Abstract
Strain MC-1 is a marine, microaerophilic, magnetite-producing, magnetotactic coccus phylogenetically affiliated with the α-Proteobacteria. Strain MC-1 grew chemolithotrophically with sulfide and thiosulfate as electron donors with HCO3−/CO2 as the sole carbon source. Experiments with cells grown microaerobically in liquid with thiosulfate and H14CO3−/14CO2 showed that all cell carbon was derived from H14CO3−/14CO2 and therefore that MC-1 is capable of chemolithoautotrophy. Cell extracts did not exhibit ribulose-1,5-bisphosphate carboxylase-oxygenase (RubisCO) activity, nor were RubisCO genes found in the draft genome of MC-1. Thus, unlike other chemolithoautotrophic, magnetotactic bacteria, strain MC-1 does not appear to utilize the Calvin-Benson-Bassham cycle for autotrophy. Cell extracts did not exhibit carbon monoxide dehydrogenase activity, indicating that the acetyl-coenzyme A pathway also does not function in strain MC-1. The 13C content of whole cells of MC-1 relative to the 13C content of the inorganic carbon source (Δδ13C) was −11.4 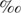 . Cellular fatty acids showed enrichment of 13C relative to whole cells. Strain MC-1 cell extracts showed activities for several key enzymes of the reverse (reductive) tricarboxylic acid (rTCA) cycle including fumarate reductase, pyruvate:acceptor oxidoreductase and 2-oxoglutarate:acceptor oxidoreductase. Although ATP citrate lyase (another key enzyme of the rTCA cycle) activity was not detected in strain MC-1 using commonly used assays, cell extracts did cleave citrate, and the reaction was dependent upon the presence of ATP and coenzyme A. Thus, we infer the presence of an ATP-dependent citrate-cleaving mechanism. These results are consistent with the operation of the rTCA cycle in MC-1. Strain MC-1 appears to be the first known representative of the α-Proteobacteria to use the rTCA cycle for autotrophy.
. Cellular fatty acids showed enrichment of 13C relative to whole cells. Strain MC-1 cell extracts showed activities for several key enzymes of the reverse (reductive) tricarboxylic acid (rTCA) cycle including fumarate reductase, pyruvate:acceptor oxidoreductase and 2-oxoglutarate:acceptor oxidoreductase. Although ATP citrate lyase (another key enzyme of the rTCA cycle) activity was not detected in strain MC-1 using commonly used assays, cell extracts did cleave citrate, and the reaction was dependent upon the presence of ATP and coenzyme A. Thus, we infer the presence of an ATP-dependent citrate-cleaving mechanism. These results are consistent with the operation of the rTCA cycle in MC-1. Strain MC-1 appears to be the first known representative of the α-Proteobacteria to use the rTCA cycle for autotrophy.
Magnetotactic bacteria are a morphologically, metabolically, and phylogenetically diverse assemblage of prokaryotes that synthesize intracellular, membrane-bound, single-magnetic-domain crystals of the minerals magnetite (Fe3O4) and greigite (Fe3S4) (4). These unique structures, called magnetosomes (3), are generally organized in chains within the cell (5) and cause the cell to passively align along magnetic field lines (21). Magnetotaxis results from this magnetic alignment combined with active cellular motility (21). The discovery of magnetotaxis 30 years ago by Blakemore (9) was based on the observation of large populations of extremely motile magnetotactic cocci all swimming in the same direction. Since then, the magnetotactic cocci have been found to be ubiquitous in marine, brackish, and freshwater habitats (20, 21, 49). The magnetotactic cocci are characterized structurally by being bilophotrichous (possessing two bundles of flagella on one side of the cell) (21, 38, 39). Cells also often possess pili, and some contain intracellular sulfur-rich globules (13, 21, 38). Magnetotactic cocci display a polar preference in their swimming direction and thus exhibit polar magneto-aerotaxis (21); this distinguishes these magnetotactic bacteria from Magnetospirillum spp., which display axial magneto-aerotaxis (21). Phylogenetically, based on 16S rRNA sequences, all known magnetotactic cocci cluster at the base of the α subdivision of the Proteobacteria, the division that contains almost all of the known Fe3O4-producing magnetotactic bacteria (15, 20, 49). Despite the ubiquity of the magnetotactic cocci and the ease of collecting cells in large numbers (39), only one strain of a magnetotactic coccus, presently called strain MC-1, has been isolated and cultivated in axenic culture (21). Strain MC-1 was isolated from a chemically stratified estuary in the Pettaquamscutt River (R.I.) and appears to be an obligate microaerophile.
We recently demonstrated that the marine magnetotactic bacterial strains MV-1 and MV-2 grow chemolithoautotrophically (6). Cell extracts of these strains show ribulose-1,5-bisphophate carboxylase/oxygenase (RubisCO) activity, and both strains possess intact form II RubisCO genes (6) and thus rely on the Calvin-Benson-Bassham (CBB) cycle for CO2 fixation and autotrophy. Cells of strain MC-1 also appeared to grow chemolithoautotrophically using sulfide and thiosulfate as electron donors (21, 37). Because strain MC-1 grows microaerophilically (21), it seemed probable that it would also use the CBB cycle for autotrophy and RubisCO for CO2 fixation as in most other aerobic chemolithoautotrophic members of the domain Bacteria. However, cell extracts of strain MC-1 did not show RubisCO activity, and typical genes of the various forms of RubisCO are not present in the draft genome sequence of strain MC-l (sequence available at the Joint Genome Institute website, http://genome.jgi-psf.org/draft_microbes/magm1/magm1.home.html) (6). Recently, we discovered open reading frames (ORFs) in the draft MC-1 genome that have high sequence identity to pyruvate:ferredoxin oxidoreductase and 2-oxoglutarate:ferredoxin oxidoreductase from autotrophic members of the Aquificales that utilize the reverse or reductive tricarboxylic acid (rTCA) cycle for CO2 fixation and autotrophy. This suggested to us that strain MC-1 might also use the rTCA cycle for CO2 fixation. The aim of this study was to further investigate this possibility.
MATERIALS AND METHODS
Bacteria and growth conditions.
The marine magnetotactic coccus strain MC-1 was isolated from water collected from the oxic-anoxic interface of the Pettaquamscutt River Estuary, R.I. (15). This strain has now been characterized and will be named shortly (D. A. Bazylinski, unpublished data). Strain MC-1 is an obligate microaerophile and may be an obligate autotroph (D. A. Bazylinski, unpublished results) although it appeared to grow with acetate heterotrophically when it was first isolated (37).
For experiments to determine the contribution of CO2 to cell carbon, cells of strain MC-1 were grown microaerobically in semisolid medium at 28°C from small inocula in [O2] gradients, with thiosulfate (S2O32−) as an electron donor and radiolabeled [14C]bicarbonate (HCO3−; American Radiolabeled Chemicals, St. Louis, Mo.) as the sole carbon source, as previously described (6).
For enzyme assays, lipid extraction, and isotopic composition measurements, strain MC-1 was grown in liquid medium. Cells were grown in 850 ml of medium in 2-liter glass bottles. The medium consisted of an artificial seawater (6) base to which was added the following (per liter), in order, prior to autoclaving: 5 ml of modified Wolfe's mineral elixir (21), 0.25 g of NH4Cl, and 100 μl of 0.2% (wt/vol) aqueous resazurin. The pH of the medium was adjusted to 7.0, 1.26 g of NaHCO3 was added per liter, and the vessel was sealed. The medium was then bubbled with 7.5% CO2 gas in N2 (flow rate about 100 ml min−1) passed over heated copper wire to remove O2 for 45 min. The medium was sealed and autoclaved. After the autoclaving and cooling steps, the following solutions were injected into the medium bottles, in order, from anaerobic stocks (except for the cysteine, which was made fresh and filter sterilized directly into the medium): 1.28 ml of 0.5 M KHPO4 buffer, pH 6.9, 0.85 ml of 0.23 M neutralized cysteine · HCl · H2O, 8.5 ml of 25% (wt/vol) Na2S2O3 · 5H2O, and 0.4 ml of vitamin solution (21). The medium was allowed to become reduced (colorless), after which 2.5 ml of 0.01 M FeSO4 dissolved in 0.2 N HCl was injected. The medium was inoculated, after which 6 ml of sterile O2 was injected (0.4% of the headspace) and the medium was carefully placed without shaking so as not to disturb the forming [O2] gradient, at 25°C. The [O2] gradient that became established was clearly evident by the fact that the surface of the medium became pink while the bottom remained colorless. Growth initiated at the pink-colorless interface near the surface, and as growth and turbidity increased, greater amounts of sterile O2 could be injected and used by growing cells.
Chlorobaculum tepidum was grown at 47°C under bright light according to the method of Wahlund et al. (54), except that 10 mM morpholinopropanesulfonic acid (MOPS) was added to the medium. Rhodospirillum rubrum was grown anaerobically at 30°C under bright light in SMN medium (per liter: 1 μg of biotin, 4 g of malic acid, 1 g of NH4Cl, 2.8 mg of H3BO3, 20 mg of Na2EDTA, 4 mg of ferric citrate, 1 mg of Na2MoO4, 0.6 g of KH2PO4, 0.9 g of K2HPO4, 0.25 g of MgSO4 • 7H2O, 0.1 g of CaCl2 • 2H2O, 1.0 g of yeast extract, 10.5 g of MOPS, 10 μM NiCl2; pH 7.0) (34) and then transferred to anaerobic RRNCO medium (per liter: 2 μg of biotin, 10 ml of chelated iron-molybdenum solution [per liter: 0.28 g of H3BO3, 2 g of Na2EDTA, 0.4 g of ferric citrate, 0.1 g of Na2MoO4], 0.25 g of MgSO4 • 7H2O, 0.132 g of CaCl2 • 2H2O, 1 g of NH4Cl, 20 μM NiCl2, 1.0 g of yeast extract 2.1 g of MOPS, 0.82 g of sodium acetate, 10 mM KHPO4 buffer, 0.01% Na2S • 9H2O, 12.5 mM NaHCO3; pH 7.1) (35) under 1 atm CO.
CO2 as source of cell carbon.
The contribution of CO2 to total cellular carbon of strain MC-1 grown autotrophically, based on protein determinations, was measured as H14CO3−/14CO2 incorporation into cell material as previously described (6).
Preparation of cell extracts.
Cell extracts of MC-1, C. tepidum, and R. rubrum were prepared by harvesting cells grown in liquid culture by centrifugation at 6,000 × g for 15 min at 4°C. MC-1 cells were washed once in artificial seawater buffered with 10 mM Tris · HCl, pH 7.0. Cells of C. tepidum were washed once in 100 mM Tris · HCl, pH 7.0; R. rubrum cells were washed once in 50 mM KHPO4 buffer, pH 7.0. Cells were recentrifuged at 6,000 × g for 15 min at 4°C. Cells were then resuspended in 50 mM KHPO4, pH 7.0, and lysed by two to three passes through a French pressure cell at 124 MPa. The lysate was kept anaerobic by releasing it into a N2-filled tube connected to the exit of the French pressure cell and by carrying out all transfers of the lysate in an anaerobic chamber (Coy Laboratory Products, Grass Lake, Mich.). Cell lysates were centrifuged anaerobically at 10,000 × g for 30 min at 4°C to remove nonlysed cells, cell debris, and magnetosomes when present. Protein in cell extracts was measured using Coomassie brilliant blue (10) with a Bio-Rad protein assay kit (Bio-Rad Laboratories, Hercules, Calif.). Lyophilized bovine serum albumin was used as a standard (Sigma-Aldrich, St. Louis, Mo.).
Analysis and carbon isotopic composition of lipids. (i) Lipid extraction.
Cells of strain MC-1 grown autotrophically were freeze-dried and extracted for lipids (55, 61, 62). Lipids were separated on a silicic acid column into fractions containing neutral lipids, glycolipids, and polar lipids (24). The polar lipids were treated using a mild alkaline methanolysis to produce fatty acid methyl esters (FAMEs). FAMEs were identified using an Agilent 6890 series gas chromatograph interfaced to an Agilent 5973 mass selective detector (61). Mass spectra were determined by electron impact at 70 eV. Methyl heneicosanoate was used as the internal standard. FAMEs were expressed as equivalent peaks against the internal standard.
(ii) Stable carbon isotopes.
Carbon isotope compositions of the FAMEs were determined using an HP 6890 gas chromatograph connected to a Finnigan MAT Delta Plus-XL mass spectrometer (Scientific Instrument Services, Ringoes, N.J) (61, 62). Carbon isotope compositions of whole cells were also determined. The 13C/12C ratios of whole cells and headspace CO2 were determined using a Delta Plus isotope ratio mass spectrometer (Scientific Instrument Services) with a precision of ± 0.2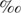 .
.
Enzyme assays.
The RubisCO activity of cell extracts was determined as described by Beudeker et al. (8), except that the dithiothreitol concentration was changed to 5 mM and the pH was adjusted to 7.0.
Pyruvate:acceptor oxidoreductase and 2-oxoglutarate:acceptor oxidoreductase activities were determined by measuring the pyruvate- or α-ketoglutarate-dependent reduction, respectively, of methyl viologen (MV) at 578 nm (ɛ578 nm = 9.8 × 103 M−1 cm−1) (59) spectrophotometrically. The reactions were carried out anaerobically in serum-stoppered cuvettes under N2 at 25°C. The standard assay mixture (final volume, 2 ml) contained 50 mM KHPO4 buffer, pH 7.0, 0.25 mM coenzyme A (CoA), 1 mM MgCl2, 5 mM MV, 1 mM dithiothreitol, and 10 mM sodium pyruvate or 10 mM sodium α-ketoglutarate as the substrate. Anaerobic cell extract was injected to initiate each reaction. Two molecules of MV are reduced per each molecule of 2-oxoacid oxidized.
Carbon monoxide dehydrogenase activity was also determined spectrophotometrically by measuring the CO-dependent reduction of MV at 578 nm. The reaction was carried out exactly as for the 2-oxoacid assays above except that CO was the substrate. Reaction mixtures were rendered anaerobic using N2 gas, after which the N2 was replaced by CO. Two molecules of MV are reduced per each molecule of CO oxidized.
Fumarate reductase activity was determined spectrophotometrically based on a procedure designed for an assay for nitrous oxide reductase (48). This assay uses photochemically reduced (rather than chemically reduced) benzyl viologen (BV; ɛ600 nm = 10.4 × 103 M−1 cm−1) (36) as the electron donor. The reaction mixture (1 ml) contained 50 mM KHPO4 buffer, pH 7.0, 0.4 mM proflavin, 1 M triethanolamine, 10 mM BV, and 2 mM sodium fumarate. The pH of the final reaction mix was 7.0, and reactions were carried out 25°C. BV was photochemically reduced to the blue cation radical by exposure to a blue fluorescent bulb (Philips F15T8/B; Atlanta Light Bulbs Inc., Tucker, Ga.) at a distance of about 10 cm. The reaction was initiated by injection of anaerobic cell extract. Two molecules of BV are oxidized per each molecule of fumarate reduced.
Two spectrophotometric assays were used to assay ATP-citrate lyase activity. In the first, acetyl-CoA formed from the ATP-dependent cleavage of citrate reacts with hydroxylamine to form acetyl-hydroxamate, which then forms a colored complex with Fe3+ that can be measured spectrophotometrically (1, 29). The alternative method is a coupled assay: malate formed by the ATP-dependent cleavage of citrate is determined by the oxidation of NADH in the presence of malate dehydrogenase (MDH) (30). The MDH-coupled assay used here was based on the method of Wahlund and Tabita (53). The reaction mixture (1 ml) contained 10 mM MgCl2, 0.1 mM sodium citrate, 2 mM NADH, 3 mM ATP, 0.9 mM CoA, 100 nkat (6.0 U) MDH, in 100 mM Tris · HCl, pH 7.0. The reaction was initiated by adding NADH. The oxidation of NADH was determined spectrophotometrically at 340 nm (ɛ = 6.22 mM−1 cm−1) at 25°C.
Thin-layer chromatography (TLC) in combination with autoradiography was also used to test for ATP-dependent citrate cleavage in cell extracts of strain MC-1. The reaction mixture (200 μl) contained 10 mM MgCl2, 0.1 mM sodium citrate, 25 kBq [1,5-14C]citric acid (American Radiolabeled Chemicals), 2 mM NADH, 3 mM ATP, 0.9 mM CoA, and 100 nkat (6.0 U) MDH in 100 mM Tris · HCl, pH 7.0. Reactions were carried out at 25°C and initiated by the addition of 50 μl of cell extract to the reaction mixture. Cellulose plates (20-by 20-cm Cellulose F; EMD Chemicals Inc., Gibbstown, N.J.) were equilibrated in a sealed glass chamber containing 1-pentanol:formic acid:water (48.8:48.8:2.4). A total of 10 μl of reaction mixture was spotted onto the plate at timed intervals. TLC proceeded until the solvent front reached the top of the plate. After the solvent had vaporized, the plates were exposed to X-ray film, left for 4 to 6 days, and then developed. The following compounds were used as references (100 nmol loaded as a 4-μl sample) to determine their Rf values (in parentheses): acetyl-CoA (0.13), citrate (0.29), CoA (0.17), malate (0.38), oxaloacetate (0.21), pyruvate (0.52), and succinate (0.56). Spots representing the presence of these compounds were visualized by spraying the plate with a bromocresol green solution, followed by a brief exposure to ammonia fumes (14).
Nucleotide sequence accession numbers.
Nucleotide sequences for genes encoding proteins thought to be involved in the operation of the rTCA cycle in strain MC-1 (see Fig. 2) are available in the GenBank database under accession number AAAN03000051.
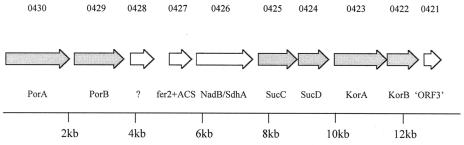
FIG. 2.
Gene cluster within contig 294 (GenBank accession no. AAAN03000051) in the draft genome of strain MC-1, showing identities of ORFs that encode putative proteins implicated in the rTCA cycle (shaded in gray). Each ORF is represented by an arrow indicating the direction of gene transcription. Above each ORF is a number provided by the U.S. Department of Energy/Joint Genome Institute draft annotated genome of strain MC-1. Identities of proteins are based on BLASTP searches and are shown beneath each ORF. The question mark indicates a protein of unknown function. PorA and PorB are the α and β subunits of pyruvate:acceptor oxidoreductase, respectively; fer2+ACS is a putative protein with an N-terminal 2Fe-2S iron-sulfur cluster binding domain (fer2) and a C-terminal acetyl-CoA synthase (ACS) domain. NadB/SdhA is a FAD-binding protein showing homology to both aspartate oxidase and succinate dehydrogenase/fumarate reductase, a flavoprotein subunit protein. SucC and SucD are the β and α subunits of succinyl-CoA synthetase, respectively. KorA and KorB are the α and β subunits of 2-oxoglutarate:acceptor oxidoreductase, respectively. “ORF3” matches an unknown putative protein that immediately follows the KorA and KorB genes in H. thermophilus.
RESULTS
Growth experiments.
Cells of strain MC-1 can now be grown routinely in liquid culture. Initiation of growth, however, is dependent on the formation of an [O2] gradient in the medium. This, in turn, was dependent on the amount of O2 that was introduced to anaerobic medium and the size of the inoculum (number of cells). Typically, the amount of O2 that allowed growth to proceed from an initial inoculum of 1% was about 0.5% O2 in the headspace. Higher amounts resulted in much longer lag periods or no growth. Growth of strain MC-1 always initiated at the oxic-anoxic interface (based on the color of the resazurin in the medium), and eventually cells formed a macroscopic biofilm on the glass at the surface of the medium. Once the medium became close to colorless, additional O2 could be introduced to the culture, which resulted in increased growth in the form of a thick layer at the surface. At this point, volumes of O2 up to 4 to 5% of the headspace could be introduced without a detrimental effect on the cells, in order to promote further growth. Growth of strain MC-1 is slow: given that a strict [O2] gradient must be maintained in order for MC-1 to grow during the first 6 to 7 days after inoculation, cell growth cannot be measured without disrupting the gradient, thereby interfering with cell growth if not completely inhibiting it. Using cell numbers before and after growth, we estimate an average doubling time of ∼30 h for strain MC-1 in these gradient cultures.
The growth medium contained S2O32− as the electron donor and no organic carbon other than a small amount of vitamins and cysteine. Cells of strain MC-1 did not grow, however, when S2O32− was omitted. The major source of carbon in the medium was CO2/HCO3−. Cells produced internal S-rich globules when grown on S2O32− (D. A. Bazylinski and B. L. Cox, unpublished data). Elemental S also precipitated external to the cell in the growth medium (data not shown).
Bicarbonate/CO2 as source of cell carbon.
Cells of strain MC-1 were grown microaerobically from small inocula in [O2] gradients with S2O32− as an electron donor and radiolabeled H14CO3−/14CO2 as the sole carbon source. For each experiment, the amount of 14C from H14CO3−/14CO2 incorporated into protein was compared with the calculated amount of protein carbon determined by protein measurements and an average carbon content of protein of 54% (16, 32) (Table 1). The results show that the average percentage (triplicate analysis) of protein carbon derived from H14CO3−/14CO2 in cells of strain MC-1 was 99.9 ± 2.5%.
TABLE 1.
Contribution of CO2 to protein carbon in cells of the magnetotactic coccus strain MC-1 grown in semisolid, oxygen-gradient medium containing 10 mM thiosulfate as the electron source and 14C-bicarbonate/CO2 as the sole carbon sourcea
| Total CO2 fixed (μg of C)b | CO2 incorporation (protein fraction [μg of C]) | Total cell protein (μg) | Total protein carbon (μg of C)c | % Protein carbon from CO2d | % Recovery of radiolabel |
|---|---|---|---|---|---|
| 27.4 ± 1.3 | 7.26 ± 0.82 (26.5 ± 2.9%)e | 13.4 ± 1.3 | 7.26 ± 0.70 | 99.9 ± 2.5 | 82.6 ± 2.9 |
Results are reported as means ± standard deviations derived from triplicate cultures.
CO2 fixation in uninoculated tubes was virtually undetectable, and counts from all fractions of uninoculated controls never exceeded 50 cpm and were subtracted from counts.
Computed from total cellular protein times 54%, the average carbon content of protein (wt/wt) from Doolittle (16) and Jukes et al. (32).
Calculated as follows: 100% × μg of C incorporated into protein fraction/calculated total protein C.
C incorporated from CO2 into protein fraction as a percentage of total CO2 fixed.
Profiles and carbon isotopic compositions of whole cells and fatty acids of strain MC-1.
The fatty acid profile of strain MC-1 was dominated by 16:1, 16:0, and 18:1 (Table 2) fatty acids. The isotopic compositions of these fatty acids were −21.8 to −24.5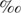 , which were 3.7 to 6.5
, which were 3.7 to 6.5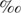 heavier (more positive) than that of whole cells (−28
heavier (more positive) than that of whole cells (−28 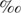 ) (Table 2).
) (Table 2).
TABLE 2.
Major phospholipid fatty acids and their isotopic compositions for cells of strain MC-1
| Parameter or component | Amount (mol%) ina:
|
δ13C ( |
|
|---|---|---|---|
| MC-1A | MC-1B | ||
| Fatty acid | |||
| 14:0 | 0.6 | 0.4 | |
| 15:0 | 0.3 | 0.1 | |
| Σ16:1c | 60.9 | 63.1 | −21.8d |
| 16:0 | 29.1 | 26.5 | −24.5 |
| Σ18:1e | 9.0 | 9.9 | −22.0f |
| 18:0 | 0.2 | 0.1 | |
| Total | 100.0 | 100.0 | |
| Whole cells | −28.1 | ||
| CO2 | −16.7 | ||
| ɛFatty acids-whole cellsg | 3.7-6.5 | ||
| ɛWhole cells-CO2g | −11.6 | ||
Values were determined in duplicate MC-1 samples designated MC-1A and MC-1B.
Values were determined for MC-1A plus MC-1B.
Dominated by 16:1ω7c.
Weighed average of 16:1ω7c and 16:1a of an unknown double-bond position.
Dominated by 18:1ω7c.
Isotopic composition for 18:1ω7c only.
ɛA-B = [(1,000 + δ13CA)/(1,000 + δ13CB) − 1] × 1,000.
Enzyme assays.
RubisCO activity was not detected in cell extracts of strain MC-1. Cell extracts of chemolithoautotrophically grown strain MV-1 were used as a positive control (6) and showed a RubisCO-specific activity of 10.2 nmol of CO2 fixed min−1 mg−1 of cell protein. Strain MV-1 is known to utilize the CBB cycle (6).
MC-1 cell extracts showed both pyruvate:acceptor (MV) and 2-oxoglutarate:acceptor (MV) oxidoreductase activity: 3.9 ± 0.9 nmol min−1 mg of cell protein−1 and 9.1 ± 5.5 nmol min−1 mg of cell protein−1, respectively (values given as mean ± standard deviation of triplicate analyses). In both cases, the reduction of MV was dependent upon the presence of the substrate, CoA, or cell extract. Activity was not detected when the cell extract was heated to 100°C for 5 min prior to its introduction in the assay mixture. Fumarate reductase activity was determined to be 21.4 ± 6.1 nmol min−1 mg of cell protein−1. Enzyme activities did not occur in the absence of substrate or cell extract or when the cell extract was heated to 100°C for 5 min prior to its introduction in the assay mixture. Cell extracts of MC-1 did not display detectable CO dehydrogenase activity. Cell extracts of R. rubrum grown anaerobically with CO were used as a positive control and showed a specific activity of 0.51 ± 0.12 nmol min−1 mg of cell protein−1.
We did not detect ATP-citrate lyase activity using the MDH-coupled oxidation of NADH in cell extracts of strain MC-1. Relatively high rates of NADH oxidation were observed in the presence of cell extract, but it was independent of exogenous ATP, CoA, or citrate. ATP-citrate lyase activity was also not detected using the hydroxamate method. Cell extracts of C. tepidum were used as a positive control and showed an ATP-citrate lyase specific activity of 101 nmol min−1 mg of cell protein−1 in the MDH-coupled NADH oxidation assay.
ATP-dependent cleavage of citrate.
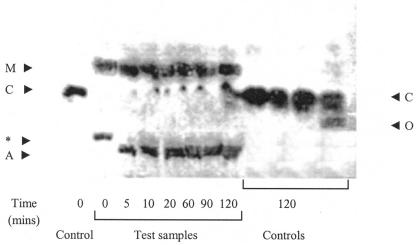
To determine the ATP-dependent cleavage of citrate, we incubated cell extract with [1,5-14C]citrate, NADH, ATP, CoA, and MDH and determined the products of this reaction over time using TLC (Fig. 1). In this experiment, the major products of ATP-dependent citrate cleavage were acetyl-CoA and malate (based on Rf values) as expected. Depletion of citrate occurred virtually instantly, with two spots forming immediately after the introduction of the cell extract (time zero), coinciding with the loss of the citrate spot. The higher of the two spots appears to represent malate (Rf of ∼0.4). The lower spot represents a compound that migrates further from the origin (has a higher Rf value) than the lower spot observed at 5 min (and thereafter), which is presumed to be acetyl-CoA based on its Rf value (∼0.13). The identity of the lower spot at time zero is unknown; it may represent an intermediate that was converted into acetyl-CoA by 5 min. Also visible was a small spot that migrated slightly above citrate that became increasingly stronger in intensity over time. The Rf value was too low to be succinate (Rf of ∼0.56); its identity is unclear, but it does not appear to be an artifact since it was not formed in control experiments. A very small, faint spot was slightly visible in the test experiment that had an Rf value of that of succinate that was not observable in the controls. Attempts to visually enhance this spot using computer software rendered the rest of the figure unclear. In the absence of cell extract, CoA, and ATP, cleavage of citrate was not observed even after 120 min. However, some depletion of citrate occurred in the absence of NADH, and a fainter spot was observed just under citrate, which possibly represents oxaloacetate based on an Rf value of ∼0.21.
FIG. 1.
TLC results, showing the ATP-dependent cleavage of 14C-citrate by cell extracts of strain MC-1. TLC was performed at 25°C on cellulose plates and visualized using autoradiography. A 10-μl sample of reaction mixture was spotted onto the plate at timed intervals (0, 5, 10, 20, 60, 90, and 120 min). Each reaction was started by the addition of cell extract (time zero). Test samples comprised cell extract, ATP, NADH, CoA, sodium citrate, 14C-citrate, MgCl2, and malate dehydrogenase in Tris · HCl buffer, pH 7.0. Five controls are shown, one in the first lane, spotted at time zero (no cell extract), and the rest in the four final lanes (no cell extract, no ATP, no CoA, and no NADH, respectively), spotted at 120 min. Spots corresponding to organic acids are as follows, based on Rf values: A (acetyl-CoA), C (citrate), M (malate), O (oxaloacetate). The spot marked with the asterisk is unknown, and was apparent only in the test sample from time zero. We discern two spots in the final lane (no-NADH control), including a diffuse upper spot (citrate).
DISCUSSION
Cells of strain MC-1 grow well microaerobically in semisolid and liquid growth medium containing an [O2] gradient and S2O32− as an electron donor. The main source of carbon is CO2/HCO3−, and the medium contained only traces of organic carbon as low amounts of vitamins and cysteine, which they cannot grow on alone. It thus seems likely that strain MC-1 grows as a chemolithoautotroph in this medium. To demonstrate autotrophy in strain MC-1, we determined both the amount of 14C from H14CO3−/14CO2 incorporated into protein and the total amount of protein in separate experimental cultures. We then calculated the amount of protein carbon from protein determinations using an average carbon content of protein of 54% (16, 32). The amount of 14C from H14CO3−/14CO2 incorporated into protein was compared to the calculated amount of protein carbon. The percentage of calculated protein carbon derived from H14CO3−/14CO2 was 99.9% for strain MC-1. We infer from this result that all cell carbon in strain MC-1 grown with S2O32− comes from H14CO3−/14CO2, and we conclude that strain MC-1 grows chemolithoautotrophically.
Because strain MC-1 is an obligate microaerophile (4, 20) and because other magnetotactic bacteria have been shown to use the CBB cycle for autotrophy, we suspected that it would also use the CBB cycle and RubisCO for CO2 fixation as do most other aerobic chemolithoautotrophic bacteria. Cell extracts of strain MC-1 did not exhibit RubisCO activity, however, and the various forms of RubisCO genes could not be found in the draft genome sequence of strain MC-l (6). We have also ruled out the possibility that strain MC-1 uses the acetyl-CoA pathway because we did not detect CO dehydrogenase activity, nor could we identify genes for this enzyme in the draft genome. We cannot unequivocally rule out operation of the 3-hydroxyproprionate pathway in addition to the rTCA cycle, although we regard this as unlikely, given that we did not find genes for key enzymes of this pathway (malonyl-CoA reductase and propionyl-CoA synthase) in the draft genome of MC-1.
In this study, we present several strong lines of evidence that MC-1 utilizes the rTCA cycle for CO2 fixation and autotrophy when grown chemolithoautotrophically. First, all chemolithoautotrophs produce biomass characterized by discrimination against 13C during CO2 fixation. Of the four known autotrophic pathways in Bacteria and Archaea, the rTCA cycle and 3-hydroxypropionate cycle typically discriminate less strongly against 13C than the CBB cycle and reductive acetyl-CoA pathway. Isotope fractionation between biomass and CO2 (−11.6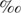 ) (Table 2) for MC-1 falls within the range of values previously obtained for the rTCA cycle (22, 41). This range is much lower than that of the CBB cycle (−20 to −26
) (Table 2) for MC-1 falls within the range of values previously obtained for the rTCA cycle (22, 41). This range is much lower than that of the CBB cycle (−20 to −26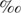 ) and the reductive acetyl-CoA pathway (−34 to −40
) and the reductive acetyl-CoA pathway (−34 to −40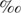 ) and is lower than the 3-hydroxypropionate cycle (−13.7
) and is lower than the 3-hydroxypropionate cycle (−13.7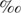 ) (26, 51). However, fractionation values around −13
) (26, 51). However, fractionation values around −13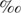 were obtained for one mesophilic ɛ-proteobacterial species (“Candidatus Arcobacter sulfidicus”) that was recently demonstrated to use the rTCA cycle (28, 56).
were obtained for one mesophilic ɛ-proteobacterial species (“Candidatus Arcobacter sulfidicus”) that was recently demonstrated to use the rTCA cycle (28, 56).
Isotopic compositions of cellular fatty acids provide additional proof of the operation of the rTCA cycle in strain MC-1. In the rTCA cycle, straight-chain fatty acids are hypothesized to be synthesized from the pool of acetyl-CoA that is generated by the first step (citrate cleavage) of the cycle (47). Thus, straight-chain lipids are enriched in 13C relative to whole cells; that is, the δ13C of straight chain lipids should be greater than that of whole cells. Strain MC-1 exhibits fractionations of 3.7 to 6.5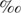 between fatty acids and whole cells (Table 2). Green and purple sulfur bacteria that use the rTCA cycle for carbon fixation produce lipids that are 2 to 16% enriched in 13C relative to biomass (50). The values for MC-1 are less than the 10 to 16
between fatty acids and whole cells (Table 2). Green and purple sulfur bacteria that use the rTCA cycle for carbon fixation produce lipids that are 2 to 16% enriched in 13C relative to biomass (50). The values for MC-1 are less than the 10 to 16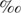 enrichment observed in the green sulfur phototroph Chlorobium limicola but higher than the range of 2 to 4
enrichment observed in the green sulfur phototroph Chlorobium limicola but higher than the range of 2 to 4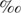 seen in Thiocapsa roseopersicina (50), Thermocrinis ruber (31), and Persephonella marina (60, 63), all of which potentially use the rTCA cycle (or a modification of this pathway) for carbon assimilation (31, 50, 63).
seen in Thiocapsa roseopersicina (50), Thermocrinis ruber (31), and Persephonella marina (60, 63), all of which potentially use the rTCA cycle (or a modification of this pathway) for carbon assimilation (31, 50, 63).
Finally, the presence of specific enzyme activities key to the rTCA cycle, as well as the presence of the genes for most of these enzymes in the genome of strain MC-1, indicates that this organism relies on the rTCA cycle for autotrophy. The activities of 2-oxoglutarate:acceptor oxidoreductase, pyruvate:acceptor oxidoreductase, and fumarate reductase were demonstrated in cell extracts of strain MC-1. Pyruvate:acceptor oxidoreductase and 2-oxoglutarate:acceptor oxidoreductase catalyze the reductive carboxylation of acetyl-CoA and succinyl-CoA to pyruvate and 2-oxoglutarate, respectively, in the rTCA cycle (17). In C. limicola, the electron donor for these reactions is ferredoxin (17). Fumarate reductase, a key enzyme of the rTCA cycle (28), catalyzes the reduction of fumarate to succinate, thereby replacing succinate dehydrogenase of the oxidative TCA cycle (17). The archaeon Thermoproteus neutrophilus, which appears to use the rTCA cycle, turns off fumarate reductase during heterotrophic growth by repression and/or inactivation of this enzyme (7, 43).
We identified putative genes in the draft genome of MC-1 for 2-oxoglutarate:acceptor oxidoreductase, pyruvate:acceptor oxidoreductase, and succinyl-CoA synthetase, a reversible enzyme also involved in the rTCA cycle. These genes are found in the same region of the genome (Fig. 2). The two putative genes that code for the α and β subunits of 2-oxoglutarate:acceptor oxidoreductase indicate that this enzyme belongs to the αβ-type enzyme family found in Hydrogenobacter thermophilus (but not Aquifex aeolicus) and some archaeal species (57, 58). The arrangement of these putative genes in strain MC-1 is similar to that in H. thermophilus in that the α and β subunit genes are immediately followed by an ORF of unknown but presumably identical function based on sequence identity.
We did not detect ATP-citrate lyase activity in MC-1 with commonly used assays and could not find evidence for genes for any ATP-dependent citrate-cleaving enzymes (ATP-citrate lyase, citryl-CoA synthetase, and citryl-CoA lyase) in the draft genome. Nevertheless, cell extracts showed ATP-dependent citrate cleavage activity. In the oxidative TCA cycle, citrate synthase catalyzes the condensation of acetyl-CoA and oxaloacetate to form citrate; for bacteria that employ the rTCA for CO2 fixation and autotrophy, this reaction is carried out in reverse: citrate is cleaved to acetyl-CoA and oxaloacetate (17). Our results for MC-1 show malate as a major product, with no detectable fumarate, and an extremely faint succinate spot (Fig. 1); these data were similar to those obtained for the archaeon T. neutrophilus, although the latter shows no visible spot representing acetyl-CoA (7). The results obtained when NADH was withheld from the reaction are problematic, given that acetyl-CoA might be expected to be a product of citrate cleavage independent of the presence of NADH. This, combined with the absence of known citrate-cleaving enzyme genes in the draft genome of strain MC-1, suggests that the citrate cleavage mechanism in MC-1 may be different from that of other bacterial species that use the rTCA cycle. Many such bacteria (e.g., representatives of Chlorobiales and ɛ-Proteobacteria) possess a two-subunit ATP-citrate lyase that catalyzes the cleavage of citrate in a single reaction (28, 33). However, H. thermophilus and A. aeolicus cleave citrate using two separate enzymes: citryl-CoA synthetase activates citrate with CoA to form citryl-CoA, which citryl-CoA lyase cleaves to produce oxaloacetate and acetyl-CoA (1, 2). In other prokaryotes demonstrated or inferred to use the rTCA cycle for carbon fixation, the mechanism for citrate cleavage is unknown. Moreover, demonstration of ATP-dependent citrate cleavage activity has not always correlated with the identification of the genes or protein(s) responsible. For example, several representatives of the Thermoproteaceae are thought to use the rTCA cycle for autotrophic CO2 fixation based on enzyme activities (e.g., T. neutrophilus [7] or Pyrobaculum islandicum [27]) or on the identification of putative genes for enzymes of the oxidative or reductive TCA cycles in the complete genomes (e.g., Pyrobaculum aerophilum [19] or Thermoproteus tenax [46]). For T. tenax, the genes responsible for the ATP-dependent cleavage of citrate could not be identified (19, 46). Siebers et al. (46) proposed an alternative mechanism for citrate cleavage in T. tenax that has yet to be verified experimentally. Thus, it seems that disparate enzymatic mechanisms for ATP-dependent citrate cleavage are represented among prokaryotes. In light of this, and given that ATP-citrate lyase is also expressed in some δ-Proteobacteria during organotrophic growth (40, 44), we concur with Hügler et al. (28) in advising caution with regard to the use of acl genes as functional markers for the presence of the rTCA cycle (11, 12).
Within the domain Bacteria, the rTCA cycle has been demonstrated for representatives of the Aquificales (1, 2, 7, 45), Chlorobiales (17, 23, 33), δ-Proteobacteria (44), and ɛ-Proteobacteria (28). This study represents the first evidence of the operation of the rTCA cycle in a member of the α-Proteobacteria group. The magnetotactic cocci show no close affinities to any other α-Proteobacteria and appear to constitute a unique lineage that diverged early from the main branch of the α-Proteobacteria (15, 18) and are only distantly related to other magnetotactic α-proteobacteria (e.g., Magnetospirillum spp., strain MV-1). Representatives of both the Bacteria and Archaea are known to employ the rTCA cycle, and it has been hypothesized by some to be the primordial metabolic cycle for CO2 fixation (42, 52). To date, all known species in the domain Bacteria that utilize the rTCA cycle for CO2 fixation and autotrophy are found in the Aquificales, Chlorobiales, and Proteobacteria (28). In contrast to the Proteobacteria clade, the Aquificales and Chlorobiales are generally regarded as deep-branching lineages within the domain Bacteria. However, the distribution of the rTCA cycle in these groups is in accord with another phylogenetic hypothesis, which holds that these groups are contained within a clade that branched off relatively late, and that the δ- and ɛ-Proteobacteria were the first subdivisions to emerge after the Proteobacteria split from the Aquificales, followed by the α-Proteobacteria, and finally the β- and γ-Proteobacteria (25). The operation of the rTCA cycle in strain MC-1 is congruent with this hypothesis, given the basal position of MC-1 within the α-Proteobacteria lineage. Alternatively, lateral gene transfer may be the major factor for the distribution of the rTCA cycle in prokaryotes.
Acknowledgments
We thank G. Fuchs, J. F. Robyt, and L. E. Nielsen for assistance with TLC; D. W. Choi, Y. Do, and A. A. DiSpirito for assistance with CO dehydrogenase assays; M. T. Madigan for cultures of C. tepidum and R. rubrum; and T. S. Brettin, D. Martinez, N. Thayer, and G. Xie of the Joint Genome Institute and Los Alamos National Laboratories for help and suggestions with regard to searching the genome of strain MC-1.
This work was supported by U.S. National Science Foundation grants MCB-9696027 and EAR-0311950 and by U.S. Department of Energy Financial Assistance Award DE-FC09-96SR18546 to the University of Georgia Research Foundation.
REFERENCES
- 1.Aoshima, M., M. Ishii, and Y. Iharashi. 2004a. A novel enzyme, citryl-CoA synthetase, catalyzing the first step of the citrate cleavage reaction in Hydrogenobacter thermophilus TK-6. Mol. Microbiol. 52:751-761. [DOI] [PubMed] [Google Scholar]
- 2.Aoshima, M., M. Ishii, and Y. Iharashi. 2004b. A novel enzyme, citryl-CoA lyase, catalyzing the second step of the citrate cleavage reaction in Hydrogenobacter thermophilus TK-6. Mol. Microbiol. 52:763-770. [DOI] [PubMed] [Google Scholar]
- 3.Balkwill, D. L., D. Maratea, and R. P. Blakemore. 1980. Ultrastructure of a magnetic spirillum. J. Bacteriol. 141:1399-1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bazylinski, D. A., and R. B. Frankel. 2004. Magnetosome formation in prokaryotes. Nat. Rev. Microbiol. 2:217-230. [DOI] [PubMed] [Google Scholar]
- 5.Bazylinski, D. A., A. J. Garratt-Reed, and R. B. Frankel. 1993. Electron microscopic studies of magnetosomes in magnetotactic bacteria. Microsc. Res. Tech. 27:389-401. [DOI] [PubMed] [Google Scholar]
- 6.Bazylinski, D. A., A. J. Dean, T. J. Williams, L. K. Long, S. L. Middleton, and B. L. Dubbels. 2004. Chemolithoautotrophy in the marine, magnetotactic bacterial strains MV-1 and MV-2. Arch. Microbiol. 182:373-387. [DOI] [PubMed] [Google Scholar]
- 7.Beh, M., G. Strauss, R. Huber, K. O. Stetter, and G. Fuchs. 1993. Enzymes of the reductive citric acid cycle in the autotrophic eubacterium Aquifex pyrophilus and in the archaebacterium Thermoproteus neutrophilus. Arch. Microbiol. 160:306-311. [Google Scholar]
- 8.Beudeker, R. F., G. C. Cannon, J. G. Kuenen, and J. M. Shively. 1980. Relations between d-ribulose-1,5-bisphosphate carboxylase, carboxysomes and CO2 fixing capacity in the obligate chemolithotroph Thiobacillus neapolitanus grown under different limitations in the chemostat. Arch. Microbiol. 124:185-189. [Google Scholar]
- 9.Blakemore, R. P. 1975. Magnetotactic bacteria. Science 190:377-379. [DOI] [PubMed] [Google Scholar]
- 10.Bradford, M. M. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72:248-254. [DOI] [PubMed] [Google Scholar]
- 11.Campbell, B. J., and S. C. Cary. 2004. Abundance of reverse tricarboxylic acid cycle genes in free-living microorganisms at deep-sea hydrothermal vents. Appl. Environ. Microbiol. 70:6282-6289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Campbell, B. J., J. L. Stein, and S. C. Cary. 2003. Evidence of chemolithoautotrophy in the bacterial community associated with Alvinella pompejana, a hydrothermal vent polychaete. Appl. Environ. Microbiol. 69:5070-5078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cox, B. L., R. Popa, D. A. Bazylinski, B. Lanoil, S. Douglas, A. Belz, D. L. Engler, and K. H. Nealson. 2002. Organization and elemental analyses of P-, S-, and Fe-rich inclusions in a population of freshwater magnetococci. Geomicrobiol. J. 19:387-406. [Google Scholar]
- 14.Dawson, R. M. C., D. C. Elliot, W. H. Elliot, and K. M. Jones. 1986. Data for biochemical research, 3rd ed. Oxford Science Publications, Clarendon Press, Oxford, United Kingdom.
- 15.DeLong, E. F., R. B. Frankel, and D. A. Bazylinski. 1993. Multiple evolutionary origins of magnetotaxis in bacteria. Science 259:803-806. [DOI] [PubMed] [Google Scholar]
- 16.Doolittle, R. F. 1981. Similar amino acid sequences: chance or common ancestry? Science 214:149-159. [DOI] [PubMed] [Google Scholar]
- 17.Evans, M. C. W., B. B. Buchanan, and D. I. Arnon. 1966. A new ferredoxin-dependent carbon reduction cycle in a photosynthetic bacterium. Proc. Natl. Acad. Sci. USA 55:928-934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fernández de Henestrosa, A. R., J. Cuñé, G. Mazón, B. L. Dubbels, D. A. Bazylinski, and J. Barbé. 2003. Characterization of a new LexA binding motif in the marine magnetotactic bacterium strain MC-1. J. Bacteriol. 185:4471-4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fitz-Gibbon, S. T., H. Ladner, U. J. Kim, K. O. Stetter, M. I. Simon, and J. H. Miller. 2002. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum. Proc. Natl. Acad. Sci. USA 99:984-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Flies, C. B., J. Peplies, and D. Schüler. 2005. Combined approach for characterization of uncultivated magnetotactic bacteria from various aquatic environments. Appl. Environ. Microbiol. 71:2723-2731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Frankel, R. B., D. A. Bazylinski, M. S. Johnson, and B. L. Taylor. 1997. Magneto-aerotaxis in marine coccoid bacteria. Biophys. J. 73:994-1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fuchs, G. 1980. Alternate pathways of autotrophic CO2 fixation, p. 365-382. In H. G. Schlegel and B. Bowien (ed.), Autotrophic bacteria. Springer-Verlag, Berlin, Germany.
- 23.Fuchs, G., E. Stupperich, and G. Eden. 1980. Autotrophic CO2 fixation in Chlorobium limicola. Evidence for the operation of the reductive tricarboxylic acid cycle in growing cells. Arch. Microbiol. 128:64-71. [Google Scholar]
- 24.Guckert, J. B., C. B. Antworth, P. D. Nichols, and D. C. White. 1985. Phospholipid ester-linked fatty acid profiles as reproducible assays for changes in prokaryotic community structure of estuarine sediments. FEMS Microbiol. Ecol. 31:147-158. [Google Scholar]
- 25.Gupta, R. S., and E. Griffiths. 2002. Critical issues in bacterial phylogeny. Theor. Popul. Biol. 61:423-434. [DOI] [PubMed] [Google Scholar]
- 26.Holo, H., and R. Sirevåg. 1986. Autotrophic growth and CO2 fixation of Chloroflexus aurantiacus. Arch. Microbiol. 145:173-180. [Google Scholar]
- 27.Hügler, M., H. Huber, K. O. Stetter, and G. Fuchs. 2003. Autotrophic CO2 fixation pathways in archaea (Crenarchaeota). Arch. Microbiol. 179:160-173. [DOI] [PubMed] [Google Scholar]
- 28.Hügler, M., C. O. Wirsen, G. Fuchs, C. D. Taylor, and S. M. Sievert. 2005. Evidence for autotrophic CO2 fixation via the reductive tricarboxylic acid cycle by members of the ɛ subdivision of proteobacteria. J. Bacteriol. 187:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ishii, M., Y. Igarashi, and T. Kodama. 1989. Purification and characterization of ATP:citrate lyase from Hydrogenobacter thermophilus TK-6. J. Bacteriol. 171:1788-1792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ivanovsky, R. N., N. V. Sintsov, and E. N. Kondratieva. 1980. ATP-linked citrate lyase activity in the green sulfur bacterium Chlorobium limicola forma thiosulfatophilum. Arch. Microbiol. 128:239-241. [Google Scholar]
- 31.Jahnke, L. L., W. Eder, R. Huber, J. M. Hope, K. U. Hinrichs, J. M. Hayes, D. J. Des Marais, S. L. Cady, and R. E. Summons. 2001. Signature lipids and stable carbon isotope analyses of Octopus Spring hyperthermophilic communities compared with those of Aquificales representatives. Appl. Environ. Microbiol. 67:5179-5189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jukes, T. H., R. Holmquist, and H. Moise. 1975. Amino acid composition of proteins: selection against the genetic code. Science 189:50-51. [DOI] [PubMed] [Google Scholar]
- 33.Kanao, T., T. Fukui, H. Atomi, and T. Imanaka. 2001. ATP-citrate lyase from the green sulfur bacterium Chlorobium limicola is a heteromeric enzyme composed of two distinct gene products. Eur. J. Biochem. 68:1670-1678. [PubMed] [Google Scholar]
- 34.Kerby, R. L., S. S. Hong, S. A. Ensign, L. J. Coppoc, P. W. Ludden, and G. P. Roberts. 1992. Genetic and physiological characterization of the Rhodospirillum rubrum carbon monoxide dehydrogenase system. J. Bacteriol. 174:5284-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kerby R. L., P. W. Ludden, and G. P. Roberts. 1995. Carbon monoxide-dependent growth of Rhodospirillum rubrum. J. Bacteriol. 177:2241-2244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kristjansson, J. K., and T. C. Hollocher. 1980. First practical assay for soluble nitrous oxide reductase of denitrifying bacteria and a partial kinetic characterization. J. Biol. Chem. 255:704-707. [PubMed] [Google Scholar]
- 37.Meldrum, F. C., S. Mann, B. R. Heywood, R. B. Frankel, and D. A. Bazylinski. 1993. Electron microscopy study of magnetosomes in a cultured coccoid magnetotactic bacterium. Proc. R. Soc. Lond. B. 251:231-236. [Google Scholar]
- 38.Moench, T. T. 1988. Bilophococcus magnetotacticus gen. nov. sp. nov., a motile, magnetic coccus. Antonie Leeuwenhoek 54:483-496. [DOI] [PubMed] [Google Scholar]
- 39.Moench, T. T., and W. A. Konetzka. 1978. A novel method for the isolation and study of a magnetotactic bacterium. Arch. Microbiol. 119:203-212. [DOI] [PubMed] [Google Scholar]
- 40.Möller, D., R. Schauder, G. Fuchs, and R. K. Thauer. 1987. Acetate oxidation to CO2 via a citric acid cycle involving an ATP-citrate lyase: mechanism for the synthesis of ATP via substrate level phosphorylation in Desulfobacter postgatei growing on acetate and sulfate. Arch. Microbiol. 148:202-207. [Google Scholar]
- 41.Preuss, A., R. Schauder, G. Fuchs, and W. Stichler. 1989. Carbon isotope fractionation by autotrophic bacteria with three different CO2 fixation pathways. Z. Naturforsch. Sect. C 44:397-402. [Google Scholar]
- 42.Romano, A. H., and T. Conway. 1996. Evolution of carbohydrate metabolic pathways. Res. Microbiol. 147:448-455. [DOI] [PubMed] [Google Scholar]
- 43.Schäfer, S., C. Barkowski, and G. Fuchs. 1986. Carbon assimilation by the autotrophic thermophilic archaebacterium Thermoproteus neutrophilus. Arch. Microbiol. 146:301-308. [Google Scholar]
- 44.Schauder, R., F. Widdel, and G. Fuchs. 1987. Carbon assimilation pathways in sulfate-reducing bacteria. II. Enzymes of a reductive citric acid cycle in the autotrophic Desulfobacter hydrogenophilus. Arch. Microbiol. 148:218-225. [Google Scholar]
- 45.Shiba, H., T. Kawasumi, Y. Igarashi, T. Kodama, and Y. Minoda. 1985. The CO2 assimilation via the reductive tricarboxylic-acid cycle in an obligately autotrophic, aerobic hydrogen-oxidizing bacterium, Hydrogenobacter thermophilus. Arch. Microbiol. 141:198-203. [Google Scholar]
- 46.Siebers, B., B. Tjaden, K. Michalke, C. Dorr, H. Ahmed, M. Zaparty, P. Gordon, C. W. Sensen, A. Zibat, H. P. Klenk, S. C. Schuster, and R. Hensel. 2004. Reconstruction of the central carbohydrate metabolism of Thermoproteus tenax by use of genomic and biochemical data. J. Bacteriol. 186:2179-2194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sirevåg, R., B. B. Buchanan, J. A. Berry, and J. H. Troughton. 1977. Mechanisms of CO2 fixation in bacterial photosynthesis studied by the carbon isotope fractionation technique. Arch. Microbiol. 112:35-38. [DOI] [PubMed] [Google Scholar]
- 48.Snyder, S. W., and T. C. Hollocher. 1987. Purification and some characteristics of nitrous oxide reductase from Paracoccus denitrificans. J. Biol. Chem. 262:6515-6525. [PubMed] [Google Scholar]
- 49.Spring, S., U. Lins, R. Amann, K. H. Schleifer, L. C. Ferreira, D. M. Esquivel, and M. Farina. 1998. Phylogenetic affiliation and ultrastructure of uncultured magnetic bacteria with unusually large magnetosomes. Arch. Microbiol. 169:136-147. [DOI] [PubMed] [Google Scholar]
- 50.van der Meer, M. T. J., S. Schouten, and J. S. Sinninghe Damsté. 1998. The effect of the reversed tricarboxylic-acid cycle on the 13C contents of bacterial lipids. Org. Geochem. 28:527-533. [Google Scholar]
- 51.van der Meer, M. T. J., S. Schouten, B. E. van Dongen, W. I. C. Rijpstra, G. Fuchs, J. S. Sinninghe Damsté, W. de Leeuw, and D. M. Ward. 2001. Biosynthetic controls on the 13C contents of organic components in the photoautotrophic bacterium Chloroflexus aurantiacus. J. Biol. Chem. 276:10971-10976. [PubMed] [Google Scholar]
- 52.Wächtershäuser, G. 1990. Evolution of the first metabolic cycles. Proc. Natl. Acad. Sci. USA 87:200-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wahlund, T. M., and F. R. Tabita. 1997. The reductive tricarboxylic acid cycle of carbon dioxide assimilation: initial studies and purification of ATP-citrate lyase from the green sulfur bacterium Chlorobium tepidum. J. Bacteriol. 179:4859-4867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wahlund, T. M., C. R. Woese, R. W. Castenholz, and M. T. Madigan. 1991. A thermophilic green sulfur bacterium from New Zealand hot springs, Chlorobium tepidum, sp. nov. Arch. Microbiol. 156:81-90. [Google Scholar]
- 55.White, D. C., W. M. Davis, J. S. Nickels, J. D. King, and R. J. Bobbie. 1979. Determination of the sedimentary microbial biomass by extractible lipid phosphate. Oceologia 40:51-62. [DOI] [PubMed] [Google Scholar]
- 56.Wirsen, C. O., S. M. Sievert, C. M. Cavanaugh, S. J. Molyneaux, A. Ahmad, L. T. Taylor, E. F. DeLong, and C. D. Taylor. 2002. Characterization of an autotrophic sulfide-oxidizing marine Arcobacter sp. that produces filamentous sulfur. Appl. Environ. Microbiol. 68:316-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yoon, K. S., M. Ishii, Y. Igarashi, and T. Kodama. 1996. Purification and characterization of 2-oxoglutarate:ferredoxin oxidoreductase from a thermophilic, obligately chemolithoautotrophic bacterium, Hydrogenobacter thermophilus TK-6. J. Bacteriol. 178:3365-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Yun, N. R., M. Yamamoto, H. Arai, M. Ishii, and Y. Igarashi. 2002. A novel five-subunit-type 2-oxoglutalate:ferredoxin oxidoreductases from Hydrogenobacter thermophilus TK-6. Biochem. Biophys. Res. Commun. 22:280-286. [DOI] [PubMed] [Google Scholar]
- 59.Zeikus, J. G., G. Fuchs, W. Kenealy, and R. K. Thauer. 1977. Oxidoreductase involved in cell carbon synthesis of Methanobacterium thermoautotropicum. J. Bacteriol. 132:604-613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, C. L., B. W. Fouke, G. Bonheyo, A. Peacock, D.C. White, Y. Huang, and C. S. Romanek. 2004. Lipid biomarkers and carbon-isotopes of modern travertine deposits (Yellowstone National Park, USA): implications for biogeochemical dynamics in hot-spring systems. Geochim. Cosmochim. Acta 68:3157-3169. [Google Scholar]
- 61.Zhang, C. L., Z. Huang, J. Cantu, R. D. Pancost, R. L. Brigmon, T. W. Lyons, and R. Sassen. 2005. Lipid biomarkers and carbon-isotope signatures of a microbial (Beggiatoa) mat associated with gas hydrates in the Gulf of Mexico. Appl. Environ. Microbiol. 71:2106-2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang, C. L., Y. Li, Q. Ye, J. Fong, A. D. Peacock, E. Blunt, J. Fang, D. R. Lovley, and D. C. White. 2003. Carbon isotope signatures of fatty acids in Geobacter metallireducens and Shewanella algae. Chem. Geol. 195:17-28. [Google Scholar]
- 63.Zhang, C. L., Q. Ye, D. Goetz, A.-L. Reysenbach, A. Peacock, D. C. White, J. Horita, D. R. Cole, J. Fong, L. Pratt, J. Fang, and Y. Huang. 2002. Carbon isotopic fractionations associated with thermophilic bacteria Thermotoga maritima and Persephonella marina. Environ. Microbiol. 4:58-64. [DOI] [PubMed] [Google Scholar]