Abstract
Identification and functional analysis of key members of bacterial communities in marine and estuarine environments are major challenges for obtaining a mechanistic understanding of biogeochemical processes. In the Baltic Sea basins, as in many other marine environments with anoxic bodies of water, the oxic-anoxic interface is considered a layer of high bacterial turnover of sulfur, nitrogen, and carbon compounds that has a great impact on matter balances in the whole ecosystem. We focused on autotrophic denitrification by oxidation of reduced sulfur compounds as a biogeochemically important process mediating concomitant turnover of sulfur, nitrogen, and carbon. We used a newly developed approach consisting of molecular analyses in stimulation experiments and in situ abundance. The molecular approach was based on single-strand conformational polymorphism (SSCP) analysis of the bacterial community RNA, which allowed identification of potential denitrifiers based on the sequences of enhanced SSCP bands and monitoring of the overall bacterial community during the experiments. Sequences of the SSCP bands of interest were used to design highly specific primers that enabled (i) generation of almost complete 16S rRNA gene sequences using experimental and environmental DNA as templates and (ii) quantification of the bacteria of interest by real-time PCR. By using this approach we identified the bacteria responsible for autotrophic denitrification as a single taxon, an epsilonproteobacterium related to the autotrophic denitrifier Thiomicrospira denitrificans. This finding was confirmed by material balances in the experiments that were consistent with those obtained with continuous cultures of T. denitrificans. The presence and activity of a bacterium that is phylogenetically and physiologically closely related to T. denitrificans could be relevant for the carbon budget of the central Baltic Sea because T. denitrificans exhibits only one-half the efficiency for carbon dioxide fixation per mol of sulfide oxidized and mol of nitrate reduced of Thiobacillus denitrificans hypothesized previously for this function.
The Baltic Sea is the world's largest brackish water environment. It is strongly influenced by anthropogenic loads of nitrogen (6, 43). The central Baltic Sea is characterized by a pronounced salinity gradient (depth, 60 to 80 m) that inhibits vertical mixing. Due to the combined effect of hydrography and anthropogenic pollution, oxygen deficiency and sulfide accumulation occur in the deep water below the halocline, which mainly affects the large basins of the central Baltic Sea, such as the Gotland Basin. Denitrification plays a major role in the nitrogen budget of the Baltic Sea (31).
The Gotland Basin is the largest basin of the central Baltic Sea, and the station in the Gotland Basin that we studied is considered representative of the central Baltic Sea. This site has been studied for many decades, and thus there is an excellent database that includes microbiological parameters and data from denitrification investigations (3, 30, 42; for online databases on the Baltic Sea see http://www.helcom.fi). The oxic-anoxic interface has been identified as a major site of biogeochemical cycling in many ecosystems with anoxic basins, such as the Black Sea, the Cariaco Trench, the Baltic Sea, and fjords (5, 8, 16, 35, 36). High turnover of carbon, nitrogen, and sulfur compounds has been demonstrated for the oxic-anoxic interfaces of the central Baltic Sea (2, 5, 8, 30). Autotrophic denitrification driven by sulfide oxidation has been shown to be a major pathway for nitrogen loss from the central Baltic Sea (2, 3) because processes at oxic-anoxic interfaces have considerable importance. In addition to the oxic-anoxic interface on top of anoxic deep water, these biogeochemical processes may occur to a much larger extent during occasional interleaving of oxidized water masses in the anoxic deep water and thus have a pronounced impact on the material balance of the Baltic Sea as a whole (24).
The first “whole-genome shotgun sequencing” analysis of open ocean microbial communities was performed recently, and it generated a plethora of genetic information (39). Understanding the biogeochemical and ecological implications of this genetic information will be one of the great challenges in the years to come. Identification of bacterial catalysts for important biogeochemical processes is particularly relevant for (i) understanding factors that regulate biogeochemical processes by relating knowledge about cultured bacteria to in situ processes and rates and (ii) using the bacterial catalysts identified as indicators of in situ processes on a global scale (25). Identification of the bacterial catalysts to the species level is needed to determine such links. Studies of the biogeochemical functions of bacterial catalysts should yield results that are most relevant for an ecosystem if they are done under natural conditions; i.e., the experimental conditions should maintain the physicochemical conditions and the microbial “background” community of the environment as much as possible.
While denitrification in aquatic environments is generally considered a heterotrophic process, autotrophic denitrification is especially relevant in aquatic environments with anoxic waters, where it can represent a significant nitrogen sink (3). The bacterial autotrophic denitrification process is currently not well understood due to a lack of knowledge regarding the bacteria that are responsible and the factors regulating the process. Consortia consisting of different bacterial taxa, as well as single taxa, are conceivable mediators. Therefore, the aims of this study were to identify the bacterial catalysts responsible for autotrophic denitrification in the Gotland Deep and to obtain insight into metabolism in this environment. To do this, the activities of autotrophic denitrifiers in water samples obtained around the oxic-anoxic interface were specifically stimulated to obtain a mechanistic understanding of the biogeochemical processes involved. Newly developed molecular tools were used to study the responsible bacterial catalysts at the species level in situ and during stimulation experiments.
MATERIALS AND METHODS
Study site, sampling, and physicochemical parameters.
All seawater samples were obtained at station BY15 (57.19°N, 20.03°E) in the Gotland Deep, a major anoxic basin in the central Baltic Sea, on 17 and 18 September 1998 using Niskin polyvinylchloride bottles mounted on a conductivity-temperature-depth rosette. The methods used for sampling, sample handling, and physicochemical analysis have been described in detail elsewhere (2, 9). Levels of nutrients, oxygen, and H2S were determined aboard RV Aranda as described by Grasshoff et al. (9) immediately after sampling. Total bacterial counts were determined as described by Weinbauer et al. (41). The bacterial biomass in the water samples was harvested by filtration with a sandwich consisting of a glass fiber filter (90 mm; Whatman GF/F) on top of a polycarbonate filter (pore size, 0.2 μm; Nuclepore) and were stored frozen (−70°C) for later analysis.
Experimental design.
Water samples obtained from the oxic-anoxic interface (138 m; 0.4 μmol liter−1 NO3−, 5.7 μmol liter−1 H2S, no O2), above the interface (120 m; 0.3 ml liter−1 O2, 8.0 μmol liter −1 NO3−, no H2S), and below the interface (175 m; 24 μmol liter−1H2S, no NO3−) were incubated at 5°C in the dark in gas-tight 1.2-liter bottles after sequential addition of 100 μmol KNO3 (at the start of incubation) plus 50 μmol−1 KNO3 (after 44 h of incubation), 100 μmol liter−1 Na2S2O3, and 25 μCi liter−1 H14CO3−. The controls did not contain substrates. All samples were handled so that changes in the in situ partial gas pressures (O2, H2S) were avoided (2); i.e., all bottles were filled until they were overflowing and capped so that they were air tight (Teflon-coated butyl rubber septa) and did not contain gas bubbles, and all solutions added were kept under a nitrogen atmosphere. Three replicates were used for each assay. The time of incubation was within the linear range of in situ denitrification rates demonstrated by Brettar and Rheinheimer (2). After 44 h and 88 h samples were analyzed to determine the levels of N compounds (NO3−, NO2−, NH4+) and H14CO3− uptake into cells and into exuded material (9, 33); the bacterial biomass was harvested by filtration as described previously (40) and was stored frozen for later analysis.
As shown in a previous study, nitrate removal can be used to estimate autotrophic denitrification in the chemocline layer of the central Baltic Sea (1, 2). At least 75% of the nitrate consumed was recovered as gaseous nitrogen compounds. N2O production was low compared to N2 production (with thiosulfate as the electron donor, the N2O production was less than 2% of the total gas production [i.e., N2O plus N2]). Nitrate reduction to ammonium did not occur, either in this study or in previous studies.
Nucleic acid extraction, community fingerprinting by SSCP analysis of 16S rRNA RT-PCR amplicons, and sequencing.
Nucleic acid extraction from frozen filters and quantification were performed by parallel extraction of RNA and DNA as described by Weinbauer et al. (40). Prior to reverse transcription (RT)-PCR, RNA extracts were purified from DNA by incubation with DNase I (Roche Diagnostics, Mannheim, Germany) for 60 min at 37°C. The primers used for 16S rRNA amplification from environmental RNA were described by Schwieger and Tebbe (32) (primer set Com1/Com2 amplifying positions 519 to 926 [Escherichia coli numbering] of the 16S rRNA gene). RT-PCR amplification was performed using the C. therm polymerase one-step RT-PCR system (Roche Diagnostics, Mannheim, Germany) and the protocol provided by the manufacturer. Ten nanograms of environmental RNA was used as the template for this RT-PCR. Generation and purification of single-stranded DNA, single-strand conformational polymorphism (SSCP) analysis, and silver staining of the gels were performed as described by Schwieger and Tebbe (32). Reamplification of individual bands excised from the SSCP gels was performed as described by Pöhler et al. (26), and this was followed by cycle sequencing (ABI Prism BigDye terminator cycle sequencing Ready Reaction kit; Applied Biosystems, Foster City, CA) using the primers described above. SSCP fingerprints were analyzed as described by Pöhler et al. (26), using the Gelcompare II software (Applied Maths, Inc., Austin, TX). For cluster analysis the Dice coefficient, fuzzy logic, and complete linkage were used.
Design of specific primers and quantitative RT-PCR.
Specific, 16S rRNA-targeted primers (Ost1F and Ost1R) for the Thiomicrospira denitrificans-like bacterium were designed based on the sequences obtained from the excised bands of the SSCP gel using the sequence between positions 610 and 850 (E.coli numbering) of the 16S rRNA molecule as described in detail by Höfle et al. (11). The Thiomicrospira denitrificans-like bacterium was represented by a double band (i.e., two bands that had the same sequence [see the supplemental material] but had slightly different migration distances on the SSCP gel). The occurrence of double bands is a well-known phenomenon on SSCP gels that is caused by different conformational states of the single-stranded DNA under nondenaturing conditions (32).
Details concerning quantification of the fraction of the Thiomicrospira denitrificans-like bacterium in the total RNA by real-time PCR have been described by Labrenz et al. (18). Briefly, 30 ng of template RNA was reverse transcribed using the Thermoscript RT-PCR system (Invitrogen) and following the manufacturer's instructions. Approximately 4 ng of the complementary 16S rRNA gene was preamplified using universal primers 27F and 1492R (19). The relative amount of the Ost1-specific 16S rRNA gene was determined by comparison with the bacterial 16S rRNA genes by a nested PCR. The Com and Ost1 primer systems, as well as 2 μl of the preamplification PCR mixture, were used in a real-time PCR to determine relative amounts of the Thiomicrospira denitrificans-like 16S rRNA in comparisons with total bacterial 16S rRNA.
Generation of the complete 16S rRNA gene sequence.
Complete sequences were generated by combining primers Ost1F and 1492R, as well as primers Ost1R and 27F (19). We used DNA extracted from in situ samples and experiments as the DNA templates. The PCR conditions were the conditions described above for the real-time PCR approach. Details concerning generation of the complete sequence have been described by Höfle et al. (11).
Phylogenetic analysis of 16S rRNA gene sequences.
Phylogenetic trees were constructed by using three different methods (BIONJ using a Kimura two-parameter correction, maximum likelihood using the Global option, and maximum parsimony). The BIONJ program from Gascuel (7) and the DNADIST, ML, and MP programs from PHYLIP (Phylogeny Inference Package, version 3.573c; distributed by J. Felsenstein, Department of Genetics, University of Washington, Seattle) were used. A tree (see Fig. 3C) was drawn from the neighbor-joining topology, and it included information for bootstrap values (1,000 replications) and confirmation of each branch by the other two methods.
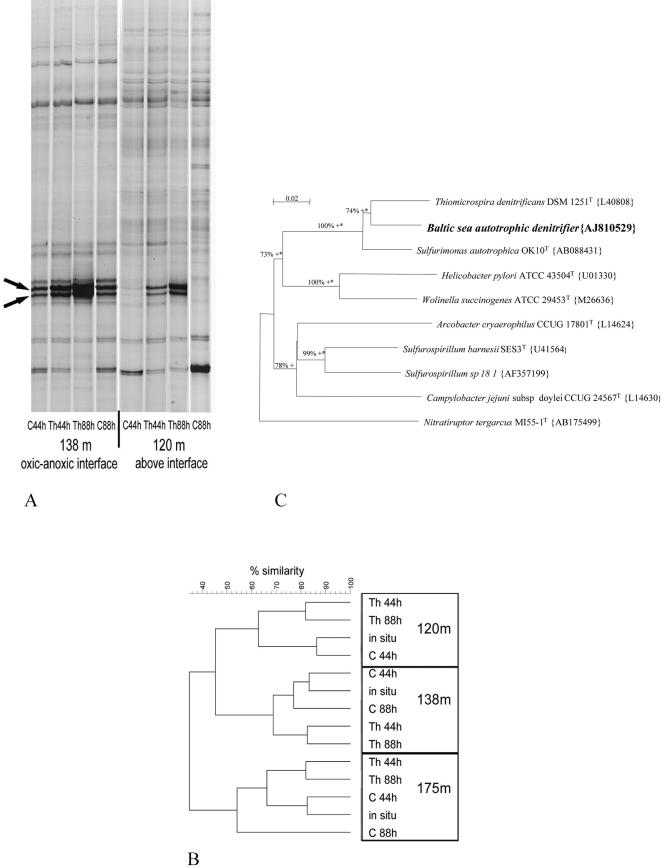
FIG. 3.
Results of molecular analyses of the bacterial community structure and composition of the original seawater and of stimulation experiments using RNA-based high-resolution SSCP fingerprints. (A) Fingerprints of the bacterial community in samples obtained from the oxic-anoxic interface (138 m) and above the interface (120 m) incubated with (Th) and without (C) addition of thiosulfate plus nitrate at the times indicated. The SSCP fingerprints show the increases in two bands, while the “background” community remained the same. The two bands that increased (arrows) had identical 16S rRNA sequences and represent an epsilonproteobacterium related to Thiomicrospira denitrificans (see panel C and the supplemental material). (B) Cluster analysis (Dice coefficient, fuzzy logic, complete linkage) of SSCP fingerprints from experiments with water samples obtained from 120 m, 138 m, and 175 m. The cluster analysis demonstrated that the overall bacterial community structure during the experiments remained comparable to the structure in the original seawater. The SSCP data were derived from the experiments whose results are shown in Fig. 2 but different gels. Th, sample to which thiosulfate and nitrate were added; C, control (sample incubated without substrate addition); 44h and 88h, incubation times of 44 and 88 h, respectively; in situ, original seawater from the depth indicated. (C) Phylogenetic position of the Baltic Sea autotrophic denitrifying bacterium among cultured Epsilonproteobacteria based on 16S rRNA gene sequence comparison. The topology of the phylogenetic tree is the result of 1,000 bootstrap replications using the neighbor-joining methods. In addition to neighbor joining, confirmation of the branches by parsimony is indicated by a plus sign, and confirmation of the branches by maximum likelihood (at P < 0.01) is indicated by an asterisk. Branches in the phylogenetic environment of the Baltic Sea autotrophic denitrifier were confirmed by all three methods and have a high degree of confidence. The almost complete 16S rRNA gene sequence of the Baltic Sea bacterium (accession no. AJ810529) was generated by designing two highly specific primers based on the sequence of the double band (indicated by arrows in panel A) and using them together with nonspecific primers for environmental and experimental DNA as templates (11).
Nucleotide sequence accession number.
The 16S rRNA gene sequence obtained for the Thiomicrospira denitrificans-like bacterium has been deposited in databases under accession number AJ810529.
RESULTS
In situ conditions during the study in the central Baltic Sea.
The physicochemical and bacterial parameters determined (Fig. 1) can be considered typical for the summer stratification in the Gotland Deep, a station representative of the central Baltic Sea, in periods when there is stagnant deep water (2). The water was well oxygenated above the halocline (enhanced salinity gradient from 60 to 80 m). Below 60 m, the oxygen level decreased, and oxygen was absent at depths below 130 m; the sulfide concentration increased from the oxic-anoxic interface down to the sediment. The nitrate level increased in the low-oxygen layer; low concentrations of nitrate were present in the layer of the chemocline containing low concentrations of sulfide (138 m; 5.7 μmol liter−1 H2S, 0.4 μmol liter−1 NO3−, no O2). Bacterial abundance was highest in the surface water and declined with depth until just above the oxic-anoxic interface, where the number of bacteria began to increase (Fig. 1B).
FIG. 1.
Depth distribution of physical, chemical, and biological background parameters and relative abundance of the Thiomicrospira denitrificans-like bacterium in the Gotland Basin (station BY15; 57.1920°N, 20.0302°E) on 17 September 1998. (A) Distribution of oxygen (O2), nitrate (NO3−), ammonium (NH4+), and sulfide (H2S). (B) Temperature, salinity, bacteria (total bacterial counts), and total RNA along the depth profile. (C) Contributions of the Thiomicrospira denitrificans-like bacterium to the total bacterial 16S rRNA (•) and the total bacterial 16S rRNA genes (▵) as determined by real-time PCR (18). The cross-hatched area represents the oxic-anoxic interface layer where both nitrate and reduced sulfur compounds occurred.
Biogeochemical stimulation experiments and bacterial community response.
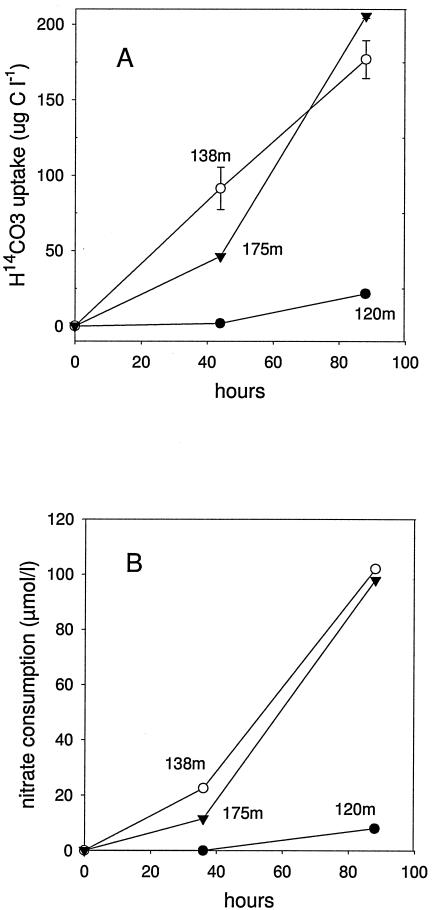
The activity and abundance of the bacterial catalysts were monitored for up to 88 h under conditions that were close to the in situ physicochemical conditions during the experiments, i.e., after addition of thiosulfate and nitrate to samples obtained from the chemocline itself (138 m; with concomitant occurrence of nitrate and reduced sulfur compounds in situ), from above the chemocline (120 m), and from below the chemocline (175 m). Nitrate removal and H14CO3− uptake (Fig. 2) were observed as indicators of autotrophic denitrification. Nitrate removal data can be considered estimates of autotrophic denitrification in the deep water of the central Baltic Sea, as shown in a previous study by Brettar and Rheinheimer (2). In samples obtained from the chemocline (138 m), carbon dioxide fixation was almost linear for the 88-h observation period, and the uptake for the total incubation period was 177 μg C liter−1. Samples obtained from 175 m showed a slightly higher level of CO2 fixation (about 206 μg C liter−1 for the total period [88 h], with lower CO2 fixation during the first 44 h). Samples obtained from 120 m showed a much lower level of CO2 fixation, 22 μg C liter−1 for the total period; in the first 44 h no CO2 fixation was detected. The high CO2 fixation rates of the samples obtained from 138 and 175 m corresponded to a high level of nitrate consumption, about 100 μmol N liter−1; the low level of CO2 fixation in samples obtained from 120 m was accompanied by a low level of nitrate consumption. Thus, samples obtained from the chemocline and below the chemocline showed much higher activity of autotrophic denitrification than the sample obtained from above the interface. Assuming that the rate for the 88-h period was linear, the CO2 fixation rates of the anoxic samples corresponded to rates of 49 μg C liter−1 day−1 (138 m) to 57 μg C liter−1 day−1 (175 m).
FIG. 2.
Results of stimulation experiments using unfiltered seawater obtained from around the oxic-anoxic interface. (A) Carbon dioxide fixation (measured as H14CO3− uptake in cell material). (B) Nitrate consumption in samples obtained from the oxic-anoxic interface (138 m), above the interface (120 m), and below the interface (175 m) after addition of nitrate plus thiosulfate incubated at the in situ temperature (5°C).
Bacterial community structure and composition were assessed by 16S rRNA-based SSCP community fingerprint analysis (Fig. 3A and B). The microbial community remained rather stable during the experiments compared to the community in the original sample, as revealed by cluster analysis of the SSCP community fingerprints (Fig. 3B). After substrate addition, only two bands showed a pronounced increase in intensity in samples from all depths (Fig. 3A). The increase was most pronounced in samples obtained from 120 m; for these samples the intensities of the bands increased from close to the level of detection to very high.
The 16S rRNA sequences of the two enhanced bands were identical (see the supplemental material). These sequences were used to design highly specific primers in order to generate a complete 16S rRNA gene sequence using DNA extracted from in situ samples and experimental samples (11). Phylogenetic analysis of the almost complete sequence (1,422 nucleotides; accession no. AJ810529) revealed that the autotrophic denitrifier was an epsilonproteobacterium related to Thiomicrospira denitrificans DSM 1251T, which was the closest cultivated neighbor (95.8% 16S rRNA gene sequence similarity) (22) (Fig. 3C). Identical complete sequences were obtained from all samples (i.e., from in situ samples obtained from different depths and from the experimental samples) (11). Another close relative is Sulfurimonas autotrophica, a sulfide oxidizer that is not able to denitrify (15). Due to the physiological similarity and phylogenetic relatedness to Thiomicrospira denitrificans, we designated the Baltic Sea bacterium the Thiomicrospira denitrificans-like bacterium.
The percentage of the Thiomicrospira denitrificans-like bacterium based on a comparison of its 16S rRNA with the total 16S rRNA of the community increased markedly after addition of thiosulfate plus nitrate in all samples, as assessed by quantification of the Thiomicrospira denitrificans-like bands from the community fingerprints (Fig. 4). The band intensities increased from initial values of 2% (120 m) to 15% (175 m) to final values of 48% (120 m) to 63% compared to all bands for samples from the three depths (average band intensities for three SCCP analyses).
FIG. 4.
Relative amounts of the Thiomicrospira denitrificans-like bacteria during the stimulation experiments. Quantification was based on the areas of the specific bands from replicate SSCP gels (circles) and quantitative RT-PCR (diamonds) as described by Labrenz et al. (18). Solid symbols indicate the results of experiments with thiosulfate and nitrate addition, and open symbols indicate the results obtained for controls with no addition. The experiments were identical to those whose results are shown in Fig. 2. (A) Results for samples obtained above the oxic-anoxic interface (120 m). (B) Results for samples obtained at the interface (138 m). (C) Results for samples obtained below the interface (175 m).
The increase in the amount of Thiomicrospira denitrificans-like RNA on the SSCP gels was also observed via quantification of the Thiomicrospira denitrificans-like RNA fraction of the total 16S rRNA by real-time RT-PCR using the specific primers mentioned above and the appropriate environmental RNA (Fig. 4). The lower estimate for the Thiomicrospira denitrificans-like RNA fraction by real-time PCR at the beginning of the experiments (identical to in situ values) is consistent with the results of our calibration experiments for the SSCP analysis that showed enhanced sensitivity for low-abundance members of the bacterial community due to PCR bias (most pronounced below 1% RNA abundance [data not shown]).
According to real-time RT-PCR quantification, the maximum level of the Thiomicrospira denitrificans-like bacterium occurred in the RNA fraction around the chemocline layer (120 to 175 m) (18) (Fig. 1C). The size of the Thiomicrospira denitrificans-like 16S rRNA fraction of the total 16S rRNA in seawater samples obtained in the vicinity of the interface layer (120 to 175 m) ranged from 0.1% (120 m) to 0.9% (138 m) (18) (Fig. 1C). By using real-time PCR, a pronounced maximum and a large contribution of the Thiomicrospira denitrificans-like bacterium to the total bacterial DNA in the vicinity of the chemocline were estimated (Fig. 1C). Based on the 16S rRNA gene fraction, maxima for the numbers of Thiomicrospira denitrificans-like cells were determined for 138 and 175 m (7.1 × 104 cells/ml at 138 m and 1.4 × 105 cells/ml at 175m, accounting for 8.3 and 15.1% of all bacterial 16S rRNA genes, respectively) (18). In summary, the major contribution of the Thiomicrospira denitrificans-like bacterium to the 16S rRNA, as well as to the 16S rRNA genes, was in the layer around the oxic-anoxic interface, with a maximum for the 16S rRNA at 138 m (i.e., in the interface layer) and a maximum for the 16S rRNA gene in the anoxic deep water below the interface (175 m).
DISCUSSION
We demonstrated that an epsilonproteobacterium closely related to Thiomicrospira denitrificans was the major bacterial catalyst for denitrification in the Gotland Basin by a combined experimental-molecular approach and study of in situ abundance. This finding was supported by concomitant observations of autotrophic denitrification driven by oxidation of thiosulfate and an increase in the abundance of the Thiomicrospira denitrificans-like 16S rRNA fraction during stimulation experiments. The increase in abundance was determined by SSCP analyses as well as by real-time PCR. An additional indicator of increased activity of the Thiomicrospira denitrificans-like cells was the 70- to 100-fold increase in the ribosome content observed in the stimulation experiments compared to the ribosome content in the original water samples obtained from 138 and 175 m (18). This indicated that there was an immediate growth and activity response after substrate addition (17).
Autotrophic denitrification at the oxic-anoxic interface in the central Baltic Sea was studied in more detail by Brettar and Rheinheimer (2). It was found that autotrophic denitrification was the major nitrate removal process in the water column, and the major fraction of nitrate consumed was reduced to dinitrogen (N2). Nitrate reduction to ammonium was never observed in the samples.
The substrate turnover, as well as the community analyses, indicated that we were able to mimic in situ conditions during our experiments. The overall bacterial community composition remained intact, and increases in abundance were observed only for the bacteria responsible for autotrophic oxidation of reduced sulfur compounds. The carbon dioxide fixation rates in our experiments ranged from 49 μg C liter−1 day−1 (138 m) to 57 μg C liter−1 day−1 (175 m), and they were very similar to previously observed in situ rates (approximately 53 μg C liter−1 day−1) for the oxic-anoxic interface of the Gotland Deep (5, 8). These rates were among the highest rates recorded for the chemocline of the Gotland Deep and coincided with the presence of nitrate and sulfide. Rates that were the same order of magnitude were reported for oxic-anoxic interfaces of the Cariaco Trench (35) and Saanich Inlet (16).
For samples obtained from above the oxic-anoxic interface (120 m), the increase in the Thiomicrospira denitrificans-like RNA in the presence of a low concentration of oxygen without detectable nitrate consumption (during the first incubation period) could be interpreted as a hint that oxygen was used in addition to nitrate as an electron acceptor for S2O32− oxidation. This layer was characterized in situ by a low oxygen concentration (0.3 ml liter−1), while samples obtained from the lower layers did not contain oxygen.
Phylogenetic identification and physiological features of the bacterial catalyst for autotrophic denitrification.
The epsilonproteobacterium responsible for autotrophic denitrification in the central Baltic Sea is phylogenetically closely related to the cultured bacterium Thiomicrospira denitrificans. It is striking that our experiments revealed physiological features for the Baltic Sea autotrophic denitrifier comparable to physiological features of Thiomicrospira denitrificans. Like Thiomicrospira denitrificans, the Baltic Sea autotrophic denitrifier was obviously able to oxidize reduced sulfur compounds, such as thiosulfate, with nitrate and oxygen as electron acceptors. Another close relative, the cultured sulfide oxidizer S. autotrophica, cannot be compared physiologically to the Baltic Sea denitrifier because it is not able to use nitrate as an electron acceptor (15).
Timmer-ten-Hoor (37) measured for autotrophic growth by denitrification (S2O32− + NO3− + CO2 → SO42− + N2 + Corg) for Thiomicrospira denitrificans ratios of 1 g organic C produced by oxidation of 0.36 mol S2O32− or H2S and consumption of 0.5 mol NO3−. Continuous cultures grown under nitrate and thiosulfate limitation conditions had the same ratios (37). The CO2 fixation per mol of nitrate consumed by the epsilonproteobacterium Thiomicrospira denitrificans is only one-half as efficient as the CO2 fixation by the betaproteobacterium Thiobacillus denitrificans. It has previously been suggested that the latter organism is a catalyst for denitrification in the Baltic Sea (2, 10). The reason for the different growth efficiency was attributed in part to the fact that Thiomicrospira denitrificans lacks adenosine phosphosulfate reductase (37). Additionally, it was shown that the energy needed for maintenance was twice as high for Thiomicrospira denitrificans (37). On the other hand, it was recently shown that Thiomicrospira denitrificans uses the more energy-efficient reverse tricarboxylic acid cycle for carbon dioxide fixation (13). Thus, the difference between the metabolic efficiencies of the two organisms is not fully understood yet.
To compare the stoichiometry for autotrophic denitrification by Thiomicrospira denitrificans with the stoichiometry observed in our experiments, the ratio of CO2 fixation to NO3− consumption was calculated for the total incubation period (88 h) for the anoxic samples obtained from 138 and 175 m. For the 120-m sample the ratio could not be calculated due to the low turnover rates and concomitant use of oxygen. To calculate the ratios, the cellular uptake shown in Fig. 2 and the exudates containing labeled carbon (on average 11% of the incorporated labeled carbon) were compared to the amount of nitrate consumed. In the experiments with samples obtained from 138 m, the percentage of nitrate consumed versus CO2 fixed was 97% of that of Thiomicrospira denitrificans; for samples obtained from 175 m the value was 110%. Thus, the ratios of CO2 fixation to nitrate consumption were similar to the ratio for Thiomicrospira denitrificans (within 10% of the stoichiometry for Thiomicrospira denitrificans, as measured by Timmer-ten-Hoor [37]) but very different from the ratio for Thiobacillus denitrificans. Thus, the phylogenetic position corresponded to the physiological features.
From an ecological point of view, it is surprising that a bacterium with a physiology very similar to that of Thiomicrospira denitrificans controls autotrophic denitrification at the redoxcline in the central Baltic Sea, because the metabolism of this organism is only one-half as efficient in terms of growth yield per mol S2O32− or H2S oxidized as the metabolism of the betaproteobacterium Thiobacillus denitrificans (37).
Sequences of Epsilonproteobacteria have been determined several times by cloning environmental DNA from ecosystems with an active sulfur cycle, such as hydrothermal vents or anoxic basins, in which their involvement in sulfur cycling was hypothesized (21, 29). Enrichment cultures of Epsilonproteobacteria have been analyzed, and involvement of these organisms in autotrophic sulfate reduction has been shown (e.g., Sulfurospirillum sp. strain 18-1) (4). Additionally, a set of Epsilonproteobacteria has recently been isolated, and some of these organisms are even able to oxidize sulfide using oxygen or nitrate (34); also, many new genera have been described recently (23), and the aerobic sulfide oxidizer S. autotrophica (15) is the closest relative of the Baltic Sea denitrifier among the new genera.
However, the specific role of members of the Epsilonproteobacteria in situ could not be demonstrated until now. To our knowledge, this is the first study that estimated the contribution of a specific member of the Epsilonproteobacteria under in situ conditions to a specific predominant biogeochemical process, such as autotrophic denitrification.
Advantages of the combined experimental-molecular approach.
We used biogeochemical experiments to stimulate a biogeochemical process of interest, and we used a set of newly developed molecular tools in the experiments to identify and quantify the bacterial catalyst of interest. The core of the molecular analyses was the 16S rRNA-based community fingerprints that allowed us to monitor the community, identify the active members due to “highlighted” fingerprint bands, and use the sequences of the bands for rough phylogenetic identification and for generation of highly specific primers. These primers were used to generate almost complete 16S rRNA gene sequences for comprehensive phylogenetic analysis (20, 28) and to quantify the species of interest by real-time PCR (11, 18). The material turnover in the stimulation experiments was used to calculate the stoichiometry of the process.
A major advantage of fingerprint techniques is that they allow workers to obtain an immediate overview of the structure and composition of bacterial communities. This overview provides insight into changes in the bacterial community during experiments, an important aspect considering that interspecies competition is a major factor for activities and survival of bacteria in nature (14). We used RNA-based community fingerprints because RNA reflects actively growing bacteria better than DNA does (17). Combined in situ analyses and biogeochemical experiments reflected rapidly increasing activities and numbers of the bacterial catalyst in the Baltic Sea water samples.
Under certain circumstances (e.g., dominance of a single specific process) the stoichiometry of a process can be calculated based on stimulation experiments. In our study, we were able to relate the stoichiometry of autotrophic denitrification (i.e., nitrate reduction and carbon dioxide fixation) to a specific taxon. For this relationship it was necessary to monitor the bacterial community to be sure that the abundance of only the specific taxon studied was increasing parallel to the biogeochemical process. Support for this relationship was obtained by quantification and assessment of the activity by real-time RT-PCR.
In conclusion, the combined experimental-molecular approach allowed us to relate the precise phylogenetic position of a single bacterial taxon to its specific biogeochemical function in situ.
General relevance of the presence of Thiomicraspira denitrificans-like bacteria in marine systems.
The presence and physiological responses of the autotrophic denitrifier at and around the oxic-anoxic interface are consistent with prior observations of in situ denitrification and carbon dioxide fixation in basins with sulfidic deep water in the central Baltic Sea (2, 5, 8). Reduced sulfur compounds were identified as a major electron donor for denitrification. This was explained by the low organic carbon supply from the euphotic layer for most of the year and by the availability of reduced sulfur compounds from the anoxic water (3). Turbulent vertical diffusion at the oxic-anoxic interface above the anoxic water, as well as lateral interleaving of nitrate-containing water masses into layers of anoxic deep water, can be perceived as mechanisms that promote intense autotrophic denitrification (2, 8, 24, 43). The presence of the autotrophic denitrifiers at the interface as well as in the anoxic deeper water layers explains the observed potential of the water masses for denitrification.
In addition to mixing of sulfide- and nitrate-containing water masses, storage of electron donors or acceptors by the autotrophic denitrifiers would increase their potential for denitrification when they come into contact with the complementary substrate. The observed high in situ CO2 fixation rates support such a storage and/or transport mechanism, as shown for the sediment-dwelling gammaproteobacteria Thioploca spp. (12). Such mechanisms and many others (e.g., grazing resistance, motility, and competitive uptake mechanisms for nitrate and reduced sulfur compounds) could be selection factors that overcome the disadvantage of the lower growth yield of a Thiomicrospira denitrificans-like bacterium. Genome sequencing of Thiomicrospira denitrificans is currently under way (http://www.jgi.doe.gov), and the results will be a step toward predictive biogeochemistry. “Whole-genome sequencing” with stimulation experiments could provide more detailed information on the in situ genomics of the catalyst, as recently demonstrated by Tyson et al. (38) for an acid mining community with reduced biodiversity.
The lower carbon fixation rate of the Thiomicrospira denitrificans-like bacterium could be very relevant for the carbon budget of the deep water and the benthos of the central Baltic Sea, if we assume that this bacterium catalyzes a major fraction of the oxidation of reduced sulfur compounds at the chemocline. Rahm (27) calculated a yearly export of 50 g C m−2 year−1 from the euphotic zone to deeper water layers. Assuming a rate of 50 mg C m−3 day−1 as reported by Gocke (8) and Dettmer et al. (5) for a 5-m layer at the oxic-anoxic interface, dark carbon dioxide fixation would increase the yearly carbon input to the deep water by 91 g C m−2. Due to the dependence of sulfur oxidation on mixing at the interface, very high variability must be taken into account, and it is difficult to predict rates. For the C budget of the Baltic Sea, it is an interesting question whether specific environmental conditions could promote replacement of the Thiomicrospira denitrificans-like bacterium by a more efficient sulfide oxidizer, such as Thiobacillus denitrificans, and thus greatly increase the amount of dark carbon dioxide fixation.
In conclusion, the combined biogeochemical-molecular approach used in this study allowed us to relate the precise phylogenetic position of a single bacterial population to its specific biogeochemical function in situ. This relationship can be used to predict the potential of a specific biogeochemical process when substantial amounts of 16S rRNA of a microorganism are detected in an environment. This study demonstrated the potential impact of a single bacterial taxon on the matter balance in a marine water column. Thus, it can serve as a good example to demonstrate the utility of identifying major bacterial catalysts in pelagic aquatic environments. In more general terms, our results indicate that ecological principles other than growth efficiency can play a dominant role as selection factors and thus influence the stoichiometry of a biogeochemical process relevant to marine carbon and nitrogen budgets.
Supplementary Material
Acknowledgments
We acknowledge the support of J. Kuparinen for the cruise on RV Aranda in September 1998. We thank the crew of RV Aranda for sampling, shipboard analysis, and a cordial atmosphere. M. G. Weinbauer is acknowledged for determining numbers of bacterial cells, and B. Engelen is acknowledged for help on the cruise. S. Pretzer, C. Höltje, and P. Westphal are acknowledged for skillful technical assistance. Comments on and discussions concerning a version of the manuscript with E. Kaplan, D. L. Kirchman, and H. W. Pearl are greatly appreciated.
This work was funded in part by the Marine Science and Technology (MAST III) Program of the European Commission within the EU project “Marine Bacterial Genes and Isolates as Sources for Novel Biotechnological Products” (MARGENES) (contract MAS3-CT97-0125).
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Brettar, I. 1991. Denitrification in the water column of the central Baltic: regulatory factors and microbiological aspects. Ph.D. thesis. University of Kiel, Kiel, Germany. Ber. Inst. Meereskd. Kiel 208:1-145. [Online.] http://www.baltic.vtt.fi/pdfs/ber208.pdf. [Google Scholar]
- 2.Brettar, I., and G. Rheinheimer. 1991. Denitrification in the central Baltic: evidence for H2S-oxidation as motor of denitrification at the oxic-anoxic interface. Mar. Ecol. Prog. Ser. 77:157-169. [Google Scholar]
- 3.Brettar, I., and G. Rheinheimer. 1992. Influence of carbon availability on denitrification in the water column of the central Baltic. Limnol. Oceanogr. 37:1146-1163. [Google Scholar]
- 4.Campbell, B. J., C. Jeanthon, J. E. Kostka, G. W. Luther III, and C. Cary. 2001. Growth and phylogenetic properties of novel bacteria belonging to the epsilon subdivision of the Proteobacteria enriched from Alvinella pompejana and deep-sea hydrothermal vents. Appl. Environ. Microbiol. 67:4566-4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dettmer, A. E., H. C. Giesenhagen, V. M. Trenkel, H. Au dem Venne, and F. J. Jochem. 1993. Phototrophic and heterotrophic pico- and nanoplankton in anoxic depths of the central Baltic Sea. Mar. Ecol. Progr. Ser. 99:197-203. [Google Scholar]
- 6.Elmgren, R. 1989. Man's impact on the ecosystem of the Baltic: energy flows today and at the turn of the century. Ambio 18:326-332. [Google Scholar]
- 7.Gascuel, O. 1997. BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol. Biol. Evol. 14:685-695. [DOI] [PubMed] [Google Scholar]
- 8.Gocke, K. 1989. Bakterielle Stoffaufnahme im aeroben und anaeroben Milieu der Ostsee. Ber. Inst. Meereskd. Kiel 188:40-47. [Google Scholar]
- 9.Grasshoff, K., M. Ehrhardt, and K. Kremling. 1983. Methods of sea water analysis, Verlag Chemie, Weinheim, Germany.
- 10.Höfle, M. G., and I. Brettar. 1995. Taxonomic diversity and metabolic activity of microbial communities in the water column of the central Baltic Sea. Limnol. Oceanogr. 40:868-874. [Google Scholar]
- 11.Höfle, M. G., S. Flavier, R. Christen, J. Bötel, M. Labrenz, and I. Brettar. 2005. Retrieval of nearly complete 16S rRNA gene sequences from environmental DNA following 16S rRNA based community fingerprinting. Environ. Microbiol. 7:670-675. [DOI] [PubMed] [Google Scholar]
- 12.Huettel, M., S. Forster, S. Klöser, and H. Fossing. 1996. Vertical migration in the sediment-dwelling sulfur bacteria Thioploca spp. in overcoming diffusion limitations. Appl. Environ. Microbiol. 62:1863-1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hügler, M., C. O. Wirsen, G. Fuchs, C. D. Taylor, and S. M. Sievert. 2005. Evidence for autotrophic CO2 fixation via the reductive tricarboxylic acid cycle by members of the epsilon subdivision of proteobacteria. J. Bacteriol. 187:3020-3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hurst, C. J. 2002. Neighborhoods and community involvement: no microbe is an island, p. 6-18. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, M. J. McInerney, and L. D. Stelzenbach (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 15.Inagaki, F., K. Takai, H. Kobayashi, K. H. Nealson, and K. Horikoshi. 2003. Sulfurimonas autotrophica gen. nov., sp. nov., a novel sulfur-oxidizing epsilon-Proteobacteria isolated from hydrothermal sediments in the Mid-Okinawa Trough. Int. J. Syst. Evol. Microbiol. 53:1801-1805. [DOI] [PubMed] [Google Scholar]
- 16.Juniper, S. K., and R. O. Brinkhurst. 1986. Water-column dark CO2 fixation and bacterial-mat growth in intermittently anoxic Saanich Inlet, British Columbia. Mar. Ecol. Progr. Ser. 33:41-50. [Google Scholar]
- 17.Kerkhof, L., and P. Kemp. 1999. Small ribosome RNA content in marine Proteobacteria during non-steady state growth. FEMS Microbiol. Ecol. 30:253-260. [DOI] [PubMed] [Google Scholar]
- 18.Labrenz, M., S. Flavier, R. Christen, J. Bötel, I. Brettar, and M. G. Höfle. 2004. Development and application of a real-time PCR approach for quantification of uncultured bacteria in the central Baltic Sea. Appl. Environ. Microbiol. 70:4971-4979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lane, D. J. 1991. 16S/23S rRNA sequencing, p. 115-175. In E. Stackebrandt and M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. John Wiley and Sons, Chichester, United Kingdom.
- 20.Liu, W. T., and D. A. Stahl. 2002. Molecular approaches for the measurement of density, diversity and phylogeny, p. 114-134. In C. J. Hurst, R. L. Crawford, G. R. Knudsen, M. J. McInerney, and L. D. Stetzenbach (ed.), Manual of environmental microbiology, 2nd ed. ASM Press, Washington, D.C.
- 21.Madrid, V. M., G. T. Taylor, M. I. Scranton, and A. Y. Chistoserdov. 2001. Phylogenetic diversity of bacterial and archaeal communities in the anoxic zone of the Cariaco Basin. Appl. Environ. Microbiol. 67:1663-1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muyzer, G., A. Teske, C. O. Wirsen, and H. W. Jannasch. 1995. Phylogenetic relationships of Thiomicrospira species and their identification in deep-sea hydrothermal vent samples by denaturing gel electrophoressis of 16S rDNA fragments. Arch. Microbiol. 164:165-172. [DOI] [PubMed] [Google Scholar]
- 23.Nakagawa,S., K. Takai F. Inagaki, K. Horikoshi, and Y. Sako. 2005. Nitratiruptor tergarcus gen. nov., sp. nov. and Nitratifractor salsuginis gen. nov., sp. nov., nitrate-reducing chemolithoautotrophs of the epsilon-Proteobacteria isolated from a deep-sea hydrothermal system in the Mid-Okinawa Trough. Int. J. Syst. Evolution. Microbiol. 55:925-933. [DOI] [PubMed] [Google Scholar]
- 24.Nehring, D. 1987. Temporal variations of phosphate and inorganic nitrogen compounds in the central Baltic Sea. Limnol. Oceaonogr. 32:494-499. [Google Scholar]
- 25.Newman, D. K., and J. F. Banfield. 2002. 2002. Geomicrobiology: how molecular-scale interactions underpin biogeochemical systems. Science 296:1071-1077. [DOI] [PubMed] [Google Scholar]
- 26.Pöhler, I., D. F. Wenderoth, K. Wendt-Potthoff, and M. G. Höfle. 2002. Bacterioplankton community structure and dynamics in enclosures during bio-remediation experiments in an acid mining lake. Water Air Soil Pollut. Focus 2:111-121. [Google Scholar]
- 27.Rahm, L. 1987. Oxygen consumption in the Baltic proper. Limnol. Oceanogr. 32:973-978. [Google Scholar]
- 28.Rappé, M. S., and S. J. Giovannoni. 2003. The uncultured microbial majority. Annu. Rev. Microbiol. 57:369-394. [DOI] [PubMed] [Google Scholar]
- 29.Reysenbach, A.-L., K. Longnecker, and J. Kirshstein. 2000. Novel bacterial and archaeal lineages from an in situ growth chamber deployed at a Mid-Atlantic Ridge hydrothermal vent. Appl. Environ. Microbiol. 66:3798-3806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rheinheimer, G., K. Gocke, and H. G. Hoppe. 1989. Vertical distribution of microbiological and hydrographic-chemical parameters in different areas of the Baltic Sea. Mar. Ecol. Prog. Ser. 52:55-70. [Google Scholar]
- 31.Savchuk, O., and F. Wulff. 2001. A model of the biogeochemical cycles of nitrogen and phosphorus in the Baltic, p. 373-415. In F. V. Wulff, L. A. Rahm, and P. Larson. (ed.), A system analysis of the Baltic Sea. Springer, Berlin, Germany.
- 32.Schwieger, F., and C. Tebbe. 1998. A new approach to utilize PCR-single-strand conformation polymorphism for 16S rRNA gene-based microbial community analysis. Appl. Environ. Microbiol. 64:4870-4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steeman Nielsen, E. 1952. The use of radioactive carbon (14C) for measuring organic production in the sea. J. Cons. Int. Explor. Mer 18:117-140. [Google Scholar]
- 34.Takai, K., F. Inagaki, S. Nakagawa, H. Hirayama, T. Nunoura, Y. Sako, K. H. Nealson, and K. Horikoshi. 2003. Isolation and phylogenetic diversity of members of previously uncultivated epsilon-Proteobacteria in deep-sea hydrothermal fields. FEMS Microbiol. Lett. 218:167-174. [DOI] [PubMed] [Google Scholar]
- 35.Taylor, G. T., M. Iabichella, T. Y. Ho, and M. I. Scranton. 2001. Chemoautotrophy in the redox transition zone of the Cariaco Basin: a significant midwater source of organic carbon production. Limnol. Oceanogr. 46:148-163. [Google Scholar]
- 36.Thunell, R. C., D. M. Sigman, F. Muller-Karger, Y. Astor, and R. Varela. 2004. Nitrogen isotope dynamics of the Cariaco Basin, Venezuela. Global Biogeochem. Cycles 18:GB3001. [Online.] doi: 10.1029/2003GB002185. [DOI] [Google Scholar]
- 37.Timmer-ten-Hoor, A. 1981. Cell yield and bioenergetics of Thiomicrospira denitrificans compared with Thiobacillus denitrificans. Antonie Leeuwenhoek 47:231-243. [DOI] [PubMed] [Google Scholar]
- 38.Tyson, G. W., J. Chapman, P. Hugenholtz, E. E. Allen, R. J. Ram, P. M. Richardson, V. V. Solovyev, E. M. Rubin, D. S. Rokhsar, and J. F. Banfield. 2004. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature (London) 428:37-43. [DOI] [PubMed] [Google Scholar]
- 39.Venter, J. C., K. Remington, J. F. Heidelberg, A. L. Halpern, D. Rusch, J. A. Eisen, D. Wu, I. Paulsen, K. E. Nelson, W. Nelson, D. E. Fouts, S. Levy, A. H. Knap, M. W. Lomas, K. Nealson, O. White, J. Peterson, J. Hoffman, R. Parsons, H. Baden-Tillson, C. Pfannkoch, Y.-H. Rogers, and H. O. Smith. 2004. Environmental genome shotgun sequencing of the Sargasso Sea. Science 304:66-74. [DOI] [PubMed] [Google Scholar]
- 40.Weinbauer, M. G., I. Fritz, D. F. Wenderoth, and M. G. Höfle. 2002. Simultaneous extraction of total RNA and DNA from bacterioplankton suitable for quantitative structure and function analyses. Appl. Environ. Microbiol. 68:1082-1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Weinbauer, M. G., I. Brettar, and M. G. Höfle. 2003. Viral infection and lysogeny of bacterioplankton in surface, deep and anoxic marine waters. Limnol. Oceanogr. 48:1457-1465. [Google Scholar]
- 42.Wulff, F. V., L. A. Rahm, and P. Larson. 2001. A system analysis of the Baltic Sea. Springer, Berlin, Germany.
- 43.Wulff, F. V., A. Stigebrandt, and L. A. Rahm. 1990. Nutrient dynamics of the Baltic Sea. Ambio 19:126-133. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.