Abstract
Salivaricin A (SalA), the first Streptococcus salivarius lantibiotic to be characterized, appears to be inhibitory to most Streptococcus pyogenes strains. A variant of the SalA structural gene (salA1) is present in more than 90% of S. pyogenes strains, but only strains of M serotype 4 and T pattern 4 produce the biologically active peptide. The present study identifies four additional variants (salA2 to salA5) of the SalA structural gene and demonstrates that each of the corresponding inhibitory peptides (SalA2 to SalA5) is produced in vitro. These variants appear to be similar to SalA and SalA1 in their inhibitory activity against Micrococcus luteus and in their ability to act as inducers of SalA production. It had previously been shown that S. pyogenes strain SF370 had a deletion (of approximately 2.5 kb) in the salM and salT genes of the salA1 locus. In the present study, several additional characteristic deletions within the salA1 loci were identified. S. pyogenes strains of the same M serotype all share the same salA1 locus structure. Since S. salivarius is a predominant member of the normal oral flora of healthy humans, strains producing anti-S. pyogenes lantibiotics, such as SalA, may have excellent potential for use as oral probiotics. In the present study, we have used a highly specific SalA induction system to directly detect the presence of SalA in the saliva of humans who either naturally harbor populations of SalA-producing S. salivarius or who have been colonized with the SalA2-producing probiotic S. salivarius K12.
Class I bacteriocins, more commonly referred to as lantibiotics because they contain the posttranslationally modified amino acids lanthionine and/or methyllanthionine, have now been shown to be produced by various members of the oral streptococcal species Streptococcus mutans (10-12), Streptococcus pyogenes (7, 26), and Streptococcus salivarius (13, 20). The first of the S. salivarius lantibiotics to be characterized was salivaricin A (SalA), coded by the structural gene salA in S. salivarius strain 20P3 (13). Strains that produce SalA exhibit specific immunity to the homologous bacteriocin. Our initial observation that the growth of all of 81 tested S. pyogenes strains was inhibited by strain 20P3 in agar-based deferred antagonism tests (2) at first appeared anomalous when it was found that all but 2 of 65 S. pyogenes strains, each representing a different M protein serotype, hybridized with a salA probe (15). The 63 salA-positive S. pyogenes strains differed, however, in that they harbored a variant (named salA1) of the SalA structural gene, encoding a SalA homologue (SalA1) that had conservative amino acid differences in residues 2 (R2K) and 7 (I7F) of the propeptide part of the molecule. Interestingly, expression of biologically active (inhibitory) levels of SalA1 by S. pyogenes was evident only in strains of serotype M4 (8, 19).
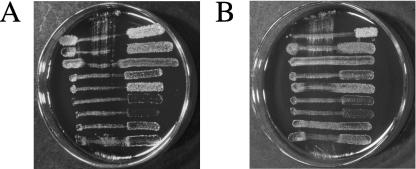
We use a blood agar-based deferred antagonism test to initially detect bacterial inhibitory activity and, until fully characterized, the inhibitory agents are referred to as bacteriocin-like inhibitory substances (BLIS). The patterns of inhibitory activity produced by the test strains against a set of nine standard indicator bacteria (I1 to I9) are, for convenience, converted to numerical codes referred to as BLIS production (P) types. For example, P type 777 represents inhibition of all nine indicators (Fig. 1A) (19). SalA1-producing S. pyogenes strains display a P type of 655 (25), corresponding to inhibition of all indicators other than I3 (Streptococcus constellatus), I5 (S. pyogenes), and I8 (S. pyogenes) (Fig. 1B). A perusal of the results of our P typing of several thousand strains of a wide variety of streptococcal species has now shown us that only a small number display P type 655, a finding consistent with inhibitory activity being composed solely of SalA-like peptides. For example, most SalA-producing S. salivarius strains exhibit the somewhat broader P-type profiles of 676 or 677 (2), presumably due to their production of some as-yet-uncharacterized BLIS, in addition to SalA. A survey of over 5,000 S. salivarius isolates from 180 subjects showed that 1% had P-type patterns of 676 or 677 (21). Very occasionally, P-type 777 S. salivarius strains have been detected and, upon further testing, it appears that at least some of these strains, such as the probiotic S. salivarius K12 (BLIS Technologies Ltd., Dunedin, New Zealand), produce the lantibiotic salivaricin B (SalB) (20) in addition to a SalA-like peptide.
FIG. 1.
(A) Deferred antagonism of S. salivarius strain K12 illustrating a P-type pattern of 777 (i.e., inhibition of all nine indicator strains). (B) Deferred antagonism test of S. pyogenes strain 148 illustrating a 655 P-type pattern (i.e., inhibition of indicators I1, I2, I4, I6, I7, and I9).
Comparison of the entire salA and salA1 loci in S. salivarius 20P3 and S. pyogenes SF370 (serotype M-1), respectively, showed that the locus in strain 20P3 was approximately 2.5 kb larger (25) than that in strain SF370. Open reading frames present in the salA locus, in addition to the structural genes, were those thought to encode prepeptide modification enzymes (salB, salC), transporters (salT), two-component response regulators (salR, salK), and proteins conferring host cell immunity to SalA (salX, salY). Present in strain 20P3 but missing in strain SF370 were salC and part of salB and salT. In that same study, it was demonstrated that SalA functions as the signal for up-regulation of its own production in cultures of strain 20P3 via the activation of the SalR/K signal transduction system (25). Moreover, culture supernatants containing either SalA or SalA1 were found capable of specifically up-regulating the transcription of either salA (in strain 20P3) or salA1 (in strain T11), this being the first demonstration of interspecies induction of lantibiotic gene expression (25).
In the present study, we report the results of our examination of the distribution and variety of SalA-like peptides produced by a selection of strains of various streptococcal species. Furthermore, we have exploited the SalA auto-regulation system when developing a highly sensitive assay to specifically detect the presence of SalA peptides in saliva and demonstrate that positive reactions occur only in specimens from individuals harboring populations of SalA-producing S. salivarius within their oral microbiota.
MATERIALS AND METHODS
Bacterial strains and culture media.
S. pyogenes SF370 (17), the standard streptococcal BLIS indicator strains (I1 to I9) (19), the set of 73 prototype S. pyogenes strains (M serotypes 1 to 81) (18), and some of the streptococci found to be producers of SalA-like peptides, including S. salivarius 20P3 (2, 13, 25), S. salivarius K12 (20), S. salivarius JH (22), S. salivarius 9 (2), Streptococcus dysgalactiae subsp. equisimilis 4003 (formerly called Streptococcus equisimilis 4003 [8, 14]), and Streptococcus agalactiae 120 (14), have been described previously. The representative strains of S. pyogenes, S. salivarius, S. agalactiae, S. dysgalactiae, Streptococcus uberis, and Streptococcus thermophilus tested for the presence and expression of salA were from the culture collection of J. R. Tagg. Unless stated otherwise, all incubation was at 37°C in a 5% CO2-in-air atmosphere. The medium for detection of BLIS production was BaCa (Columbia agar base; Life Technologies Ltd., Paisley, United Kingdom) supplemented with human blood (5%, vol/vol) and CaCO3 (0.1%, wt/vol). The representative serotype M4 S. pyogenes strain 148 was incubated aerobically at 30°C in order to enhance its production of SalA1 (8). Other culture media utilized in the present study were Ba (Columbia agar base supplemented with human blood [5%, vol/vol]), Todd Hewitt broth (THB; Difco, Becton Dickinson and Co., Sparks, Md.), and THB supplemented with 1.8 μM CaCO3 (THBCa) and sometimes also with 0.1% (wt/vol) glucose (THBCaGlu).
Assay of SalA peptide inhibitory activity.
Wells were cut in BaCa agar medium using a hollow glass rod (6-mm diameter). The base of each well was then sealed with 20 μl of molten bacteriological agar (Scientific Supplies Ltd., Auckland, New Zealand). Samples (50 μl) of preparations to be tested for inhibitory activity were deposited into the wells, and the plate was left to dry at 37°C. The surface of the medium was then sterilized by exposure to chloroform vapor for 30 min followed by airing for a further 30 min. The SalA-sensitive Micrococcus luteus T18 (standard indicator strain I1), pregrown for 18 h in THB, was then applied evenly over the surface of the agar using a cotton swab. Following incubation at 37°C in air, the bacteriocin titer (in arbitrary units [AU] per ml) was taken to be the reciprocal of the highest dilution (of a series of doubling dilutions) to show definite inhibitory activity.
DNA extraction.
Chromosomal DNA for Southern and dot blot analyses was extracted using the method of Upton et al. (24). Streptococcal DNA for use as template in PCRs was isolated as described elsewhere (1). One-microliter aliquots of DNA-containing supernatant were used as template for each 50-μl PCR mixture.
DNA manipulations.
Detection of salA was by application of a PCR of 30 cycles, consisting of a denaturing temperature of 94°C for 30 s followed by an annealing temperature of 55°C for 30 s and an elongation time of 30 s at 65°C, using the primer pair SalAUS (5′-GTAGAAAATATTTACTACATACT) and SalADS (5′-GTTAAAGTATTCGTAAAACTGATG) (corresponding to positions 544 to 556 and positions 859 to 882, respectively, within the sal locus of S. salivarius strain 20P3 [GenBank accession no. AY005472]). The products derived from the PCRs were purified and sequenced directly with a Perkin-Elmer ABI 377A sequencer. Primary sequence data were collated with SeqEd sequencer software, and sequence alignments, translation, and general analyses were performed using either DNAMAN (Lynnon Biosoft, Vaudreuil, Canada) or Lasergene 99 expert sequence analysis software (DNASTAR, Inc., Madison, Wis.). The consensus sequences derived were compared to those in DNA and protein sequence databases using the BLAST facilities on the National Center for Biotechnology Information (NCBI) server (http://www.ncbi.nlm.nih.gov) and the University of Oklahoma server (http://www.genome.ou.edu/strep.html).
Long-template PCR.
PCR amplification of entire sal loci was done using the PCR primer pair SalAF (positions 604 to 629, GATATTTTGAACAATGCTATCGAAGA) and SalRR (positions 10411 to 10391, 5′TCAACATAATCCTGAGATTCG) with an annealing temperature of 60°C by using an Expand Long Template PCR kit (Roche) following the manufacturer's instructions. PCR products were analyzed by electrophoresis through a 1% agarose gel to allow determination of locus size.
Selected S. pyogenes strains of different M serotypes were tested to determine the completeness of their salM and salT genes with a PCR of 30 cycles, consisting of a denaturing temperature of 94°C for 30 s followed by an annealing temperature of 55°C for 30 s and an elongation time of 3.5 min at 65°C, using the primer pair S.pyodelFwd (5′-ATATACCCTCATTCAGTCTTC) and S.pyodelRev (5′-GTTATACATCACATCCCCATCAA) (corresponding to positions 1415 to 1435 and positions 5326 to 5304, respectively, of the salA locus of S. salivarius strain 20P3). Representatives of each PCR product type (based on product size) were sequenced as described above.
salA detection using dot blots.
The distribution of salA in sets of strains representative of various streptococcal species was determined by dot blotting. DNA was extracted and applied to the membranes as described previously (26). The membranes were then probed with a digoxigenin-dUTP (Roche Diagnostics, Ltd., Lewes, England)-labeled salA probe derived with the use of the PCR primers salAUS and salADS described above.
Deferred antagonism method.
The method of deferred antagonism originally described by Tagg and Bannister (19) was used, either to determine the patterns (P type) of BLIS activity of the test strains or to compare the relative susceptibilities of different bacterial strains to the BLIS activities produced in agar media. The test strain was inoculated diametrically across the surface of the BaCa medium as a 1-cm-wide streak. After incubation, the visible growth of the test strain was removed using a glass slide, and the surface of the agar was sterilized by exposure to chloroform vapors for 30 min. The plate was then aired for 15 min prior to inoculating 18-h THB cultures of the indicator strains across the line of the original producer growth. The plates were then incubated as before for 24 h and examined for zones of interference with the indicator growth. Definite inhibition of indicator growth was recorded as +. For the purposes of P typing, the inhibitory activity against the nine standard indicators was recorded in code form (the P type) by considering the indicators to be three triplets (i.e., I1, I2, I3; I4, I5, I6; and I7, I8, I9). Inhibition of the first member of an indicator triplet was given a score of 4, that for the second a score of 2, and that for the third a score of 1. No inhibition of an indicator was scored as 0. The complete P-type code was recorded as a sequence of three numbers representing the sum of each triplet. All tests were performed in duplicate, and further testing was undertaken if significant discrepancies were detected in the inhibition patterns that were obtained.
Purification and characterization of SalA-like peptides.
Initial preparations of the SalA-like peptides were obtained by extracting the cells obtained from 1-liter THBCa cultures of the producer strains with 200 ml of 95% methanol (adjusted to approximately pH 2 by the addition of 2 ml of concentrated HCl) at 4°C for 18 h. After centrifugation to pellet the cells, the supernatant was subjected to rotary evaporation to remove the methanol. Portions (4 ml) of the residual aqueous preparations (titer of 64 to 128 AU/ml against indicator I1) were fractionated by C8 reversed-phase chromatography using an acetonitrile gradient of 0 to 80% over 60 min. Inhibitory activity against indicator I1 generally eluted in three 1-ml fractions at approximately 33% (vol/vol) acetonitrile. These fractions were pooled, vacuum concentrated to approximately 1 ml, and then refractionated using C18 reversed-phase chromatography with an acetonitrile gradient of 20 to 50% over 50 min. SalA-associated inhibitory activity was typically detected for a single 0.5-ml fraction eluting at 34 to 35% acetonitrile. Fractions were subjected to mass spectrometry analysis (6) and N-terminal sequencing (5) at the Protein Microchemistry Facility, Department of Biochemistry, University of Otago, as described previously.
Assay of the auto-inducing and cross-inducing activities of SalA-like peptides.
THB cultures (3 ml) of the SalA-producing test strain (e.g., S. pyogenes strain 148) were grown for 18 h at 37°C in 5% CO2 in air. These cultures were centrifuged (15,300 × g for 1 min), and the pellet was washed three times in saline (0.85% NaCl [wt/vol]) to reduce background SalA prior to resuspension of the cells in a volume of saline equivalent to that of the original culture. A 20-ml THBCaGlu broth was inoculated with 100 μl of washed cells, and then 180-μl aliquots of this suspension were dispensed into wells in a microtiter plate. A total of four wells were used for each sample to be tested for salA-inducing activity. Two wells of each set were designated controls (i.e., uninduced) and two as tests (i.e., the experiment was performed in duplicate). To each of the test wells, 20 μl of the sample was added, and the microtiter plate was incubated for 18 h (at 30°C in air for S. pyogenes strain 148 or at 37°C in 5% CO2 in air for other producer strains). Following incubation, 20 μl of each sample was added to the two control wells in the tray (i.e., for each pair of test wells, there is a pair of control wells which are exactly the same as the test wells except that the sample is added following the 18-h incubation). Samples (50 μl) from the test and control wells were then tested for inhibitory activity against indicator I1 using the agar well diffusion assay. Induction of SalA production is demonstrated by an inhibitory zone surrounding the test well, and not the corresponding control well, in the agar diffusion assay.
Saliva collection and quantitation of SalA-producing S. salivarius.
Nonstimulated samples (ca. 2 ml) of freshly collected saliva were obtained from subjects either known to be naturally colonized with SalA-producing S. salivarius or those who had just been colonized with the SalA-producing S. salivarius strain K12. Assessment of the population levels of SalA-producing S. salivarius in the saliva was achieved by spiral plating a 10−4 dilution (in saline) of the saliva on Mitis-Salivarius agar (Difco). Following incubation, the S. salivarius count (CFU/ml) was estimated on the basis of the number of characteristic (large, soft) colonies. One hundred of these putative S. salivarius colonies were then tested for production of BLIS activity by stabbing into a freshly seeded lawn of indicator I1 on BaCa (to more specifically identify S. salivarius K12, colonies were also picked into a freshly seeded lawn of indicator I3). Colonies producing definite inhibition of indicator I1 were considered presumptive SalA producers (for K12 identification, colonies were required to be inhibitory to both I1 and I3). A selection of representative inhibitory isolates were then P typed, and those yielding patterns 655, 676, 677, or 777 (together with a positive PCR product with the salAUS and salADS primer pair) were considered to be confirmed SalA producers. This information was then used to help estimate the number of CFU/ml of SalA producers in the original saliva specimens. For the detection of SalA using the induction assay, the freshly collected saliva samples were first clarified by centrifugation and then boiled for 15 min to kill the natural bacterial population, after which 20-μl aliquots were used in each assay (carried out in duplicate).
Nucleotide sequence accession numbers.
The new salivaricin A-variant DNA sequences (salA1 to salA5) described in this paper were submitted to GenBank and assigned the following accession numbers: DQ217832, salA1 (S. pyogenes 148); DQ217837, salA1 (S. dysgalactiae subsp. equisimilis 4003); DQ217836, salA1 (S. agalactiae 120); DQ217838, salA2 (S. salivarius K12); DQ217835, salA3 (S. salivarius JH); DQ217833, salA4 (S. salivarius 9); and DQ217834, salA5 (S. salivarius H21f).
RESULTS
Occurrence and in vitro expression of salA and its variants in oral streptococci.
A PCR-dot blot hybridization screen for salA was conducted with a selection of streptococcal strains representative of different oral species. These strains were also tested by deferred antagonism for inhibitory activity against indicators I1 to I9 and classified into three groups, (i) P type 655 (activity typical of SalA alone), (ii) P types of less than 655 (not consistent with SalA production), and (iii) P types greater than 655 (possibly sometimes attributable to SalA plus additional BLIS having activity against indicators not affected by SalA) (Table 1). A comparison of the frequency of salA in each species with the number of strains having a P-type pattern consistent with SalA production showed that the species having the highest frequency of salA-positive strains (97%) was S. pyogenes, but the only strains found to produce SalA were of serotype M4. On the other hand, only 2 S. agalactiae strains out of 16 and 1 S. dysgalactiae strain out of 13 were salA positive and, in each case, their BLIS activity profiles (P type 655) were consistent with production of SalA only. In the tested S. salivarius strains, the situation appeared more complex. Of the 36 strains having P types greater than 655 (e.g., 677, 777, etc.), 28 (77%) were salA positive, as was the sole P-type 655 isolate. In contrast to the situation for S. pyogenes, only 5 (17%) of the 30 S. salivarius isolates with P types less than 655 were salA positive. None of the 14 S. thermophilus and 12 S. uberis strains tested were salA positive.
TABLE 1.
Relationship between salA and P-type pattern in representative strains of different streptococcal species
| Species | P-type pattern | No. of strains | No. of strains having salA |
|---|---|---|---|
| S. pyogenes | >655a | 12 | 11 |
| 655 | 11 | 11 | |
| <655b | 50 | 49 | |
| S. salivarius | >655 | 36 | 28 |
| 655 | 1 | 1 | |
| <655 | 30 | 5 | |
| S. agalactiae | >655 | 0 | 0 |
| 655 | 2 | 2 | |
| <655 | 14 | 0 | |
| S. dysgalactiae | >655 | 0 | 0 |
| 655 | 1 | 1 | |
| <655 | 12 | 0 | |
| S. uberis | >655 | 10 | 0 |
| 655 | 0 | 0 | |
| <655 | 2 | 0 | |
| S. thermophilus | >655 | 0 | 0 |
| 655 | 0 | 0 | |
| <655 | 14 | 0 |
P-type pattern with a numerical value higher than 655, e.g., 677 or 777.
P-type pattern with a numerical value lower than 655, e.g., 204, 004, or 000.
An examination of the sequences of the PCR products of the salA-positive strains revealed several new variants of salA coding for amino acid differences in the propeptide parts of their putative SalA-like products (Table 2). Variations within the nucleotide sequence inferring amino acid differences within the SalA leader peptide regions were not taken into consideration when novel variants of the SalA peptides were defined. Inhibitory peptides corresponding to the propeptide forms of SalA1 to SalA5 were purified from culture supernatants of representative host bacteria, and it was established by mass spectrometry that each peptide had a molecular mass closely matching that predicted to be encoded by the form of salA detected in the host producer strain (Table 2). Interestingly, salA1 was the only form of the SalA structural gene detected in the P-type 655 strains of S. agalactiae and S. dysgalactiae and (with the exception of the M5 strain) also in the set of M-prototype strains of S. pyogenes. The M5 strain contained a S17L variation within the predicted SalA1 propeptide. By contrast, the form present in the sole P-type 655 S. salivarius strain was salA2. Only the serotype M11 and M37 strains of S. pyogenes had no detectable SalA structural gene (see below).
TABLE 2.
Propeptide sequences and masses of SalA variants
| Species | Prototype strain | SalA form | Predicted propeptide sequencea | Predicted mass (Da)b | Mass of purified peptide (Da)c |
|---|---|---|---|---|---|
| S. salivarius | 20P3 | SalA | KRGSGWIATITDDCPNSVFVCC | 2,316 | 2,315d |
| S. pyogenes | 148 | SalA1 | KKGSGWFATITDDCPNSVFVCC | 2,322 | 2,327 |
| S. dysgalactiae | 4003 | SalA1 | |||
| S. agalactiae | 120 | SalA1 | |||
| S. salivarius | K12 | SalA2 | KRGTGWFATITDDCPNSVFVCC | 2,364 | 2,368 |
| S. salivarius | JH | SalA3 | KKGPGWIATITDDCPNSIFVCC | 2,312 | 2,319 |
| S. salivarius | 9 | SalA4 | KRGPGWIATITDDCPNSIFVCC | 2,340 | 2,342 |
| S. salivarius | H21f | SalA5 | KRGPGWIATITDDCPNSVFVCC | 2,328 | 2,329 |
Residues that differ from SalA are underlined and in boldface type.
Based on the predicted translation product of the corresponding salA structural gene.
As determined by matrix-assisted laser desorption ionization-time of flight mass spectrometry.
From Ross et al. (13).
Previous studies had indicated that both SalA and SalA1 could up-regulate production of either SalA or SalA1 when added to cells containing the appropriate functional locus (25). By extending the induction assay to also include representative strains encoding SalA2 to SalA5, it was found that preparations containing each of the newly identified SalA variants were capable of inducing production of both the homologous and heterologous forms of SalA (Table 3). Specific induction of SalA3 production by strain JH could not be detected, since this strain also produces large amounts of an as-yet-uncharacterized BLIS (22) that masked the activity attributable to SalA3. The high specificity of the induction reaction for SalA and its variants was evident in that there was no cross-induction of any of the SalA variants brought about by the heterologous lantibiotics nisin and SA-FF22 (Table 3). In addition, it was found (results not shown) that there was no cross-induction effected in the production of SalA1 by S. pyogenes strain 148 brought about by exposure to preparations of the streptococcal lantibiotics streptin and salivaricin B. Hence, it is evident that the induction assay is highly specific for the SalA lantibiotic group of peptides.
TABLE 3.
Auto- and cross-inducing activity of SalA variants
| Source of SalA | Putative inducing moleculea | P-type pattern | Induction of inhibitor production in indicated strainc
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| 20P3 | 148 | 4003 | 120 | K12 | JH | 9 | FF22 | A5 | |||
| S. salivarius 20P3 | SalA | 677 | + | + | + | + | + | NDb | + | − | − |
| S. pyogenes 148 | SalA1 | 655 | + | + | + | + | + | ND | + | − | − |
| S. dysgalactiae 4003 | SalA1 | 655 | + | + | + | + | + | ND | + | − | − |
| S. agalactiae 120 | SalA1 | 655 | + | + | + | + | + | ND | + | − | − |
| S. salivarius K12 | SalA2 | 777 | + | + | + | + | + | ND | + | − | − |
| S. salivarius JH | SalA3 | 677 | + | + | + | + | + | ND | + | − | |
| S. salivarius 9 | SalA4 | 677 | + | + | + | + | + | ND | + | − | − |
| S. salivarius H25 | SalA5 | 777 | + | + | + | + | + | ND | + | − | − |
| S. pyogenes FF22 | SA-FF22 | 436 | − | − | − | − | − | ND | − | + | − |
| Lactobacillus lactis A5 | Nisin | 777 | − | − | − | − | − | ND | − | − | + |
All SalA-containing preparations were crude extracts that had been adjusted to a titer of 1 AU/ml against indicator I1 and were subinhibitory to the producer strains. Twenty microliters of extract per test well was used in the assay.
ND, not determined due to the production of multiple BLIS molecules by this strain, some of which interfered with the SalA induction assay.
+, induction detected; −, no induction detected.
Differential expression of salA and variants by streptococci.
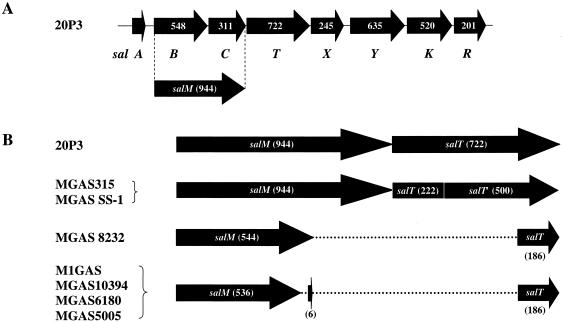
Previous studies (25) showed that a portion of salB and of salT and all of salC appeared to have been deleted from the SalA1 locus of S. pyogenes strain SF370. It should be noted here, however, that due to an error in the sequencing of the S. salivarius 20P3 locus, the lantibiotic processing gene(s) (now designated salM) was at that time designated as two distinct genes, salB and salC. PCR primers S.pyodelFwd and S.pyodelRev were used to amplify the DNA in this region from each of the M-serotype S. pyogenes prototype strains. The only S. pyogenes strains negative for salA and salMT (Table 4) were the M serotype 11 and M serotype 37 reference strains. Based on the PCR product size resulting from amplification reactions using the S.pyodelFwd and S.pyodelRev primer pair, five structural presentations of the salA1 locus were evident (Table 4). The various S. pyogenes strains appeared either to have (i) an intact salMT region (e.g., 20P3 and MGAS315) (Fig. 2), (ii) a single deletion in salMT (e.g., MGAS 8232) (Fig. 2), (iii) two deletions in salMT (e,g. M1GAS) (Fig. 2), (iv) no salMT region, although retaining remnants of salYKR (detected using the PCR primer pair salAF and salRR), or (v) no detectable component of the salA locus (as assessed by PCR using, in combination, the primers salAF, salAR, S.pyodelFwd, S.pyodelRev, and salRR).
TABLE 4.
Distribution of categories of salMT in M-prototype strains of S. pyogenes
| salMT category | M serotype(s) of S. pyogenes |
|---|---|
| Complete salMT | 3, 4,a 8, 15, 19, 24, 34, 41, 43, 52, 57,a 74, 79, 80, MGAS 315 (M3), MGAS SS-1 (M3)b |
| One deletion in salMT | 5, 17, 18, 36, 38, 89, MGAS 8232 (M18)b |
| Two deletions in salMT | 1, 2, 9, 12,a 25,a 27, 28,a 29, 32, 39, 42, 48, 49, 52,a 55, 58, 59, 60, 61, 62, 63, 64, 65, 66, 67, 68, 69, 71, 75, 77, 81, M1GAS (M1),b MGAS 5005 (M1),b MGAS 10394 (M6),b MGAS 6180 (M28)b |
| Large deletions in salA locus including salA1 | 11 |
| No detectable salA1 locus | 37 |
In addition to the M-prototype strain, there were 11 M4, 11 M57, 8 M12, 1 M25, 2 M28, and 1 M52 strains.
Genome sequence strains with M serotype shown in parentheses.
FIG. 2.
(A) Genetic structure of the salA locus in S. salivarius strain 20P3 illustrating that the modification genes previously described as salB and salC are now designated salM (modified from the work of Upton et al., reference 25). (B) Schematic representation of a comparative alignment of the putative salM and salT gene products of the seven S. pyogenes genome strains (MGAS315 [GenBank accession no. AE014074], MGAS SS-1 [GenBank accession no. BA000034], MGAS 8232 [GenBank accession no. AE009949], M1GAS [GenBank accession no. AE004092], MGAS10394 [GenBank accession no. CP00003], MGAS6180 [GenBank accession no. CP000056], and MGAS5005 [GenBank accession no. CP000017]), with the S. salivarius strain 20P3 salM and salT gene products as sequenced in the present study (GenBank accession no. AY005472). Three of the five different variations observed for salMT within S. pyogenes are shown; the other two variations (not illustrated) consist of the complete absence of any portion of the locus or a significant loss including the genes salA, salM, and salT and part of salY.
Interestingly, when additional apparently epidemiologically unrelated strains of S. pyogenes of M serotypes 4, 12, 25, 28, 52, and 57 were tested, they all clustered in the same salMT category as the homologous M prototype (Table 4). Furthermore, when the salA1 loci of seven genome-sequenced strains of S. pyogenes were aligned with the salA locus of S. salivarius strain 20P3, three different salMT locus categories were represented (Fig. 2); in each case, the category corresponded to that of the homologous M-prototype strain. A single-base-pair deletion in the salT gene resulting in a frame shift truncating the putative SalA1 transporter to a shortened (222-amino-acid) form was observed with 15 of 16 S. pyogenes strains tested. This natural mutation may account for the observed lack of production of SalA1 in many of the S. pyogenes strains that otherwise appear to have complete SalA1 loci.
The detection of specific SalA-inducing activity in the saliva of human subjects subsequent to their colonization with SalA-producing S. salivarius.
Our preliminary studies indicated that when 50-μl samples of serially (twofold) diluted purified preparations of SalA were assayed (i) by the well diffusion method for inhibitory activity against M. luteus or (ii) for auto-induction of SalA production by S. pyogenes strain 148, the latter was at least eightfold more sensitive. In addition, it had the benefit of high specificity, in that only molecules closely homologous to those encoded by the target bacterium were detected. It was decided that the induction assay would be used to detect the presence of SalA peptides in human saliva.
Eight subjects were used in the first study (Table 5). Four had SalA-producing S. salivarius present in their saliva at levels of at least 2.6 × 105 CFU/ml. Freshly collected saliva samples from each of these subjects effected induction of SalA1 production in the detector strain S. pyogenes 148. On the other hand, no SalA1-inducing activity was detected in the saliva of the four subjects who did not have detectable levels of SalA-producing S. salivarius (i.e., below the detection threshold of ca. 1 × 104 CFU/ml). In the second study, a 3-day course of 12 lozenges containing S. salivarius K12 (BLIS Technologies Ltd.), an oral probiotic strain known to produce both SalA2 and SalB when grown in vitro, was given according to the manufacturer's directions. Saliva samples taken just prior to commencement of the course were tested for their content of SalA-producing S. salivarius and for SalA1-inducing activity to ensure that there were no nonspecific saliva components capable of inducing SalA1 production (Table 6). One and seven days after the course of lozenges, further saliva samples were obtained and tested as for the presamples. SalA1-inducing activity was detected in the saliva samples of subjects in which significant levels of strain K12 colonization was achieved (enumerated as S. salivarius inhibitory to indicator I1 and also to indicator I3, which is sensitive to SalB but not SalA). The lowest salivary count of strain K12 leading to production of SalA2 at levels able to be detected in the induction assay appeared to be ca. 8 × 105 CFU per ml.
TABLE 5.
Detection of SalA in the saliva of subjects having natural salA-positive populations of S. salivarius
| Test specimen | CFU/ml of putative SalA-producing S. salivarius | Induction of SalA1 production in strain 148 |
|---|---|---|
| Saliva A | 4.0 × 106 | Yes |
| Saliva B | 1.4 × 107 | Yes |
| Saliva C | 2.6 × 105 | Yes |
| Saliva D | 1.3 × 106 | Yes |
| Saliva E | 0 | No |
| Saliva F | 0 | No |
| Saliva G | 0 | No |
| Saliva H | 0 | No |
| Positive controla | NAc | Yes |
| Negative controlb | NA | No |
Purified SalA (1 AU/ml).
Freeze-thaw extract from salA-negative S. salivarius strain Pre18.
NA, not applicable.
TABLE 6.
Use of the induction assay to detect the presence of SalA2 in the saliva of subjects on days 1 and 7 following colonization with the oral probiotic S. salivarius K12
| Subject | Counts of SalA2-producing S. salivarius and SalA1-inducing activity in saliva specimens
|
|||||
|---|---|---|---|---|---|---|
| Presample 1
|
Day 1 postcolonization
|
Day 7 postcolonization
|
||||
| Count (CFU/ml) | Induction | Count (CFU/ml) | Induction | Count (CFU/ml) | Induction | |
| 1 | 0 | No | 3.9 × 106 | No | 4.1 × 107 | Yes |
| 2 | 0 | No | 2.9 × 105 | No | 4.1 × 107 | Yes |
| 3 | 0 | No | 1 × 106 | Yes | 2.7 × 106 | Yes |
| 4 | 0 | No | 1 × 106 | Yes | 1.9 × 105 | No |
| 5 | 2.2 × 105 | No | 8 × 105 | Yes | 2.0 × 105 | No |
| 6 | 3.2 × 105 | No | 7 × 106 | Yes | 6.6 × 107 | Yes |
| 7 | 0 | No | 6.7 × 107 | Yes | 4.1 × 107 | No |
| 8 | 0 | No | 4.4 × 107 | Yes | 2.2 × 107 | No |
| Positive controla | NAc | Yes | NA | Yes | NA | Yes |
| Negative controlb | NA | No | NA | No | NA | No |
Purified SalA (1 AU/ml).
Freeze-thaw extract from salA-negative S. salivarius strain Pre18.
NA, not applicable.
DISCUSSION
In the present study, it has been shown that streptococci apparently producing only the lantibiotic SalA or its variants exhibit P-type pattern 655 when tested in standardized deferred antagonism tests on agar medium. The BLIS indicator strains not inhibited by SalA are a S. constellatus (I3), a serotype M4 S. pyogenes (itself a producer of SalA1) (I5), and a serotype T6 S. pyogenes (I8). Indicator I8 contains two deletions in the salMT section of its salA1 locus (results not shown) and, hence, is incapable of expressing SalA1 inhibitory or inducing activity. The precise mode of action of SalA is not known; therefore, further investigation is required to reveal the reason(s) for the apparent relative in vitro insensitivity of some S. pyogenes strains to SalA.
In the present study, only one strain of S. salivarius was found to be P type 655 and, in this case, it was the SalA2 form of the bacteriocin that was produced. All of the other S. salivarius strains found to produce SalA-type peptides also appeared to express additional BLIS activities, accounting for their broader activity spectra (e.g., P types 676, 677, and 777; results not shown). The expression of SalA1 was found to occur in the widest range of species (S. pyogenes, S. dysgalactiae, and S. agalactiae) and, in each case, it appeared to be the sole BLIS produced. All of the SalA1-producing strains of S. agalactiae and S. dysgalactiae were derived from nonhuman sources (14).
The sal locus was not detectable in the S. pyogenes strain representative of serotype M37 and was severely degraded in the strain representative of serotype M11. The M11 strain is an A-variant S. pyogenes strain, thought to have lost the ability to assemble intact group A carbohydrate during the course of prolonged serial subculture in vitro (D. Johnson, personal communication). The lack of an obvious selective advantage associated with SalA immunity for S. pyogenes strains grown for prolonged periods as laboratory monocultures could favor loss of immunity-related components of the locus. In addition, the M37 strain is quite unusual in that no other examples of strains of this serotype appear to have been isolated (D. Johnson, personal communication). Both of these observations are consistent with a survival advantage for S. pyogenes being linked to retention of at least the immunity-related components of the sal locus. The results from the present study support our previous observation (8) that all tested serotype M4 S. pyogenes strains appear capable of expressing SalA1. However, the association appears only to be with M4 (emm4) strains that also have T antigen 4. S. pyogenes strains having other M or T antigens combined with either the M4 or T4 antigen were inhibitor negative (8). Moreover, the SalA1-expressing S. dysgalactiae (8) and S. agalactiae (results not shown) strains neither contained emm4 nor expressed T4.
Whereas S. pyogenes strains appear almost exclusively to contain only the salA1 variant, S. salivarius strains exhibit a much greater diversity of SalA structural genes, but these apparently do not include salA1. Four new variants (salA2, salA3, salA4, and salA5) have been reported in this study. The amino acid differences, in general, reflect conservative changes that do not appear to affect induction activity or inhibitor activity of the peptides. The Ser-Pro differences in residue 4 of the propeptide could potentially have an impact on the peptide conformation. However, this Ser residue is not dehydrated in SalA (13), and Kyte-Doolittle plots (data not shown) indicate no significant predicted differences in the hydrophobicity of the SalA peptides containing either Ser or Pro. Two types of naturally occurring gene disruptions leading to abrogation of expression of biologically active SalA were also detected in strains of S. salivarius (data not shown): (i) S. salivarius strain MPS has a single-base-pair mutation in salA, resulting in the formation of a stop codon at residue 6 of the SalA propeptide, and (ii) S. salivarius strain H16H has a 16-bp insertion between bp 24 and 25 of salA, resulting in a frameshift disrupting the leader sequence of the SalA propeptide.
All five forms of SalA were purified, established to have an Mr consistent with that predicted from the expression of the corresponding structural gene, and shown to have inhibitory activity against indicator I1 and auto- and cross-inducing activity. We propose that inhibitory peptides that are established to have closely similar amino acid sequences and also to exhibit both specific cross-immunity and cross-inducing activities be considered members of the same bacteriocin cluster—in this case, the SalA cluster. Hence, we have classified SalA-like peptides containing amino acid changes within the propeptide region that do not have an impact on both (i) the cross- and auto-immunity and (ii) the induction specificities of the molecule as subtypes of that bacteriocin cluster (e.g., SalA, SalA1, and SalA2).
The presence of the SalA1 locus in all but one of the tested representatives of 53 different M serotypes of S. pyogenes indicates that this locus was acquired early in the establishment of the species. The general absence, however, of SalA1 production in these strains was initially ascribed to the deletion of a portion of the salB, salC, and salT (now salM and salT) region of the salA1 loci in most S. pyogenes strains (25). In the present study, more-detailed investigation of the salA1 loci in the M-prototype strains and also seven genome-sequenced S. pyogenes strains indicated that there are at least four possible mutations within salA1 loci that could adversely affect production of biologically active SalA1. These mechanisms consist of three different deletions within salMT that appear to be conserved within groups of M-serotype S. pyogenes strains and one frame shift in salT which would lead to truncation of the protein, presumably abrogating transport and leader peptide cleavage of SalA1. PCR-based screening of the salM/salT regions of additional representatives of M serotypes 4, 12, 25, 28, and 52 indicated that within M serotypes, the deletion type of the salA locus appears to be strongly conserved and, as such, may prove to be a useful marker of the evolutionary development of the species. Potentially, members belonging to a particular group will be more closely related to others within their own group than to those of other groups. At present, there appears to be no obvious correlation between the salMT type and the opacity factor status, tissue site preference (9), or emm pattern (4) of the strains. Horizontal gene transfer followed by intergenomic recombination appears to be the major cause of gene variation in S. pyogenes strains (3), which tends to negate phylogenetic signal from gene trees (9), indicating that any direct links between the salA1 locus type and the emm type of a strain may weaken over time. It is interesting, however, that the SalA locus occurs downstream of the same genes in each of the seven S. pyogenes genomes, indicating that the acquisition of the SalA locus was an early event in the establishment of S. pyogenes as a species.
S. salivarius is a primary and predominant colonizer of oral mucosal surfaces in humans and does not initiate infections in healthy individuals (16). On the other hand, the presence of large numbers of S. pyogenes in the oral cavity usually correlates with acute pharyngeal infection. S. pyogenes strains are typically very susceptible to growth inhibition by SalA when tested in vitro (13, 15), and the SalA-producing S. salivarius strain K12 (BLIS Throat Guard; BLIS Technologies) has recently been developed for use as an oral probiotic. However, although S. salivarius colonization of the oral cavity can be effected, direct evidence for the in situ production and detection of SalA in saliva has not been reported. Some indirect evidence for the oral production and activity of SalA was obtained by showing that the population levels of indigenous α-hemolytic cocci exhibiting resistance to SalA were significantly higher in samples of the oral microbiota from subjects who were naturally colonized with large numbers of SalA-producing S. salivarius (23). The inference was that sufficient SalA had been produced in the oral cavity to select for a relatively resistant population. In the present study, we have used a highly specific autoinduction assay to directly detect the presence of SalA in the saliva of subjects having SalA-producing S. salivarius as part of their normal flora, or following their use of a commercial product containing the probiotic SalA2-positive strain S. salivarius K12. It is important to note, however, that the production of salivaricin A to levels detectable in the saliva is extremely varied from individual to individual, with the number of S. salivarius K12 per ml of saliva required to produce detectable levels apparently ranging from 8 × 105 to 6.7 × 107 CFU per ml (Table 6). This variability may result in part either from variable saliva flow rates, which may differentially dilute the SalA, or from adsorption of SalA by cells of the normal flora, either of which could potentially differ significantly from individual to individual. This is, to our knowledge, the first demonstration of in situ production of a lantibiotic at a level that may be capable of influencing either the survival or genetic regulation of other species within the oral cavity.
Acknowledgments
We thank N. Heng for help with preparation of the manuscript.
This work was supported by grant UO0605 from the Marsden Fund, Royal Society of New Zealand, and also by a research grant from the University of Otago.
REFERENCES
- 1.Beall, B., R. Facklam, and T. Thompson. 1996. Sequencing emm-specific PCR products for routine and accurate typing of group A streptococci. J. Clin. Microbiol. 34:953-958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dempster, R. P., and J. R. Tagg. 1982. The production of bacteriocin-like substances by the oral bacterium Streptococcus salivarius. Arch. Oral Biol. 27:151-157. [DOI] [PubMed] [Google Scholar]
- 3.Feil, E. J., E. C. Holmes, D. E. Bessen, M. S. Chan, N. P. Day, M. C. Enright, R. Goldstein, D. W. Hood, A. Kalia, C. E. Moore, J. Zhou, and B. G. Spratt. 2001. Recombination within natural populations of pathogenic bacteria: short-term empirical estimates and long-term phylogenetic consequences. Proc. Natl. Acad. Sci. USA 98:182-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hollingshead, S. K., J. Arnold, T. L. Readdy, and D. E. Bessen. 1994. Molecular evolution of a multigene family in group A streptococci. Mol. Biol. Evol. 11:208-219. [DOI] [PubMed] [Google Scholar]
- 5.Hubbard, M. J., N. J. McHugh, and D. L. Carne. 2000. Isolation of ERp29, a novel endoplasmic reticulum protein, from rat enamel cells: evidence for a unique role in secretory-protein synthesis. Eur. J. Biochem. 267:1945-1957. [DOI] [PubMed] [Google Scholar]
- 6.Hubbard, M. J., and N. J. McHugh. 1996. Mitochondrial ATP synthase F1-β-subunit is a calcium-binding protein. FEBS Lett. 391:323-329. [DOI] [PubMed] [Google Scholar]
- 7.Jack, R. W., A. Carne, J. Metzger, S. Stefanovic, H. G. Sahl, G. Jung, and J. Tagg. 1994. Elucidation of the structure of SA-FF22, a lanthionine-containing antibacterial peptide produced by Streptococcus pyogenes strain FF22. Eur. J. Biochem. 220:455-462. [DOI] [PubMed] [Google Scholar]
- 8.Johnson, D. W., J. R. Tagg, and L. W. Wannamaker. 1979. Production of a bacteriocine-like substance by group-A streptococci of M-type 4 and T-pattern 4. J. Med. Microbiol. 12:413-427. [DOI] [PubMed] [Google Scholar]
- 9.McGregor, K. F., B. G. Spratt, A. Kalia, A. Bennett, N. Bilek, B. Beall, and D. E. Bessen. 2004. Multilocus sequence typing of Streptococcus pyogenes representing most known emm types and distinctions among subpopulation genetic structures. J. Bacteriol. 186:4285-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Novak, J., P. W. Caufield, and E. J. Miller. 1994. Isolation and biochemical characterization of a novel lantibiotic mutacin from Streptococcus mutans. J. Bacteriol. 176:4316-4320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qi, F., P. Chen, and P. W. Caufield. 2000. Purification and biochemical characterization of mutacin I from the group I strain of Streptococcus mutans, CH43, and genetic analysis of mutacin I biosynthesis genes. Appl. Environ. Microbiol. 66:3221-3229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Qi, F., P. Chen, and P. W. Caufield. 1999. Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthesis genes. Appl. Environ. Microbiol. 65:3880-3887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ross, K. F., C. W. Ronson, and J. R. Tagg. 1993. Isolation and characterization of the lantibiotic salivaricin A and its structural gene salA from Streptococcus salivarius 20P3. Appl. Environ. Microbiol. 59:2014-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schofield, C. R., and J. R. Tagg. 1983. Bacteriocin-like activity of group B and group C streptococci of human and of animal origin. J. Hyg. 90:7-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Simpson, W. J., N. L. Ragland, C. W. Ronson, and J. R. Tagg. 1995. A lantibiotic gene family widely distributed in Streptococcus salivarius and Streptococcus pyogenes. Dev. Biol. Stand. 85:639-643. [PubMed] [Google Scholar]
- 16.Smith, D. J., J. M. Anderson, W. F. King, J. Van Houte, and M. A. Taubman. 1993. Oral streptococcal colonization of infants. Oral. Microbiol. Immunol. 8:1-4. [DOI] [PubMed] [Google Scholar]
- 17.Suvorov, A. N., and J. J. Ferretti. 1996. Physical and genetic chromosomal map of an M type 1 strain of Streptococcus pyogenes. J. Bacteriol. 178:5546-5549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tagg, J. R. 1984. Production of bacteriocin-like inhibitors by group A streptococci of nephritogenic M types. J. Clin. Microbiol. 19:884-887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tagg, J. R., and L. V. Bannister. 1979. “Fingerprinting” beta-haemolytic streptococci by their production of and sensitivity to bacteriocine-like inhibitors. J. Med. Microbiol. 12:397-411. [DOI] [PubMed] [Google Scholar]
- 20.Tagg, J. R., K. P. Dierksen, and M. Upton. August. 2004. Lantibiotic. U.S. patent 6,773,912B1.
- 21.Tagg, J. R., V. Pybus, L. V. Phillips, and T. M. Fiddes. 1983. Application of inhibitor typing in a study of the transmission and retention in the human mouth of the bacterium Streptococcus salivarius. Arch. Oral Biol. 28:911-915. [DOI] [PubMed] [Google Scholar]
- 22.Tompkins, G. R., and J. R. Tagg. 1987. Bacteriocin-like inhibitory activity associated with beta-hemolytic strains of Streptococcus salivarius. J. Dent. Res. 66:1321-1325. [DOI] [PubMed] [Google Scholar]
- 23.Tompkins, G. R., and J. R. Tagg. 1989. The ecology of bacteriocin-producing strains of Streptococcus salivarius. Microb. Ecol. Health Dis. 2:19-28. [Google Scholar]
- 24.Upton, M., P. E. Carter, M. Morgan, G. F. Edwards, and T. H. Pennington. 1995. Clonal structure of invasive Streptococcus pyogenes in Northern Scotland. Epidemiol. Infect. 115:231-241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Upton, M., J. R. Tagg, P. Wescombe, and H. F. Jenkinson. 2001. Intra- and interspecies signaling between Streptococcus salivarius and Streptococcus pyogenes mediated by SalA and SalA1 lantibiotic peptides. J. Bacteriol. 183:3931-3938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wescombe, P. A., and J. R. Tagg. 2003. Purification and characterization of streptin, a type A1 lantibiotic produced by Streptococcus pyogenes. Appl. Environ. Microbiol. 69:2737-2747. [DOI] [PMC free article] [PubMed] [Google Scholar]