeptide transporters, based solely on sequence similarities and not on experimental evidence (34). T. maritima has not been reported to grow using polypeptides as sole sources of carbon and nitrogen, so we tested this phenotype. We measured growth after 48 h (given here as optical densities at 600 nm, averages of two measurements) on various peptide sources and found that Bacto peptone (0.066), tryptone (0.070), and polypeptone (0.089) did not support growth based upon a comparison with measurements of cell densities following growth in a medium lacking ammonium chloride and a carbon source (0.056). Maltose medium with ammonium chloride supported good growth (0.757) and Phytone-containing medium supported reasonable growth (0.287). Phytone, like yeast extract, contains significant amounts of carbohydrate, so this likely allowed those cells to grow. The sources of peptides used in these studies are reported to contain the following amounts of total carbohydrates (in mg/g): Phytone peptone, 392.9; Bacto peptone, 6.29; polypeptone peptone, 8.06; and Bacto tryptone, 4.30 (Bionutrient technical manual, 2nd ed., BD Diagnostics, Franklin Lakes, N.J., 2004). T. maritima may enhance its growth on sugars by using oligopeptides, but this large number of putative oligopeptide transporters seems unnecessary for that purpose (14).
TABLE 1.
Substrate specificities and affinities of substrate-binding proteins as determined by fluorescence spectroscopy
| Type of SBP | Locus | Substrate(s) bound | Kd (μM)a | Neighboring enzyme
|
|
|---|---|---|---|---|---|
| Locus | Activityb | ||||
| Archaea-related oligosaccharide SBPsg | TM0031 (bglE) | Cellobiose | 0.9 ± 0.2 | TM0024 | Laminarinase (LamA, extracellular) |
| Laminaribiose | 0.8 ± 0.2 | TM0025 | β-Glucosidase (BglB, cytoplasmic) | ||
| TM0071 (xloE) | Xylobiosec | + | TM0061 | Endo-β-1,4-xylanase A (XylA, extracellular) | |
| Xylotriosec | TM0069 | Mannonate hydrolase (cytoplasmic) | |||
| TM0070 | Endo-β-1,4-xylanase B (XynB, extracellular) | ||||
| TM0076 | Xylosidase (XloA, cytoplasmic) | ||||
| TM0300 (gloE) | Xyloglucan hepta-, octa-, and nonasaccharides with β-1,4 tetraglucosyl backbones | + | TM0305 | Endoglucanase/xyloglucanase (extracellular) | |
| TM0306 | α-Fucosidase (cytoplasmic) | ||||
| TM0308 | α-Xylosidase (XylQ, cytoplasmic) | ||||
| TM0310 | β-Galactosidase (cytoplasmic) | ||||
| TM0460 | No tested sugars bind | None | |||
| TM1150 | No tested sugars bind | None | |||
| TM1199 | No tested sugars bind (pectin hydrolysate) | TM1192 | α-Galactosidase (cytoplasmic) | ||
| TM1193 | β-Galactosidase (cytoplasmic) | ||||
| TM1195 | β-Galactosidase (cytoplasmic) | ||||
| TM1201 | Arabinogalactan endo-β-1,4-galactosidase (extracellular) | ||||
| TM1223 (manE) | β-1,4-Mannobiose | 13 ± 1 | TM1227 | Endo-β-1,4-mannosidase (extracellular) | |
| TM1226 (manD) | β-1,4-Mannobiose | 15 ± 2 | TM1227 | Endo-β-1,4-mannosidase (extracellular) | |
| β-1,4-Mannotriose | 1.05 ± 0.4 | ||||
| β-1,4-Mannotetraose | 0.38 ± 0.1 | ||||
| β-1,4-Galactosyl mannobiose | 10 ± 1 | ||||
| Cellobiose | 9.5 ± 1 | ||||
| Sugar SBPs | TM0114 (xylE) | Xylose | 0.0135 ± 0.001 | TM0113 | Xylanase (XylU, extracellular) |
| TM0116 | Putative xylulokinase | ||||
| TM0418 (inoE) | myo-Inositol | 24.0 ± 1 | None | ||
| TM0432 (aguE) | α-1,4-Digalacturonic acid | 0.25 ± 0.05 | TM0433 | Pectate lyase (PelA, extracellular) | |
| TM0434 | α-Glucuronidase (Agu, cytoplasmic) | ||||
| TM0437 | Exo-α-galacturonidase (cytoplasmic) | ||||
| TM0595 | No tested sugars bind | None | |||
| TM0810 | No tested sugars bind | TM0809d | β-N-Acetylglucosaminidase (cytoplasmic) | ||
| TM0958 (rbsB) | Ribose | 0.4 ± 0.5e | None | ||
| TM1204 (malE1)f | Maltose | 24 ± 1 | TM1192 | α-Galactosidase (cytoplasmic) | |
| Maltotriose | 0.008 ± 0.0005 | TM1193 | β-Galactosidase (cytoplasmic) | ||
| β-1,4-Mannotetraose | 38 ± 1 | TM1195 | β-Galactosidase (cytoplasmic) | ||
| TM1201 | Arabinogalactan endo-β-1,4-galactosidase (extracellular) | ||||
| TM1839 (malE2)f | Maltose | 8.4 ± 1 | TM1834 | α-Glucosidase (cytoplasmic) | |
| Maltotriose | 11 ± 1.5 | TM1835 | Cyclomaltodextrinase (cytoplasmic) | ||
| Trehalose | 9.5 ± 1 | TM1840 | α-Amylase (extracellular) | ||
| TM1845 | Pullulanase (extracellular) | ||||
| TM1855 | No tested sugars bind (xanthan gum) | TM1851 | α-Mannosidase (cytosolic) | ||
+, a change in fluorescence is detectable, but the data do not fit a simple model to determine a Kd value. Values are shown ±1 standard deviation. Dissociation constants were measured at 20°C, except for that of RbsB, which was measured at 55°C.
Enzymes whose genes encode potential signal peptides are listed as “extracellular,” while those that do not are listed as “cytoplasmic.”
Ninety-five-percent pure; remainder, other xylooligosaccharides.
TM0809 is similar (E value of 10−178) to a β-N-acetylglucosaminidase from T. neapolitana (CbsA, GenBank accession number AF34391).
Value determined by equilibrium dialysis using radiolabeled ribose, as described in the text.
Data for substrate affinities for MalE1 and MalE2 were published previously (33).
Annotated as oligopeptide SBPs.
The archaeal putative oligopeptide-binding proteins bind oligosaccharides.
Several of the binding protein-encoding genes listed in Table 1 are among those that are said to have arrived in the T. maritima genome or that of its ancestors via HGT from Archaea (34). Most were annotated as encoding oligopeptide-binding proteins, but TM0418 was called a sugar-binding protein-encoding gene. Another ORF, TM0460, has a sequence similar to those of some putative archaeal oligopeptide transporter genes. Although there is no evidence for its inheritance by horizontal transfer, we included it in our analyses for completeness.
We measured ligand binding by recombinant proteins encoded by these oligopeptide SBP genes and found that several of them bind sugars (Table 1). Over 40 sugars, including monosaccharides, oligosaccharides, polysaccharides, and neutralized polysaccharide hydrolysates, were used to test these proteins for ligand binding (see Table S1 in the supplemental material). We identified substrates for five of the eight archaeal SBPs that we examined and found that the ligand specificities of those proteins correlate well with the specificities of sugar hydrolases encoded near their genes. We did not attempt to measure oligopeptide binding by these proteins, so we cannot rule out the possibility that they also bind oligopeptides. However, we will hereafter refer to these proteins as oligosaccharide-binding proteins.
Based upon the measured affinities of the SBPs for sugars, we propose that the SBP-encoding genes be named to describe their substrate affinities in accord with accepted rules of genetic nomenclature (13). Since genes encoding ABC transporters are typically clustered in operons encoding two or three of the transporter proteins (SBP, membrane-spanning protein [MSP], and ATP-binding protein [ABP]), we propose naming the adjacent putative MSP- and ABP-encoding genes as well. We used the E. coli mal operon as our template because that nomenclature leaves the letters A to D available for designating associated sugar hydrolase genes. In several cases, some of those letters have already been so assigned. The letters K and L were chosen for the ABP-encoding genes because K is often used for ABP genes. Since several of the T. maritima ABC operons contain two ABP genes, we chose to use L to designate the second gene instead of using the nonstandard genetic nomenclature K1 and K2. Nonstandard nomenclature was retained for the two T. maritima mal operons because it is already widely used in the literature. Our proposed gene names and their rationales are summarized in Table 2 below. A recently proposed ABC transporter gene nomenclature (12) is cross-indexed there. The nomenclature proposed by Conners et al. (12) used nonstandard gene symbols (13) and does not reflect some of the substrates that we found to be bound by these SBPs.
TABLE 2.
Proposed nomenclature for genes encoding ABC transporters, based upon substrate specificities of binding proteins
| Transporter locus | Annotation | Directiona | Gene name proposed here | Mnemonice | Rationale | Nomenclature proposed by Conners et al. (12) |
|---|---|---|---|---|---|---|
| TM0027 | ABP | bglL | β-Glucoside transport | Abbreviation of substrates and TM0025, bglB (β-glucosidase [34]) | bgtpE | |
| TM0028 | ABP | bglK | bgtpD | |||
| TM0029 | MSP | bglG | bgtpC | |||
| TM0030 | MSP | ↑ | bglF | bgtpB | ||
| TM0031 | SBP | bglE | bgtpA | |||
| TM0071 | SBP | xloE | Xylose oligosaccharide transport | TM0076, xloA (xylosidase [homolog of GenBank U58632.2]) | xtpAb | |
| TM0072 | MSP | xloF | xtpB | |||
| TM0073 | MSP | xloG | xtpC | |||
| TM0074 | ABP | ↓ | xloK | xtpD | ||
| TM0075 | ABP | xloL | xtpF | |||
| TM0112 | MSP | xylF | Xylose transport | Abbreviation of substrate | rbsC2b | |
| TM0114 | SBP | ↓ | xylE | rbsA2 | ||
| TM0115 | ABP | xylK | rbsB2 | |||
| TM0300 | SBP | gloE | Glucose oligosaccharide transport | Abbreviation of substrate | dppAb | |
| TM0301 | MSP | gloF | dppB | |||
| TM0302 | MSP | gloG | dppC | |||
| TM0303 | ABP | ↓ | gloK | dppD | ||
| TM0304 | ABP | gloL | dppF | |||
| TM0418 | SBP | inoE | myo-Inositol transport | TM1419, ino1 (myo-inositol-1-P-synthase [1]) | malE3b | |
| TM0419 | MSP | inoF | malF3 | |||
| TM0420 | MSP | ↓ | inoG | ugpE | ||
| TM0421 | ABP | inoK | malK2 | |||
| TM0430 | MSP | aguG | α-Galactouronate transport | TM0434, agu (α-glucuronidase [53]) | ugpBb | |
| TM0431 | MSP | ↑ | aguF | ugpA | ||
| TM0432 | SBP | aguE | ugpE | |||
| TM0955 | MSP | rbsC | Ribose transport | Annotated by TIGR (34) | rbsC1 | |
| TM0956 | ABP | rbsA | rbsA1 | |||
| TM0958 | SBP | ↑ | rbsB | rbsB1 | ||
| TM0959 | MSP | rbsD | rbsD | |||
| TM1202 | MSP | malG1 | Maltose transportc | Annotated by TIGR (34) | malG1 | |
| TM1203 | MSP | ↑ | malF1 | malF1 | ||
| TM1204 | SBP | malE1 | malE1 | |||
| TM1219 | ABP | manL | Mannoside transport | Abbreviation of substrate | cbtF | |
| TM1220 | ABP | manK | cbtD | |||
| TM1221 | MSP | manG | cbtC | |||
| TM1222 | MSP | manF | cbtB | |||
| TM1223 | SBP | ↑ | manE | cbtA | ||
| TM1226 | SBP | manD | mbtA | |||
| TM1836 | MSP | malG2 | Maltose transport | Annotated by TIGR (34) | malF2 | |
| TM1837d | MSP | ↑ | malF2 | Unnamed | ||
| TM1839 | SBP | malE2 | malE2 |
Arrows indicate directions of transcription of the genes.
The correspondence between the locus number and the mnemonic for these operons was inferred from the information in Table 2 of reference 12.
The mnemonic is retained even though data indicate MalE1 is involved in mannooligosaccharide transport (33).
Contains an authentic frameshift.
Mnemonics are underlined.
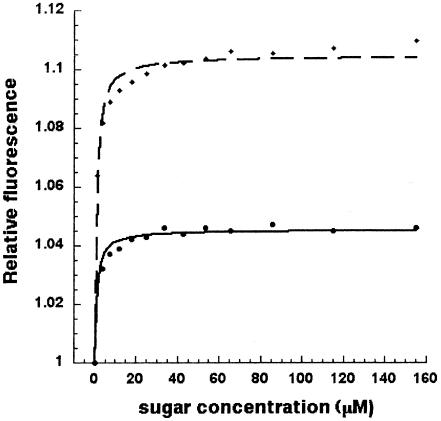
The ORFs TM0027 to TM0031 appear to encode a β-glucoside transporter (bgl, abbreviation of the glucoside substrates and name of a nearby gene, bglB). BglE (TM0031) binds the β-glucose disaccharides cellobiose and laminaribiose with equal affinities (Fig. 1). Genes adjacent to bglE encode enzymes that hydrolyze cellobiose (BglB, TM0025) and laminaribiose (LamA, TM0024) (57, 58). BglE, in contrast to ManD (see below), appears to bind only glucans, which are also the substrates for the adjacently encoded enzymes.
FIG. 1.
Relative fluorescence of BglE in response to the addition of increasing amounts of laminaribiose (dashed line) and cellobiose (solid line). The protein concentration was 0.3 μM in 50 mM sodium phosphate (pH 7.0). For this measurement, the excitation wavelength was 280 nm and the emission wavelength was 335 nm.
The transporters encoded by TM0071 to TM0075 (xlo, abbreviation of the xylose oligosaccharide substrate and the adjacent xloA gene) and TM0112, TM0114, and TM0115 (xyl, xylose, as named for E. coli [49]) transport similar substrates. The Xyl transporter is described below. Both XloE (TM0071) and GloE (TM0300) bind xylosides, but XloE binds xylans while GloE appears to bind larger xyloglucans (see below). We were unable to measure the affinity constants for XloE and GloE because the available xylosides are mixtures of sugars. We estimate that their affinities are in the micromolar range, based upon responses observed during titrations with these mixtures. The substrate specificity of XloE resembles that of Streptomyces thermoviolaceus BxlE, a xylobiose- and xylotriose-binding SBP. BlxE binds xylans from 2 to 6 residues long, with highest affinities (in the nanomolar range) for xylobiose and xylotriose (54). Two putative periplasmic endoxylanases (XylA [TM0061] and XynB [TM0070]) likely produce short-chain xylans that can be transported by XloEFGKL.
Although the specific ligands of the SBP encoded in the TM0300 to TM0304 transporter (glo, glucose oligosaccharides) were not among those we tested, GloE bound only β-1,4 tetraglucosyl xyloglucans and not cellobiose (β-1,4-Glu-Glu) or isoprimeverose (α-1,6-Xyl-Glu). A clue to its native substrates may lie in the substrate specificity of TM0305 (Cel74), a periplasmic endoglucanase that hydrolyzes β-glucans (including xyloglucans) into oligosaccharides six or more units in size (8). GloE may facilitate transport of large β-linked glucose oligosaccharides released by the activity of TM0305. Consequently, we may have observed binding only to our xyloglucans because these were the only purified β-glucans we used that were large enough to be recognized by GloE.
The genes manE and manD are separated by genes encoding a putative transcriptional regulator (TM1224) and an unknown protein (TM1225). manE and manD are highly homologous genes, one of which likely arose by a gene duplication or an acquisition by HGT. These genes and the adjoining ORFs (TM1219 to TM1222) appear to encode a mannoside (man) transporter. These two SBPs have diverged in function, as shown by their different substrate affinities. Both bind β-1,4-mannobiose, but ManD binds several longer mannosides and cellobiose. ManD has a 10-fold-lower affinity for cellobiose than BglE. Both ManE and ManD bind β-1,4 pyranose sugars, but ManD appears to do so with a more relaxed specificity. ManE has homology to the Pyrococcus furiosus cellobiose-binding protein CbtA (25), and it has been suggested that TM1223 be named CbtA (12). Although ManE binds cellobiose, BglE does too and with a 10-fold-higher affinity (Table 1). Although ManE is expressed in response to growth on carboxymethyl cellulose (9) (the expression of BglE under these conditions was not examined), ManE has higher affinities for mannosides and is highly expressed in response to growth on mannans (12). Consequently, we believe that the name ManE better describes the function of this SBP.
We were unable to identify ligands for TM0460, TM1150, and TM1199. We may not have detected binding because expression of these proteins in E. coli cells grown in a complex medium may have produced recombinant proteins already bound to substrates derived from the growth medium. Cells grown in defined media, however, also did not produce proteins that bound any of our test substrates. No sugar hydrolase genes are encoded in the vicinities of TM0460 and TM1150, but several are near TM1199, including a putative periplasmic arabinogalactan endo-β-1,4-galactosidase (TM1201) (Table 1). TM1199 did not bind β-1,3-arabinogalactose; β-1,3-digalacturonic acid; or various galactosides. We observed a small decrease in fluorescence of TM1199 in the presence of a neutralized partial hydrolysate of citrus pectin. This change was reproducible, and no other polysaccharide partial hydrolysates, including that from larch wood arabinogalactans, elicited this response. As described elsewhere, MalE1, encoded upstream of TM1199, binds β-1,4-linked mannooligosaccharides, perhaps including galactosyl mannooligosaccharides (33). Proteins encoded in the region of malE1 and TM1199 appear to be involved in catabolism of galactans like guar gum and pectin.
Most of the putative sugar-binding proteins bind different ligands than do the archaeal SBPs.
We expressed nine of the ORFs assigned as encoding sugar SBPs (34). We detected binding of specific substrates for six of these SBPs (Table 1). The results for the two maltose-binding proteins were reported previously (33).
XylE (TM0114) is a xylose-binding protein with a very high affinity for xylose, but it does not bind xylosides. The substrate specificity of the putative xylanase encoded nearby (TM0113) has not been reported, but its apparent extracellular location would suggest it could produce xylose for XylE. TM0113 shares sequence similarity with Clostridium thermocellum XynA, a family 11 extracellular xylanase that hydrolyzes long-chain xylosides to xylobiose and xylotriose and minor amounts of xylose (21). XylE did not bind xylobiose or xylotriose, but XloE did (see above). Downstream from xylE is TM0116, which likely encodes a xylulokinase (BLAST expected value of 2 × 10−114 with a Thermoanaerobacter ethanolicus homolog).
Among the most surprising results was the identification of the archaeal TM0418 as a myo-inositol-binding protein (InoE, ino, as used for TM1419 [1]). T. maritima uses di-2-O-β-mannosyl-di-myo-inositol-1,1′(3,3′)-phosphate and di-myo-inositol-1,3′-phosphate as compatible solutes under normal growth conditions (28). The putative T. maritima myo-inositol transporter operon is near the gene encoding a putative myo-inositol dehydrogenase (TM0414) but distant from other myo-inositol metabolism genes encoding an inositol monophosphatase (TM1415) (7) and myo-inositol-1-P synthase (ino1, TM1419) (1, 36). Most myo-inositol transporters belong to the major facilitator superfamily or the solute:sodium symporter family of transporters (TransportDB; www.membranetransport.org). The fact that T. maritima appears to transport myo-inositol using an ABC transporter is unusual, though genes encoding a myo-inositol ABC transporter have been found in Rhizobium leguminosarum (18).
Although inoE was originally listed among the archaeal genes (34), it has significant sequence similarity to many bacterial homologs, so its phylogenetic affinity is unclear (1). The myo-inositol-1-P synthase gene (TM1419, ino1) was acquired by HGT from archaea (36), but several interdomain HGT events of ino1 homologs have occurred, resulting in a complex evolutionary pattern. Perhaps the inoE gene has a similarly complex evolutionary history.
The ORFs TM0430 to TM0432 encode an apparent α-galacturonate (agu, as used for TM0434 [53]) transporter. AguE (TM0432) appears to function in transport of pectin hydrolysis products. An extracellular pectate lyase encoded nearby (PelA, TM0433) hydrolyzes polygalacturonate primarily to unsaturated trigalacturonate but also to digalacturonate (24, 42). These are likely transported by the AguEFG transporter. Further intracellular hydrolysis of these might be carried out by TM0437, an apparent exogalacturonidase. An α-glucuronidase, Agu (TM0434), encoded nearby was not tested for its ability to hydrolize α-galacturonates (53). Unlike the E. coli ExuT hexuronate transporter (35), AguE did not bind galacturonate. This may reflect the fact that PelA does not produce galacturonate as a product (24). We do not know if AguE binds α-glucuronates, substrates of the cytoplasmic glucuronidase Agu (53).
TM0958 (RbsB) was annotated as a ribose-binding protein, based upon its sequence similarity to this family of SBPs (34). We could not detect binding to any ligands, including ribose, by observing its fluorescence. Unlike the other recombinant SBPs we examined, RbsB has little intrinsic fluorescence, due to a relatively low aromatic amino acid content. This phenomenon is also observed for the E. coli ribose-binding protein because it has no tryptophan residues involved in ribose binding (30). We measured ribose binding by RbsB by using radiolabeled ribose in a filter-binding assay that we previously used to measure sugar binding by native T. maritima periplasmic-binding proteins (32). This method revealed that RbsB binds ribose with high affinity, 0.4 ± 0.5 μM, at 55°C. For comparison, the E. coli ribose-binding protein binds ribose with an affinity of 0.13 μM (4). α-d[3H]ribose binding by RbsB was completely inhibited by competition with unlabeled ribose. It was inhibited to lesser extents by the addition of other unlabeled sugars: glucose (30% inhibition), arabinose (26%), and fructose (24%). Galactose and xylose did not affect radiolabeled ribose binding.
Three of the bacterial SBPs (TM0595, TM0810, and TM1855) did not bind any of the sugars tested in the fluorescence assay. Neither its genomic context nor its sequence similarities suggest possible ligands for the product of TM0595. TM0810 is adjacent to a gene encoding a putative N-acetylglucosamidase (TM0809), an enzyme involved in chitin catabolism. TM0809 is 80% identical to a gene that was cloned from Thermotoga neapolitana and whose product was assigned this function (Khan-Koticha et al., unpublished results) (GenBank accession number AF343913). We found no change in the fluorescence of TM0810 in the presence of neutralized, partial hydrolysates of chitin or chitosan; nor was a change observed when acetylchitotriose was tested.
TM1855 has a putative mannosidase encoded nearby (TM1851), but no mannooligosaccharides elicited any change in the fluorescence of TM1855. Addition of a neutralized, partial hydrolysate of yeast mannan also elicited no change in its fluorescence. We did observe a 1% quenching of fluorescence by a neutralized, partial hydrolysate of xanthan gum (another potential source of mannans). This hydrolysate likely contained a complex mixture of oligosaccharides. Xanthan gum is composed of trisaccharide units of mannose-β-1,4-glucuronic acid-β-1,2-mannose α-linked to a cellulosic (β-1,4-glucose) backbone. Neither cellobiose (glucose-β-1,4-glucose) nor mannobioses (β-1,4; α-1,4; and α-1,6) elicited fluorescence changes, so TM1855 may bind a mannose-glucuronic acid oligosaccharide. The nearby cytosolic α-mannosidase, TM1851, has a wide substrate specificity for alpha-linked mannosides, but its activity with mannose-glucuronates is unknown (31).
Gene expression patterns support these assigned functions.
The regulation of expression of several of these ABC transporter genes has been examined using DNA microarrays, Northern blotting, real time-PCR, and proteomics. Their transcription has been measured in response to growth on different carbon sources (9, 10, 12, 33, 39), biofilm formation (44), growth in continuous culture (48), heat shock (45), and coculture with Methanococcus jannaschii (23). A summary of the relevant results from those published studies is provided in Table S3 in the supplemental material. In those studies, the data describing the transcriptional response of ABC transporter genes are incomplete either because a small number of carbon sources were examined, carbon sources were not used to elicit transcriptional responses, an incomplete subset of genes was examined, or only some of the results from transporter genes were reported. The available expression data generally show that the sugar transporters' genes are expressed in response to the binding ligands identified here.
Transcription of bglE is increased by growth on substrates like those it binds, including laminarin, pustulan (a β-1,6 glucan), and barley glucans. The adjoining lamA, bglB, and bgl transporter genes are all upregulated in response to laminarin and barley glucan, but pustulan does not affect expression of the hydrolase genes (12). bglE is also expressed on other sugars that do not affect adjoining genes, but the genes encoding SBPs are often expressed differently than the adjoining ABC transporter genes (38, 51).
Growth on xylose and birchwood xylan induces xloE transcription, consistent with the function proposed based upon the affinities of XloE. The nearby genes xynB, xloA, and axeA are also expressed in cells grown on these substrates. xloA and axeA are downstream from the xlo transporter genes, but xynB is immediately upstream and transcribed in the opposite direction. Presumably, these genes have overlapping transcriptional control sites between axeA and xloA.
All of the ORFs from TM0110 to TM0116, except TM0113 (xylU), are expressed in lactose-grown cells (39). The coexpression of the putative xylulokinase (TM0116) with XylE is consistent with the proposed function of XylE in xylose transport. However, the ORFs TM0116 and TM0113 (xylU, xylanase) are not expressed in response to growth on xylose and xylan, and the expression of xlyE under these conditions was not reported (12).
GloE is expressed during growth on an unusual variety of sugars, including carboxymethyl cellulose, mannose, and xylose. Adjacent, downstream genes are expressed primarily on carboxymethyl cellulose and glucomannans. Expression of GloE during growth on carboxymethyl cellulose is consistent with its proposed role in transport of β-1,4 glucose oligosaccharides. Expression of adjacent and possibly cotranscribed genes TM0305 and TM0306 during growth on glucomannan is also consistent with this hypothesis. The physiological rationale for increased expression of gloE during growth on mannose and xylose is not obvious. Neither TM0305 nor TM0306 was induced under those conditions.
The expression of the myo-inositol SBP gene inoE has been reported only from examinations of cells grown on glucose, maltose, and lactose, and its expression was the same during growth on all three sugars (39). The compatible solutes di-myo-inositol-1,3′-phosphate and di-2-O-beta-mannosyl-di-myo-inositol-1,1′(3,3′)-phosphate accumulate in heat-stressed Thermotoga neapolitana cells (28). Genes encoding compatible solute formation were induced in heat-shocked cells of Pyrococcus furiosus (49). One would expect inoE expression to increase in heat-shocked T. maritima cells to facilitate uptake of compatible solutes, but this gene was not examined in the study of gene expression in heat-shocked T. maritima cells (45). Transcription of inoE reportedly increased when cells were growing in a biofilm compared to planktonically growing cells (44). Although those biofilm cells were presumably growing in fully hydrated biofilms, they may have been responding to a matric stress (6, 55) by accumulating compatible solutes from outside the cells. Alternatively, they may have enabled transport of myo-inositol from lysed cells for use as a carbon source since adjacent genes encoding myo-inositol catabolizing enzymes were also induced.
Transcription of aguE increases during growth on glucomannan and β-xylan. The divergently transcribed pectate lyase (PelA) is also expressed under these conditions. Transcription of these two genes appears to be coordinately regulated, perhaps allowing a general response to the availability of hydrolysis products of complex polysaccharide mixtures.
The ribose-binding protein-encoding gene rbsB is highly expressed in cells grown on ribose and xylose and less so when grown on arabinose (12). These results are consistent with its role as a ribose transporter.
TM1199 is expressed in lactose-grown cells, as is the nearby gene malE1, though they do not appear to belong to the same operon (39). malE1 is also expressed in cells grown on guar gum (33). The neighboring galactosidases are also induced in lactose-grown cells as well as cells grown on carboxymethyl cellulose and galactomannans. The general pattern seems to be expression under conditions in which galactans are available, consistent with the suggested roles of malE1 and TM1199.
The transcription of manD and manE is upregulated in response to growth on mannose, konjac glucomannans, and carob galactomannans (9, 12). manE and TM1227 (encoding a periplasmic endomannosidase) are expressed in response to growth on mannose, mannans, and carboxymethyl cellulose. Transcription of manD is likely coupled with TM1227, though its expression in response to growth on carboxymethyl cellulose has not been reported. The fact that ManD binds cellobiose could explain its utility in cells growing on carboxymethyl cellulose.
malE2 is expressed in cells grown on starch, maltose, and lactose, while the nearby hydrolase-encoding genes are expressed only in starch-grown cells. Presumably, the presence of oligomers of α-1,4-glucose elicits transcription of these genes.
The available expression data for the remaining SBPs do not provide evidence for their physiological roles. Interestingly, growth with M. jannaschii increased the expression of TM0595 and TM0810 (23).
DISCUSSION
Several genes in the genome of T. maritima appear to have been acquired from archaea (29, 34). This is not surprising since members of Thermotogales, unlike most bacteria, likely live in environments disproportionately inhabited by archaea. The fact that these archaeal genes now function in bacteria raises questions about how foreign genes functionally integrate into a new genome. All newly acquired genes must adjust to any differences in G+C content and codon bias in their new hosts, but archaeal genes must also acquire control by different kinds of promoters (2, 50). Intergenic, noncoding DNA can mutate to function as regulatory control sites in bacteria (46, 52), but it is unlikely that archaeal promoters, which are unlike bacterial promoters, could accumulate sufficient point mutations to function effectively in a bacterium. The archaeal genes now found in the genome of T. maritima more likely recombined behind existing bacterial promoters.
The selective pressures that dictate where these acquired archaeal genes recombined may reflect the importance of their immediate benefit to their host. The model of Berg and Kurland (3) suggests that positive selection is necessary for such genes to persist following HGT. Novozhilov et al., however, modeled a scenario in which such genes could persist in populations without positive selection (41). If immediate positive selection is required for fixation, then the novelty of function of the newly acquired gene is important. This gene must also recombine behind a promoter that allows this new function to be expressed under physiologically relevant conditions along with genes that complement the new function. This tremendous selective pressure would limit the sites at which these genes could successfully recombine in the host's genome. Alternatively, if neutral selective pressures operate, then new foreign genes could encode functions that overlap those that already exist in the host. As long as the new genes do not seriously compromise the host, they can mutate to become more integrated into the new physiological milieu, perhaps by acquiring new sites for binding a different set of transcription factors (46, 52) or acquiring new and unique functions.
To understand how these archaeal ABC transporter genes came to be expressed within their new bacterial host, we must first know their functions. These genes were originally annotated as encoding oligopeptide transporters based on similarities to other transporter sequences. Since these transporter genes are found near sugar hydrolase genes and similarly assigned oligopeptide transporters were found to transport oligosaccharides in some archaea, these genes have been presumed to encode oligosaccharide transporters. Experimental evidence for this function was lacking until this report.
We measured the affinities for sugars for eight recombinant SBPs encoded by archaeal genes and nine SBPs encoded by apparent bacterial genes. We were able to measure binding to specific classes of sugars for 11 of these 17 proteins. In many cases, the sugars they bind are also substrates for hydrolases that are encoded nearby.
Five of the archaeal SBPs bind oligosaccharides. The substrates of these SBPs are known growth substrates for T. maritima, including cellobiose, laminaribiose, xylan, and mannans (summarized in reference 22). The archaeal and bacterial transporters appear to play nearly unique roles in the cell. With one exception, the archaeal SBPs bind different ligands than do the bacterial SBPs. Only MalE1 and ManD both bind beta-1,4-mannotetraose, though ManD does so with a 100-fold-higher affinity. There is no obvious pattern to the chemical nature of substrates bound by the two classes of proteins, though only the bacterial SBPs in T. maritima bind monosaccharides. The fact that the T. maritima archaeal SBPs bind only oligosaccharides is consistent with the suggestion that the oligosaccharide-binding proteins in archaea, like oligopeptide-binding proteins, may have larger ligand-binding pockets that accommodate oligomers (17, 25).
Previously we detected a glucose-binding activity in periplasmic extracts of T. maritima (32). In this study we found no glucose-specific binding protein. Glucose can compete with maltose and ribose by binding to their SBPs, so glucose may be transported as a low-affinity substrate of one or more SBPs. Curiously, no hexoses bound to any of the SBPs we examined. Only pentose monosaccharide-binding proteins were found, those for ribose and xylose. There are other putative oligopeptide, amino acid, and sugar ABC transporters annotated in the genome sequence (34). Perhaps hexose-binding proteins are encoded in one or more of those ABC transporters.
Based upon the currently available gene expression studies that include the transporter genes, transcription of the archaeal oligosaccharide transporter genes appears to be coordinately regulated with that of adjoining bacterial hydrolase genes. Attempts to make correlations between transcriptional regulation and transporter function have sometimes provided general features of the ligands recognized by SBPs (9, 10, 39, 44). Extensive surveys of gene expression in response to growth substrates generally supported many of the substrate assignments made here (9, 12). Gene expression studies are vital to understanding the function and evolution of these transporters, but the relationship between inducing substrates and transporter specificities is complex. Although names for transporter genes based on their expression patterns were proposed in an earlier study (12), we suggest that the names proposed here, based upon ligand affinities, more closely represent the functions of the SBPs. A table reconciling these two nomenclatures is provided in Table 2.
In the study by Chhabra et al., the ABC transporter encoded by TM1746 to TM1750 was proposed to transport galactomannans (9, 12). That transporter was originally assigned a role as a bacterial oligopeptide transporter, so we did not examine it in our survey (34). We found that the archaeal SBP ManD binds β-1,4-galactosyl mannobiose. If TM1746 also binds galactomannans, then there may be an overlap in specificities for these archaeal and bacterial oligosaccharide transporters.
There are other examples of similar, if not overlapping, substrate specificities between the archaeal and bacterial transporters examined here. XloE and XylE are involved in transport of the products of xylan hydrolysis, and their genes are among genes encoding xylan catabolism. AguE likely transports galacturonates derived from pectin hydrolysis. Although we could not define the specific substrates of TM1199, it appears to respond to some component in pectin hydrolysates. The expression patterns of genes adjoining these two transporters are different, perhaps indicating different selective pressures that allow the maintenance of two transporters that recognize similar substrates.
The substrate binding data presented here contribute to the foundation of information necessary to understand how these horizontally acquired genes have evolved. We do not yet know the substrate affinities of the corresponding SBPs in archaea that are the closest homologs of these T. maritima proteins, so we do not know if they may have changed their function after their transfer to Thermotogales. With only one genome sequence from this family, we do not know if these transporter genes are found in all members of the order Thermotogales or if their functions are the same in all species that have them. If these operons are in other Thermotogales species, it is of interest to ask if they are found in the same locations in those genomes and are expressed under the same conditions. The answers to these questions will provide us with important information for understanding how genes adapt to new physiological contexts.
Supplementary Material
Acknowledgments
We thank Carol Teschke for her generous assistance with and advice about the fluorescence-based binding studies and Milton Saier for helpful comments on nomenclature.
This work was supported by funds from the NASA exobiology program (NAG5-12367).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Bachhawat, N., and S. C. Mande. 2000. Complex evolution of the inositol-1-phosphate synthase gene among archaea and eubacteria. Trends Genet. 16:111-113. [DOI] [PubMed] [Google Scholar]
- 2.Bell, S. D., C. P. Magill, and S. P. Jackson. 2001. Basal and regulated transcription in Archaea. Biochem. Soc. Trans. 29:392-395. [DOI] [PubMed] [Google Scholar]
- 3.Berg, O. G., and C. G. Kurland. 2002. Evolution of microbial genomes: sequence acquisition and loss. Mol. Biol. Evol. 19:2265-2276. [DOI] [PubMed] [Google Scholar]
- 4.Binnie, R. A., H. Zhang, S. Mowbray, and M. A. Hermodson. 1992. Functional mapping of the surface of Escherichia coli ribose-binding protein: mutations that affect chemotaxis and transport. Protein Sci. 1:1642-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Calteau, A., M. Gouy, and G. Perriere. 2005. Horizontal transfer of two operons coding for hydrogenases between bacteria and archaea. J. Mol. Evol. 60:557-565. [DOI] [PubMed] [Google Scholar]
- 6.Chang, W. S., and L. J. Halverson. 2003. Reduced water availability influences the dynamics, development, and ultrastructural properties of Pseudomonas putida biofilms. J. Bacteriol. 185:199-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen, L., and M. F. Roberts. 1999. Characterization of a tetrameric inositol monophosphatase from the hyperthermophilic bacterium Thermotoga maritima. Appl. Environ. Microbiol. 65:4559-4567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chhabra, S. R., and R. M. Kelly. 2002. Biochemical characterization of Thermotoga maritima endoglucanase Ce174 with and without a carbohydrate binding module (CBM). FEBS Lett. 531:375-380. [DOI] [PubMed] [Google Scholar]
- 9.Chhabra, S. R., K. R. Shockley, S. B. Conners, K. L. Scott, R. D. Wolfinger, and R. M. Kelly. 2003. Carbohydrate-induced differential gene expression patterns in the hyperthermophilic bacterium Thermotoga maritima. J. Biol. Chem. 278:7540-7552. [DOI] [PubMed] [Google Scholar]
- 10.Chhabra, S. R., K. R. Shockley, D. E. Ward, and R. M. Kelly. 2002. Regulation of endo-acting glycosyl hydrolases in the hyperthermophilic bacterium Thermotoga maritima grown on glucan- and mannan-based polysaccharides. Appl. Environ. Microbiol. 68:545-554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Childers, S. E., M. Vargas, and K. M. Noll. 1992. Improved methods for cultivation of the extremely thermophilic bacterium Thermotoga neapolitana. Appl. Environ. Microbiol. 58:3949-3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Conners, S. B., C. I. Montero, D. A. Comfort, K. R. Shockley, M. R. Johnson, S. R. Chhabra, and R. M. Kelly. 2005. An expression-driven approach to the prediction of carbohydrate transport and utilization regulons in the hyperthermophilic bacterium Thermotoga maritima. J. Bacteriol. 187:7267-7282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Demerec, M., E. A. Adelberg, A. J. Clark, and P. E. Hartman. 1966. A proposal for a uniform nomenclature in bacterial genetics. Genetics 54:61-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Detmers, F. J. M., F. C. Lanfermeijer, and B. Poolman. 2001. Peptides and ATP binding cassette peptide transporters. Res. Microbiol. 152:245-258. [DOI] [PubMed] [Google Scholar]
- 15.Doolittle, W. F. 1999. Phylogenetic classification and the universal tree. Science 284:2124-2128. [DOI] [PubMed] [Google Scholar]
- 16.Eftink, M. R. 1997. Fluorescence methods for studying equilibrium macromolecule-ligand interactions. Methods Enzymol. 278:221-257. [DOI] [PubMed] [Google Scholar]
- 17.Elferink, M. G. L., S. V. Albers, W. N. Konings, and A. J. M. Driessen. 2001. Sugar transport in Sulfolobus solfataricus is mediated by two families of binding protein-dependent ABC transporters. Mol. Microbiol. 39:1494-1503. [DOI] [PubMed] [Google Scholar]
- 18.Fry, J., M. Wood, and P. S. Poole. 2001. Investigation of myo-inositol catabolism in Rhizobium leguminosarum bv. viciae and its effect on nodulation competitiveness. Mol. Plant-Microbe Interact. 14:1016-1025. [DOI] [PubMed] [Google Scholar]
- 19.Geiduschek, E. P., and M. Ouhammouch. 2005. Archaeal transcription and its regulators. Mol. Microbiol. 56:1397-1407. [DOI] [PubMed] [Google Scholar]
- 20.Gill, S. C., and P. H. von Hippel. 1989. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 182:319-326. [DOI] [PubMed] [Google Scholar]
- 21.Hayashi, H., M. Takehara, T. Hattori, T. Kimura, S. Karita, K. Sakka, and K. Ohmiya. 1999. Nucleotide sequences of two contiguous and highly homologous xylanase genes xynA and xynB and characterization of XynA from Clostridium thermocellum. Appl. Microbiol. Biotechnol. 51:348-357. [DOI] [PubMed] [Google Scholar]
- 22.Huber, R., and M. Hanning. 31 July 2003, posting date. Thermotogales, In M. Dworkin (ed.), The prokaryotes: an evolving electronic resource for the microbiological community, 3rd ed., release 3.14. Springer-Verlag, New York, N.Y. [Online.] http://link.springer-ny.com/link/service/books/10125.
- 23.Johnson, M. R., C. I. Montero, S. B. Conners, K. R. Shockley, S. L. Bridger, and R. M. Kelly. 2005. Population density-dependent regulation of exopolysaccharide formation in the hyperthermophilic bacterium Thermotoga maritima. Mol. Microbiol. 55:664-674. [DOI] [PubMed] [Google Scholar]
- 24.Kluskens, L. D., G. van Alebeek, A. G. J. Voragen, W. M. de Vos, and J. van der Oost. 2003. Molecular and biochemical characterization of the thermoactive family 1 pectate lyase from the hyperthermophilic bacterium Thermotoga maritima. Biochem. J. 370:651-659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koning, S. M., M. G. L. Elferink, W. N. Konings, and A. J. M. Driessen. 2001. Cellobiose uptake in the hyperthermophilic archaeon Pyrococcus furiosus is mediated by an inducible, high-affinity ABC transporter. J. Bacteriol. 183:4979-4984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kurland, C. G., B. Canback, and O. G. Berg. 2003. Horizontal gene transfer: a critical view. Proc. Natl. Acad. Sci. USA 100:9658-9662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Logsdon, J. M., Jr. 1999. Evolutionary genomics: Thermotoga heats up lateral gene transfer. Curr. Biol. 9:R747-R751. [DOI] [PubMed] [Google Scholar]
- 28.Martins, L. O., L. S. Carreto, M. S. Da Costa, and H. Santos. 1996. New compatible solutes related to di-myo-inositol-phosphate in members of the order Thermotogales. J. Bacteriol. 178:5644-5651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Medrano-Soto, A., G. Moreno-Hagelsieb, P. Vinuesa, J. A. Christen, and J. Collado-Vides. 2004. Successful lateral transfer requires codon usage compatibility between foreign genes and recipient genomes. Mol. Biol. Evol. 21:1884-1894. [DOI] [PubMed] [Google Scholar]
- 30.Mowbray, S. L., and L. B. Cole. 1992. 1.7 A X-ray structure of the periplasmic ribose receptor from Escherichia coli. J. Mol. Biol. 225:155-175. [DOI] [PubMed] [Google Scholar]
- 31.Nakajima, M., H. Imamura, H. Shoun, and T. Wakagi. 2003. Unique metal dependency of cytosolic alpha-mannosidase from Thermotoga maritima, a hyperthermophilic bacterium. Arch. Biochem. Biophys. 415:87-93. [DOI] [PubMed] [Google Scholar]
- 32.Nanavati, D., K. M. Noll, and A. H. Romano. 2002. Periplasmic maltose- and glucose-binding protein activities in cell-free extracts of Thermotoga maritima. Microbiology 148:3531-3537. [DOI] [PubMed] [Google Scholar]
- 33.Nanavati, D. M., T. N. Nguyen, and K. M. Noll. 2005. Substrate specificities and expression patterns reflect the evolutionary divergence of maltose ABC transporters in Thermotoga maritima. J. Bacteriol. 187:2002-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nelson, K. E., R. A. Clayton, S. R. Gill, M. L. Gwinn, R. J. Dodson, D. H. Haft, E. K. Hickey, J. D. Peterson, W. C. Nelson, K. A. Ketchum, L. McDonald, T. R. Utterback, J. A. Malek, K. D. Linher, M. M. Garrett, A. M. Stewart, M. D. Cotton, M. S. Pratt, C. A. Phillips, D. Richardson, J. Heidelberg, G. G. Sutton, R. D. Fleischmann, J. A. Eisen, O. White, S. L. Salzberg, H. O. Smith, J. C. Venter, and C. M. Fraser. 1999. Evidence for lateral gene transfer between Archaea and Bacteria from genome sequence of Thermotoga maritima. Nature 399:323-329. [DOI] [PubMed] [Google Scholar]
- 35.Nemoz, G., J. Robert-Baudouy, and F. Stoeber. 1976. Physiological and genetic regulation of the aldohexuronate transport system in Escherichia coli. J. Bacteriol. 127:706-718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nesbo, C. L., S. L'Haridon, K. O. Stetter, and W. F. Doolittle. 2001. Phylogenetic analyses of two “archaeal” genes in Thermotoga maritima reveal multiple transfers between Archaea and Bacteria. Mol. Biol. Evol. 18:362-375. [DOI] [PubMed] [Google Scholar]
- 37.Nesbo, C. L., K. E. Nelson, and W. F. Doolittle. 2002. Suppressive subtractive hybridization detects extensive genomic diversity in Thermotoga maritima. J. Bacteriol. 184:4475-4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newbury, S. F., N. H. Smith, and C. F. Higgins. 1987. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell 51:1131-1143. [DOI] [PubMed] [Google Scholar]
- 39.Nguyen, T. N., A. D. Ejaz, M. A. Brancieri, A. M. Mikula, K. E. Nelson, S. R. Gill, and K. M. Noll. 2004. Whole-genome expression profiling of Thermotoga maritima in response to growth on sugars in a chemostat. J. Bacteriol. 186:4824-4828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nielsen, H., J. Engelbrecht, S. Brunak, and G. von Heijne. 1997. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 10:1-6. [DOI] [PubMed] [Google Scholar]
- 41.Novozhilov, A. S., G. P. Karev, and E. V. Koonin. 2005. Mathematical modeling of evolution of horizontally transferred genes. Mol. Biol. Evol. 22:1721-1732. [DOI] [PubMed] [Google Scholar]
- 42.Parisot, J. L., A. Ghochikyan, V. Langlois, V. Sakanyan, and C. Rabiller. 2002. Exopolygalacturonate lyase from Thermotoga maritima: cloning, characterization and organic synthesis application. Carbohydr. Res. 337:1427-1433. [DOI] [PubMed] [Google Scholar]
- 43.Paulsen, I. T., L. Nguyen, M. K. Sliwinski, R. Rabus, and M. H. Saier, Jr. 2000. Microbial genome analyses: comparative transport capabilities in eighteen prokaryotes. J. Mol. Biol. 301:75-100. [DOI] [PubMed] [Google Scholar]
- 44.Pysz, M. A., S. B. Conners, C. I. Montero, K. R. Shockley, M. R. Johnson, D. E. Ward, and R. A. Kelly. 2004. Transcriptional analysis of biofilm formation processes in the anaerobic, hyperthermophilic bacterium Thermotoga maritima. Appl. Environ. Microbiol. 70:6098-6112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pysz, M. A., D. E. Ward, K. R. Shockley, C. I. Montero, S. B. Conners, M. R. Johnson, and R. M. Kelly. 2004. Transcriptional analysis of dynamic heat-shock response by the hyperthermophilic bacterium Thermotoga maritima. Extremophiles 8:209-217. [DOI] [PubMed] [Google Scholar]
- 46.Rajewsky, N., N. D. Socci, M. Zapotocky, and E. D. Siggia. 2002. The evolution of DNA regulatory regions for proteo-gamma bacteria by interspecies comparisons. Genome Res. 12:298-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Richarme, G., and A. Kepes. 1983. Study of binding protein-ligand interaction by ammonium sulfate-assisted adsorption on cellulose esters filters. Biochim. Biophys. Acta 742:16-24. [DOI] [PubMed] [Google Scholar]
- 48.Shockley, K. R., K. L. Scott, M. A. Pysz, S. B. Conners, M. R. Johnson, C. I. Montero, R. D. Wolfinger, and R. M. Kelly. 2005. Genome-wide transcriptional variation within and between steady states for continuous growth of the hyperthermophile Thermotoga maritima. Appl. Environ. Microbiol. 71:5572-5576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shockley, K. R., D. E. Ward, S. R. Chhabra, S. B. Conners, C. I. Montero, and R. M. Kelly. 2003. Heat shock response by the hyperthermophilic archaeon Pyrococcus furiosus. Appl. Environ. Microbiol. 69:2365-2371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Soppa, J. 1999. Transcription initiation in Archaea: facts, factors and future aspects. Mol. Microbiol. 31:1295-1305. [DOI] [PubMed] [Google Scholar]
- 51.Stern, M. J., E. Prossnitz, and G. F. Ames. 1988. Role of the intercistronic region in post-transcriptional control of gene expression in the histidine transport operon of Salmonella typhimurium: involvement of REP sequences. Mol. Microbiol. 2:141-152. [DOI] [PubMed] [Google Scholar]
- 52.Stone, J. R., and G. A. Wray. 2001. Rapid evolution of cis-regulatory sequences via local point mutations. Mol. Biol. Evol. 18:1764-1770. [DOI] [PubMed] [Google Scholar]
- 53.Suresh, C., A. Abu Rus'd, M. Kitaoka, and K. Hayashi. 2002. Evidence that the putative alpha-glucosidase of Thermotoga maritima MSB8 is a pNP alpha-d-glucuronopyranoside hydrolyzing alpha-glucuronidase. FEBS Lett. 517:159-162. [DOI] [PubMed] [Google Scholar]
- 54.Tsujibo, H., M. Kosaka, S. Ikenishi, T. Sato, K. Miyamoto, and Y. Inamori. 2004. Molecular characterization of a high-affinity xylobiose transporter of Streptomyces thermoviolaceus OPC-520 and its transcriptional regulation. J. Bacteriol. 186:1029-1037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.van de Mortel, M., and L. J. Halverson. 2004. Cell envelope components contributing to biofilm growth and survival of Pseudomonas putida in low-water-content habitats. Mol. Microbiol. 52:735-750. [DOI] [PubMed] [Google Scholar]
- 56.Worning, P., L. J. Jensen, K. E. Nelson, S. Brunak, and D. W. Ussery. 2000. Structural analysis of DNA sequence: evidence for lateral gene transfer in Thermotoga maritima. Nucleic Acids Res. 28:706-709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zverlov, V. V., I. Y. Volkov, T. V. Velikodvorskaya, and W. H. Schwarz. 1997. Highly thermostable endo-1,3-beta-glucanase (laminarinase) LamA from Thermotoga neapolitana: nucleotide sequence of the gene and characterization of the recombinant gene product. Microbiology 143:1701-1708. [DOI] [PubMed] [Google Scholar]
- 58.Zverlov, V. V., I. Y. Volkov, T. V. Velikodvorskaya, and W. H. Schwarz. 1997. Thermotoga neapolitana bglB gene, upstream of lamA, encodes a highly thermostable beta-glucosidase that is a laminaribiase. Microbiology 143:3537-3542. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.