Abstract
Through the fixation of atmospheric nitrogen and photosynthesis, marine diazotrophs play a critical role in the global cycling of nitrogen and carbon. Crocosphaera watsonii is a recently described unicellular diazotroph that may significantly contribute to marine nitrogen fixation in tropical environments. One of the many factors that can constrain the growth and nitrogen fixation rates of marine diazotrophs is phosphorus bioavailability. Using genomic and physiological approaches, we examined phosphorus scavenging mechanisms in strains of C. watsonii from both the Atlantic and the Pacific. Observations from the C. watsonii WH8501 genome suggest that this organism has the capacity for high-affinity phosphate transport (e.g., homologs of pstSCAB) in low-phosphate, oligotrophic systems. The pstS gene (high-affinity phosphate binding) is present in strains isolated from both the Atlantic and the Pacific, and its expression was regulated by the exogenous phosphate supply in strain WH8501. Genomic observation also indicated a broad capacity for phosphomonoester hydrolysis (e.g., a putative alkaline phosphatase). In contrast, no clear homologs of genes for phosphonate transport and hydrolysis could be identified. Consistent with these genomic observations, C. watsonii WH8501 is able to grow on phosphomonoesters as a sole source of added phosphorus but not on the phosphonates tested to date. Taken together these data suggest that C. watsonii has a robust capacity for scavenging phosphorus in oligotrophic systems, although this capacity differs from that of other marine cyanobacterial genera, such as Synechococcus, Prochlorococcus, and Trichodesmium.
Phytoplankton, including cyanobacteria from the genera Prochlorococcus, Synechococcus, Crocosphaera, and Trichodesmium, significantly contribute to overall marine primary production and thereby play a key role in the global cycling of carbon (C). Within the cyanobacteria, diazotrophs, such as Trichodesmium, are particularly important because of their ability to fix atmospheric nitrogen (N). Although Trichodesmium is perhaps the best-known and best-studied marine diazotroph, recent research (18, 31) has highlighted the biogeochemical importance of small, unicellular diazotrophs (3 to 10 μm), such as Crocosphaera watsonii, which can introduce a substantial fraction of new nitrogen to the euphotic zone in tropical systems where it occurs (3, 5, 9, 18, 31).
Much of our knowledge regarding unicellular diazotrophs, such as C. watsonii, has been driven by recent molecular work to assess the expression of nifH, a dinitrogenase reductase-encoding gene responsible for N2 fixation. This work has led to the identification of two open-ocean nifH sequence types, group A and group B. These sequence groups phylogenetically cluster with unicellular cyanobacteria (9, 10, 31). Group A nifH DNA phylotypes are most closely related to the genus Cyanothece, while group B nifH phylotypes are most similar to Crocosphaera watsonii WH8501 (5, 31). Quantitative PCR to detect nifH has found these phylotypes to be abundant and, at times, more abundant than Trichodesmium spp. in oligotrophic waters (5). While Trichodesmium is typically most abundant in the upper euphotic zone (4), the unicellular diazotrophs which make up groups A and B have been reported to be more uniformly distributed through the euphotic zone (17, 18). They also differ from Trichodesmium in that they fix nitrogen maximally at night, rather than during the day (31). Ultimately, the diversity, abundance, and widespread distribution of unicellular diazotrophs culminate in high rates of N2 fixation.
The factors that control the growth and N2 fixation rates of marine diazotrophs have been intensively studied. One of these factors is the bioavailability of phosphorus. Two major ocean biomes, the North Pacific Subtropical Gyre and the Sargasso Sea, have very low inorganic phosphate concentrations (<1 nM in some cases), high levels of dissolved inorganic N:P, and elevated total dissolved N:P (30). Under these conditions the availability of trace concentrations of dissolved inorganic phosphate (DIP) and the bioavailability of the larger but poorly characterized and chemically heterogeneous pool of dissolved organic phosphorus (DOP) could dramatically influence diazotroph production and N2 fixation. Several recent studies have identified P bioavailability as a possible controlling factor for the physiology of the diazotroph Trichodesmium (7, 23); however, P physiology has not yet been examined with cultures or field populations of the unicellular diazotrophs, such as Crocosphaera.
Marine cyanobacteria have evolved a number of different strategies for survival in low-DIP marine systems, such as the North Pacific Subtropical Gyre and the Sargasso Sea. Two common strategies for survival in low-phosphate environments include the induction of high-affinity phosphate scavenging systems and the up-regulation of enzymes to hydrolyze DOP into phosphate. Perhaps the best characterized of these strategies is high-affinity phosphate transport. For example, the gene cluster pstSCAB has been identified in Synechococcus sp. strain WH8102 (26) and Prochlorococcus spp. (19). These genes are up-regulated by P deficiency in Synechocystis sp. strain PCC6803 (27). Also, the presence of the PstS protein can be detected in field populations under low-P conditions (24). Notably absent in the marine cyanobacterial genomes examined to date are any low-affinity phosphate permeases (e.g., pitA) (19, 26).
Two of the dominant bond classes of oceanic high-molecular-weight DOP are phosphomonoesters and phosphonates (13). The hydrolysis of phosphomonoesters is mediated by enzymes such as alkaline phosphatase (phoA), the activity of which is commonly up-regulated by P deficiency in cyanobacteria (22). The transport of phosphonates is mediated by phnCDE. This gene cluster is present in all the available Synechococcus and Prochlorococcus genomes (19) and in all the Trichodesmium species examined to date (6). Hydrolysis of phosphonates can be mediated by multiple enzyme systems of different substrate specificities (14). Phosphonate hydrolysis in marine cyanobacteria has not been comprehensively examined, but evidence from Synechococcus sp. strain WH8102 (21, 26) growth studies and Trichodesmium erythraeum gene expression analysis (6) suggests that some marine cyanobacteria may be able to metabolize exogenous phosphonate compounds.
Despite these general advances in our understanding of P metabolism in marine cyanobacteria, little is known about how unicellular diazotrophs, such as C. watsonii, scavenge phosphorus. Here we used a combination of genomic observations and physiological studies to examine P-scavenging strategies of different strains of C. watsonii.
MATERIALS AND METHODS
Cell culturing.
Axenic Crocosphaera watsonii WH8501, previously designated Synechocystis sp. strain WH8501, and several other Crocosphaera sp. strains were obtained from John B. Waterbury at Woods Hole Oceanographic Institution (Table 1). Cultures were grown at 27.5°C using a 14:10-h light-dark cycle provided by cool white fluorescent bulbs with 65 μmol quanta m−2 s−1. Cultural purity was confirmed by testing for growth of contaminating organisms with a tryptone-fortified medium (2). Triplicate P-replete (+P) cultures were grown in SN medium (28), with a 0.2-μm-filtered Sargasso seawater base and 45 μM K2HPO4. SO medium was prepared as described above without added nitrogen. Low-phosphate (−P) medium was prepared by reducing the SN/SO medium K2HPO4 concentration to 1 μM. For growth experiments on organic phosphorus compounds, K2HPO4 was omitted from the base medium, and a suite of organic compounds (dl-α-glycerophosphate, AMP, phytic acid, phosphonoacetic acid, phosphonoformic acid, and 2-aminoethylphosphonate) were added to media at final concentrations of 45 μM. Cells used as the inoculum for all treatments were centrifuged initially for 10 min at 7,000 rpm and resuspended in medium without added phosphate to restrict carryover. Growth of all cultures was monitored in triplicate by relative fluorescence with a Turner Designs 10-AU fluorometer.
TABLE 1.
Crocosphaera watsonii strains examined in this study
| C. watsonii strain | Location | Yr isolated | Axenic |
|---|---|---|---|
| WH8501a | Tropical S. Atlantic | NAb | Yes |
| WH0401 | Tropical S. Atlantic | 2003 | Yes |
| E12 | Tropical S. Atlantic | 1985 | No |
| WH0002 | Hawaii | 2000 | Yes |
| H12 | Hawaii | 2000 | No |
| H13 | Hawaii | 2000 | No |
| OFF | Hawaii | 2000 | No |
Genome strain.
NA, not available.
Gene expression.
To examine gene expression patterns, RNA was extracted from cultures with differing P physiology. Replete (+P) samples were extracted at mid-log phase, and P-starved (−P) samples were extracted at the onset of stationary phase. In one experiment, −P cultures were refed to replete phosphate levels (45 μM). In this case RNA was extracted 24 h after the addition of P. Cells from each condition were harvested by filtering 8 ml onto 0.2-μm polycarbonate filters (25 mm), and the RNA was extracted using a RiboPure-Bacteria kit (Ambion, Austin, TX) according to the manufacturer's instructions. RNA concentrations were obtained with a NanoDrop ND-1000 (Nanodrop Technologies, Wilmington, DE) spectrophotometer. Total RNA was reverse transcribed to single stranded cDNA using an iScript cDNA synthesis kit (Bio-Rad, Hercules, CA), and additional reactions for each sample were set up without reverse transcriptase (RT) to ensure the absence of genomic DNA in no-RT controls. PCR primer sets for a putative alkaline phosphatase (5′AGTGGTGAGGCTGAAGGAGA; 3′ACCCACATTACCTGGAGCAG, gene object identifier = 400846230), pstS (5′TTGTGCAACTCAACACAGCA; 3′TTGGGATCATTCCAGTTGGT, gene object identifier = 400876370), and a putative phosphate permease (5′CCCCATCTTTTGAGTTTGGA; 3′CACCACTAATGCACCCACAG, gene object identifier = 400856070) were designed based on the sequenced genome of C. watsonii WH8501 (http://img.jgi.doe.gov/pub/main.cgi; Taxon object identifier = 400330000). Amplicon sizes are noted in Table 2. The PCR primers used to amplify 16S rRNA were the prokaryote-specific 515F (5′GTGCCAGCMGCCGCGGTAA) and 907R (3′CCGYCAATTCMTTTRAGTTT), which yielded a 392-bp product. PCR was performed using 0.2 ng cDNA in a final reaction mixture (15 μl) containing 1× PCR buffer (Bio-Rad.), 0.25 mM deoxynucleoside triphosphates (Bio-Rad), 0.3 μM of each gene-specific 5′ and 3′ primer, and 1 unit of Taq DNA polymerase (Bio-Rad). Reactions were cycled with an iCycler (Bio-Rad) using a temperature profile of 95°C for 5 min, 95°C for 1 min, annealing temperature for 1 min, and 72°C for 1 min (30 cycles), and a final extension of 72°C for 7 min. Annealing temperatures for the putative alkaline phosphatase, pstS, the putative phosphate permease, and 16S rRNA were 58°C, 59.5°C, 61.3°C, and 47°C, respectively. PCR products were resolved on a 2% agarose gel, stained with ethidium bromide, and imaged with a Gel Logic 440 imaging system (Kodak, Rochester, NY). The results were semiquantified by normalizing the band intensities to those obtained from the 16S rRNA gene using 1D image analysis software (Kodak), allowing for assessments of the presence or absence of the target gene but not relative abundance.
TABLE 2.
Putative phosphorus scavenging genes present in the Crocosphaera watsonii WH8501 genome
| Category | IMG gene object identifier | Putative function | % Similarity to reference | Reference |
|---|---|---|---|---|
| Low-affinity phosphate transport | 400856070 | Phosphate permease (transporter) low affinity | 51.96 | Shewanella frigidimarina NCMIB400 |
| Unknown function | 400885240 | PhoU family—involved in phosphate transport | 65.45 | Synechocystis sp. strain PCC6803 |
| 400856060 | PhoU family—involved in phosphate transport | 27.07 | Shewanella frigidimarina NCMIB400 | |
| 400872090 | Phosphate starvation-inducible protein, phoH | 76.73 | Synechocystis sp. strain PCC6803 | |
| Hydrolysis of phosphate esters | 400846230 | Alkaline phosphatasea | 27.44 | Deinococcus geothermalis DSM11300 |
| 400841270 | Phosphoesterase | 38.1 | Synechocystis sp. strain PCC6803 | |
| 400869600 | Metallophosphoesterase | 53.7 | Trichodesmium erythraeum IMS101 | |
| 400867140 | Alkaline phosphatase-like, dedA | 59.61 | Synechocystis sp. strain PCC6803 | |
| High-affinity phosphate transport | 400890860 | Phosphate binding, pstS | 33.33 | Methanothermobacter thermautotrophicus Delta H |
| 400876370 | Phosphate binding, pstS | 65 | Synechocystis sp. strain PCC6803 | |
| 400878470 | Phosphate binding, pstS | 50.45 | Synechocystis sp. strain PCC6803 | |
| 400876380 | Phosphate permease, pstC | 57.63 | Synechocystis sp. strain PCC6803 | |
| 400876390 | Phosphate permease, pstA | 29.82b | Methanosarcina mazei Gol | |
| 400876400 | Phosphate permease, pstA | 69.23b | Synechocystis sp. strain PCC6803 | |
| 400876410 | ATPase component, pstB | 76.78 | Synechocystis sp. strain PCC6803 | |
| 400876420 | ATPase component, pstB | 64.23 | Synechocystis sp. strain PCC6803 | |
| Polyphosphate metabolism | 400890460 | Polyphosphate kinase, ppK | 74.86 | Synechocystis sp. strain PCC6803 |
| 400849750 | Exopolyphosphatase, ppX | 62.84 | Synechocystis sp. strain PCC6803 | |
| 400852200 | Inorganic pyrophosphatase, ppA | 82.71 | Nostoc punctiforme PCC73102 |
Although much smaller, this gene is 48.46% similar to the atypical alkaline phosphatase characterized in Synechococcus PCC7942 (22).
Percent similarity over 60% of the ORF; all other similarities reported are over 80% of the ORF.
Sequencing.
Genomic DNA extracts for several C. watsonii strains were obtained from I. Ehrenreich (Table 1). The extraction procedure is described elsewhere (8). PCR amplification of each gene amplicon was conducted using 1 μl of each DNA extract (diluted 1:100) with primer sequences and reaction conditions as detailed above. The putative alkaline phosphatase, pstS, and the putative phosphate permease fragments were identified and gel purified using the QIAquick gel extraction kit (QIAGEN, Valencia, CA). Sequencing of PCR products was performed at either the sequencing facility at the Bay Paul Center at the Marine Biological Laboratory, Woods Hole, MA, or at the University of Maine, Orono, ME. All sequence data were analyzed manually using the programs Sequencher (Gene Codes Corporation, Ann Arbor, MI), MacVector (Accelrys, Burlington, MA), and BLASTN (http://www.ncbi.nlm.nih.gov/BLAST).
Genome observations.
Genomic topology was visualized with the publicly available Integrated Microbial Genomes (IMG) interface on the Department of Energy-Joint Genome Institute web page (http://img.jgi.doe.gov/cgi-bin/pub/main.cgi). Homology searches were also performed using this interface. Genes were assigned a putative function or gene name based on percent identity over at least 80% of the open reading frame (ORF). In two cases, the gene was annotated based on 60% of the ORF where the topology supported the gene call. The evidence for these annotations and the complete sequences can be viewed with the IMG gene object identifier (Table 2) at the above URL.
Nucleotide sequence accession numbers.
C. watsonii sequences determined in this study have been deposited in GenBank with the following accession numbers: putative alkaline phosphatase, DQ297912 to DQ297914; putative phosphate permease, DQ297915 to DQ297920; and pstS, DQ297921 to DQ297924.
RESULTS
Genome observations.
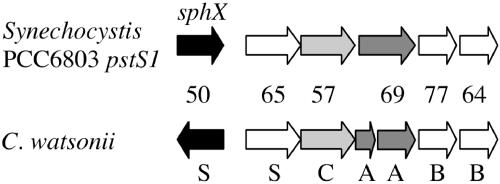
Observations from the C. watsonii WH8501 genome suggest that this diazotroph may have both low- and high-affinity phosphate transport systems. A putative low-affinity phosphate permease is present next to a putative copy of the phosphate transport regulator phoU (Table 2). A complete cluster of genes encoding the high-affinity transport of phosphate (pstSCAB) are also present in the C. watsonii WH8501 genome (Table 2). There are multiple, unique pstS orthologs distributed across different contigs, although the genes 400876370 and 400878470 are immediately adjacent to a complete phosphate transport system (pstABC) (Fig. 1). In this gene cluster, there are duplications of full-length pstB and truncated pstA (Fig. 1).
FIG. 1.
Topology of the genes encoding high-affinity phosphate transport in Crocosphaera watsonii and Synechocystis PCC6803. The genes pstS, -A, -B, and -C are designated with single letters; sphX, a member of the same gene family as pstS (1, 16), is also designated. Shading indicates the genes being compared with the percent similarity of each pair.
One common P stress strategy is to up-regulate enzymes that will hydrolyze DOP into bioavailable phosphate. There are no clear homologs of the phnCDE phosphonate transport system that is present in Synechococcus WH8102 (SYNW1168, 1169, 1170 at http://genome.ornl.gov/microbial/syn_wh/). There are also no apparent homologs of phnWX, the genes thought to encode for phosphonate hydrolysis in Synechococcus WH8102 (26). Further, there are no homologs of the phn gene cluster that encode the transport and hydrolysis of phosphonates in the diazotroph T. erythraeum (6). In contrast, C. watsonii appears to have a broad spectrum of genes with a putative role in phosphomonoester hydrolysis (Table 2).
Three genes with a potential role in polyphosphate metabolism were identified (Table 2). The putative ppK gene encodes a polyphosphate kinase for the synthesis of polyphosphate (15). The putative ppX gene encodes an exopolyphosphatase that sequentially releases phosphate from long-chain polyphosphate (11, 15). A third putative gene, ppA, encodes a soluble inorganic pyrophosphatase (11). None of these ORFs are contiguous in the current assembly of the C. watsonii WH8501 genome.
Gene expression.
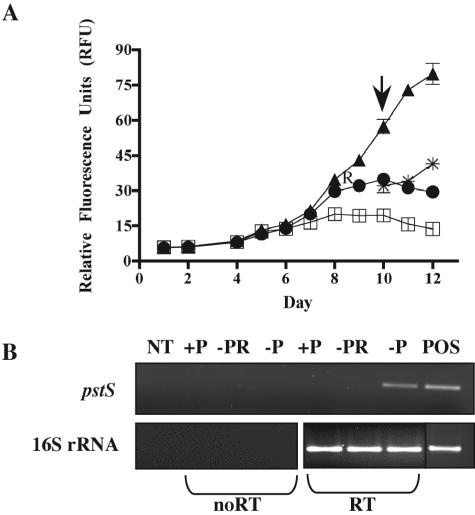
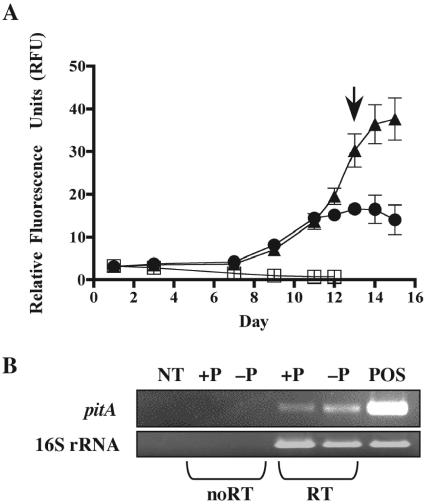
The expression of pstS, the putative phosphate permease, and the putative alkaline phosphatase were examined in axenic cultures of C. watsonii WH8501 grown under different conditions. Expression of pstS was detected in −P cultures grown as described above but not in +P cultures (Fig. 2). No pstS expression was detected in a -P culture refed (PR) to replete phosphate levels (Fig. 2). The expression of the putative phosphate permease was detected both in −P cultures grown as described above and in +P cultures (Fig. 3). To date, no consistent expression of the putative alkaline phosphatase could be detected. The 16S rRNA expression was very consistent across all treatments in both experiments (Fig. 2 and Fig. 3) and infrequently required normalization.
FIG. 2.
Expression of pstS (400876370). (A) A growth curve, plotted as relative fluorescence units, of axenic Crocosphaera watsonii WH8501 in SN medium with 45 μM (triangle), 1 μM (circle), or no added (square) phosphate. The “R” indicates where the 1 μM culture was refed (*) to 45 μM. Arrow indicates where samples for gene expression were harvested. Error bars denote the standard error from the mean (n = 3). (B) Expression of the 289-bp pstS amplicon and the 392-bp 16S rRNA amplicon. Lane designations are as follows: NT (no template), +P (cDNA from a P-replete culture, harvested as designated), −PR (cDNA from a P-starved culture refed to replete levels of P, harvested as designated), −P (cDNA from a P-starved culture, harvested as designated), POS (DNA from a P-replete culture). Samples transcribed without or with reverse transcriptase are designated noRT or RT, respectively.
FIG. 3.
Expression of a putative phosphate permease (400856070). (A) A growth curve, plotted as relative fluorescence units, of axenic Crocosphaera watsonii WH8501 in SN medium with 45 μM (triangle), 1 μM (circle), or no added (square) phosphate. Arrow indicates where samples for gene expression were harvested. Error bars denote the standard error from the mean (n = 3). (B) Expression of the 294-bp putative phosphate permease amplicon (indicated as pitA) and the 392-bp 16S rRNA amplicon. Lane designations are as follows: NT (no template), +P (cDNA from a P-replete culture, harvested as designated), −P (cDNA from a P-starved culture, harvested as designated), POS (DNA from a P-replete culture). Samples transcribed without or with reverse transcriptase are designated noRT or RT, respectively.
Strain comparison.
Strains isolated from both the Pacific (WH0002, H12, H13, and OFF) and the Atlantic (WH0401 and E12) were examined for the presence of pstS, the putative phosphate permease, and the putative alkaline phosphatase. The putative alkaline phosphatase and pstS genes were amplified and sequenced from strains OFF (pstS only), WH0401, H12, and WH0002 (3). Sequence alignments of the pstS and phoA gene fragments showed high sequence similarity to strain WH8501, with no consistent differences between Pacific and Atlantic isolates (Table 3). Nucleotide sequence alignments comparing C. watsonii WH8501 pstS to the Synechococcus WH8102 and Trichodesmium IMS101 genes were 27% and 35% similar, respectively (data not shown). A series of strains was also examined for the putative low-affinity phosphate transporter, and all the strains tested have the gene. Sequence identity across the 294-bp amplicon was 99 to 100% (Table 3).
TABLE 3.
Percent similarities of gene sequences related to P scavenging recovered from various Crocosphaera watsonii strains to orthologous sequences from C. watsonii WH8501
| Strain | % Similarity to WH8501 sequencea
|
||
|---|---|---|---|
| pstS (289 bp) | Putative alkaline phosphatase gene (246 bp) | Putative phosphate permease gene (294 bp) | |
| WH0401 | 100 | 98 | 100 |
| E12 | 100 | ‡ | 99 |
| WH0002 | 100 | 98 | 100 |
| H12 | 100 | 99 | 100 |
| H13 | ‡ | ‡ | 100 |
| OFF | 100 | ‡ | 100 |
Amplicon sizes are given in parentheses. ‡, gene fragment did not amplify.
Growth studies.
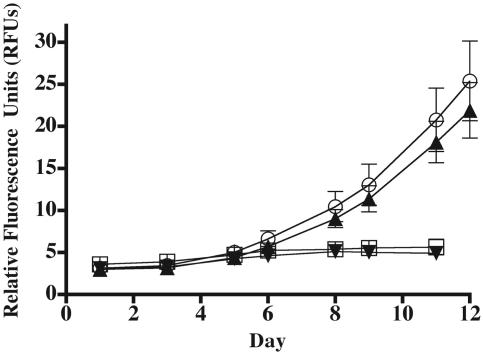
Axenic cultures of C. watsonii WH8501 (Atlantic isolate) and WH0002 (Pacific isolate) were able to grow on a variety of phosphomonoesters as a sole source of added phosphorus. Specifically, C. watsonii WH8501 growth occurred on phytic acid, AMP (Table 4), and glycerophosphate (Fig. 4). Conversely, there was no growth relative to the control (no P added) on the phosphonate, phosphonoformic acid (Fig. 4), and there has been no growth observed on any phosphonate compounds tested to date (Table 4). Of the compounds tested, similar growth patterns were observed for C. watsonii WH0002 (Table 4). Further, growth patterns were similar with (SN) or without (SO) an N source added to the medium (data not shown).
TABLE 4.
Phosphorus sources used for growth studies of Crocosphaera watsonii
| Phosphorus source | Abbreviation | Bond class | Growth
|
|
|---|---|---|---|---|
| WH8501 | WH0002 | |||
| dl-α-Glycerophosphate | GP | C-O-P | Yes | Yes |
| Adenosine monophosphate | AMP | C-O-P | Yes | Yes |
| Phytic acid | PA | C-O-P | Yes | NDa |
| Phosphonoacetic acid | PN | C-P | No | ND |
| Phosphonoformic acid | PFA | C-P | No | ND |
| 2-Aminoethylphosphonate | 2-AEP | C-P | No | No |
ND, not determined.
FIG. 4.
A representative growth curve, plotted as relative fluorescence units, of axenic Crocosphaera watsonii WH8501 in SN medium with 45 μM phosphate (triangle: potassium diphosphate), 45 μM phosphomonoester (circle: glycerophosphate), 45 μM phosphonate (reverse triangle: phosphonoformic acid), or no phosphate (square) added. Error bars denote the standard error from the mean (n = 3).
DISCUSSION
The factors that constrain nitrogen fixation in the open ocean remain a topic of intense study and debate among oceanographers. Several recent studies have highlighted the importance of phosphorus to the growth and nitrogen fixation rates of the diazotroph Trichodesmium (7, 23), but little is known about the phosphorus physiology of the unicellular photosynthetic diazotrophs, such as C. watsonii, and the extent to which phosphorus bioavailability may influence growth and nitrogen fixation in this group.
Using the publicly available sequence of the C. watsonii WH8501 genome, we identified gene homologs related to P metabolism, including genes related to the metabolism of polyphosphate, phosphate transport, and the hydrolysis of DOP. The presence of genes encoding proteins involved in the synthesis (e.g., ppK) and degradation (e.g., ppX) of polyphosphate in C. watsonii WH8501 is consistent with the presence of these genes in the other marine picocyanobacteria (19). Some strains of Prochlorococcus and Synechococcus can grow on polyphosphate as a sole source of P (19): as such, polyphosphate could be an important P source for C. watsonii in oligotrophic systems.
High-affinity phosphate transport may be a key adaptation of marine cyanobacteria that allows them to persist in low-environmental-DIP conditions. In the marine cyanobacterial genomes examined to date, multiple distinct pstS sequences, encoding the high-affinity phosphate binding protein PstS, have been identified (19, 27). This is also the case with C. watsonii, where there are three copies of pstS. Two of these copies of pstS are clustered together with a complete transport complex (pstABC), which has a duplication in both pstB and pstA. These pst genes are similar to genes of Synechocystis sp. strain PCC6803, which also has a duplication in pstB. Synechocystis sp. strain PCC6803 has two high-affinity phosphate transport systems, Pst1 and Pst2 (27), and the C. watsonii WH8501 complex is most similar to the Pst1 system. In microarray-based regulation studies, the Pst1 system was induced more rapidly than the Pst2 system in response to P stress (27). Although we have not examined the timing of expression in the different copies of pstS or the different components of the pstABC system, one of the pstS genes in this cluster (400876370) is clearly regulated by P supply in culture. This P regulation is consistent with studies in Synechocystis PCC6803 (27).
The expression of the PstS protein has been used as a target for assays of P stress in field populations of marine Synechococcus and Prochlorococcus (24). Considering the strong P regulation of C. watsonii WH8501 pstS expression, it may be possible to use this gene as a target for RT-PCR assays of P physiology. Although the sequence is identical over our amplicon in different C. watsonii strains, there is substantial sequence heterogeneity between C. watsonii and the pstS sequences of other picocyanobacteria, suggesting that gene expression studies in the field should be able to distinguish C. watsonii sequences from those of other cyanobacteria. RT-PCR assays of C. watsonii pstS may be used to examine the extent to which P supply constrains the growth and nitrogen fixation rates of Crocosphaera in the field.
One of the more striking, and unexpected, observations from the C. watsonii WH8501 genome is the presence of a putative low-affinity phosphate permease. C. watsonii has an ORF of 372 amino acids that is 25% similar to the 5′ region of the 499-amino-acid pitA gene involved in phosphate transport in Escherichia coli (reference 12 and references therein). The C. watsonii ORF is also 51% similar to a gene encoding a 347-amino-acid product in Shewanella frigidimarina (NCMIB400) that is annotated as a phosphate transporter, although it has not been experimentally characterized. No homologs of pitA or low-affinity phosphate permeases have been identified in the marine cyanobacterial genomes examined to date (19, 26), and others have suggested that the lack of pitA in the other marine cyanobacterial genomes is because the Km of E. coli pitA would not be efficient in marine systems, where DIP is often quite low (26). Yet we have been able to identify the gene in all of the C. watsonii strains we have tested. In addition, the putative phosphate permease gene is expressed in phosphate-replete axenic cultures of C. watsonii WH8501. Also of note is the presence of two putative copies of phoU, a modulator of inorganic phosphate transduction, which, like the putative phosphate permease, is not detectable in any of the marine picocyanobacteria with published genomes (19). The widespread presence of the putative phosphate permease gene in Crocosphaera and its expression suggest that this gene is serving a function for this genus. Further work is warranted to determine if the gene is indeed being used for the low-affinity transport of phosphate.
Phosphorus bioavailability is in part determined by the presence or absence of P scavenging systems within a cell, and genomic observations suggest that C. watsonii strains have a robust capacity to hydrolyze phosphomonoesters in the marine DOP pool. Genome annotation identified at least four genes that appear to encode phosphomonoesterases. These genomic observations are consistent with data presented herein, where axenic C. watsonii WH8501 was able to grow on a variety of phosphomonoesters as a sole source of added phosphorus. We were unable to consistently detect expression of the putative alkaline phosphatase (400846230) that we examined, and it may be that one or more of the other putative phosphomonoesterases listed in Table 2 are mediating this organism's growth on this bond class of DOP.
Homologs of phnCDE, encoding proteins involved in phosphonate transport, have been documented in numerous marine cyanobacteria including Synechococcus sp. strain WH8102 (19, 21, 26), Prochlorococcus sp. strain MIT9313 (19), and T. erythraeum IMS101 (6). Although there are many ABC transporters in the C. watsonii genome, we are unable to identify clear homologs of phnCDE. Phosphonate hydrolysis in heterotrophic bacteria (e.g., Pseudomonas stutzeri) is typically controlled by a suite of phn genes (phnF to -P) that encode a C-P lyase complex (29). The phnF to -P genes of the C-P lyase complex have not been identified in any of the picocyanobacterial genomes examined to date (19). However, Synechococcus sp. strain WH8102 will grow on the phosphonates 2-aminoethylphosphonate and ethylphosphonate, and here phosphonate hydrolysis is thought to be mediated by a different mechanism (putatively phnWX), since the genes phnF to -P are absent (26). In C. watsonii, there are no clear homologs of either phnF to -M or phnWX, suggesting this genus may be lacking in the ability to metabolize exogenous phosphonates as a phosphorus source. Growth studies corroborate these genomic observations in that growth for two strains was observed on phosphomonoesters but not on any of the phosphonates tested to date. This apparent lack of phosphonate metabolism is particularly striking relative to the diazotroph T. erythraeum, which has a complete C-P lyase complex, likely allowing this organism to metabolize a variety of exogenous phosphonate compounds (6).
Genes potentially involved in phosphorus scavenging (e.g., phosphate transport and DOP hydrolysis) have been described for several genera of marine cyanobacteria, including Prochlorococcus spp. (19, 25) and Synechococcus spp. (19, 26). These studies and our ongoing research with Trichodesmium spp. (20) have highlighted that there can be substantial heterogeneity in the presence and regulation of P-scavenging genes in different genera, and even between the presence and absence of these genes in strains of the same genus. For example, the genome of Prochlorococcus sp. strain MED4 has a gene encoding a putative alkaline phosphatase (phoA), whereas Prochlorococcus sp. strain MIT9313 does not (19, 25). Genes putatively encoding a high-affinity phosphate binding protein (pstS) and an alkaline phosphatase were examined in multiple C. watsonii strains isolated from different years and from both the Pacific and the Atlantic. The pstS sequence is identical over the 289-bp amplicon in all the strains examined. The putative alkaline phosphatase gene is 98 to 99% similar over the 246-bp amplicon. Although strains from the Atlantic as well as the Pacific appear to have these two P-scavenging-related genes, there are a few strains that we have been unable to amplify using the primer sets reported here. The reasons for this are unclear, but they hint at the possibility of sequence divergence or potentially the absence of these genes in some strains.
There is a growing body of research on unicellular diazotrophs, such as Crocosphaera, although much of this work has been focused on their distribution and N physiology. Our data suggest that this genus shares some of the adaptations to low DIP commonly found in marine cyanobacteria, including the capacity for the high-affinity transport of phosphate and the hydrolysis of phosphomonoesters. Despite these similarities, our data also suggest some fundamental differences between Crocosphaera and other marine cyanobacteria, including the apparent lack of phosphonate utilization and the presence of a putative low-affinity phosphate permease. If these differences are borne out in field populations, Crocosphaera occupies a different niche with regard to competition for P than other marine cyanobacteria.
Acknowledgments
We thank John Waterbury for helpful comments on the manuscript and for generously providing the Crocosphaera strains used in this study. Jonathan Urbach volunteered his time for monitoring cultures, and thanks also go to Eric Webb and Ian Ehrenreich for providing DNA samples. The Crocosphaera watsonii WH8501 genome was produced by the U.S. Department of Energy Joint Genome Institute (http://www.jgi.doe.gov/).
Funding for this research was provided by the NSF OCE Biological Oceanography Program and the Woods Hole Oceanographic Institution Ocean Life Institute.
REFERENCES
- 1.Aiba, H., and T. Mizuno. 1994. A novel gene whose expression is regulated by the response-regulator, SphR, in response to phosphate limitation in Synechococcus species PCC7942. Mol. Microbiol. 13:25-34. [DOI] [PubMed] [Google Scholar]
- 2.Anderson, R. A., D. M. Jacobson, and J. P. Sexton. 1991. Provasoli-Guillard Center for Culture of Marine Phytoplankton: catalogue of strains. Provasoli-Guillard Center for Culture of Marine Phytoplankton, West Boothbay Harbor, Maine.
- 3.Capone, D. G., J. P. Zehr, H. W. Paerl, B. Bergman, and E. J. Carpenter. 1997. Trichodesmium, a globally significant marine cyanobacterium. Science 276:1221-1229. [Google Scholar]
- 4.Carpenter, E. J., A. Subramaniam, and D. G. Capone. 2004. Biomass and primary productivity of the cynobacterium Trichodesmium spp. in the tropical N Atlantic ocean. Deep-Sea Res. 51:173-203. [Google Scholar]
- 5.Church, M. J., B. D. Jenkins, D. M. Karl, and J. P. Zehr. 2005. Vertical distributions of nitrogen-fixing phylotypes at Stn ALOHA in the oligotrophic North Pacific Ocean. Aquat. Microb. Ecol. 38:3-14. [Google Scholar]
- 6.Dyhrman, S. T., P. D. Chappell, S. T. Haley, J. W. Moffett, E. D. Orchard, J. Waterbury, and E. A. Webb. 2005. Phosphonate utilization by the globally important marine diazotroph Trichodesmium. Nature 439:68-71. [DOI] [PubMed] [Google Scholar]
- 7.Dyhrman, S. T., E. Webb, D. M. Anderson, J. Moffett, and J. Waterbury. 2002. Cell specific detection of phosphorus stress in Trichodesmium from the Western North Atlantic. Limnol. Oceanogr. 47:1823-1836. [Google Scholar]
- 8.Ehrenreich, I. M., E. A. Webb, and J. Waterbury. 2005. The distribution and diversity of natural product genes in marine and freshwater cyanobacterial cultures and genomes. Appl. Environ. Microbiol. 71:7401-7413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Falcón, L. I., E. J. Carpenter, F. Cipriano, B. Bergman, and D. G. Capone. 2004. N2 fixation by unicellular bacterioplankton from the Atlantic and Pacific oceans: phylogeny and in situ rates. Appl. Environ. Microbiol. 70:765-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Falcón, L. I., F. Cipriano, A. Y. Chistoserdov, and E. J. Carpenter. 2002. Diversity of diazotrophic unicellular cyanobacteria in the tropical North Atlantic Ocean. Appl. Environ. Microbiol. 68:5760-5764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gómez-García, M. R., M. Losada, and A. Serrano. 2003. Concurrent transcriptional activation of ppa and ppx genes by phosphate deprivation in the cyanobacterium Synechocystis sp. strain PCC 6803. Biochem. Biophys. Res. Commun. 302:601-609. [DOI] [PubMed] [Google Scholar]
- 12.Harris, R. M., D. C. Webb, S. M. Howitt, and G. B. Cox. 2001. Characterization of PitA and PitB from Escherichia coli. J. Bacteriol. 183:5008-5014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kolowith, L. C., E. D. Ingall, and R. Benner. 2001. Composition and cycling of marine organic phosphorus. Limnol. Oceanogr. 46:309-320. [Google Scholar]
- 14.Kononova, S. V., and M. A. Nesmeyanova. 2002. Phosphonates and their degradation by microorganisms. Biochemistry (Moscow) 67:220-233. [DOI] [PubMed] [Google Scholar]
- 15.Kornberg, A., N. N. Rao, and D. Ault-Riché. 1999. Inorganic polyphosphate: a molecule of many functions. Annu. Rev. Biochem. 68:89-125. [DOI] [PubMed] [Google Scholar]
- 16.Mann, N. H., and D. J. Scanlan. 1994. The SphX protein of Synechococcus species PCC 7942 belongs to family of phosphate-binding proteins. Mol. Microbiol. 14:595-596. [DOI] [PubMed] [Google Scholar]
- 17.Mazard, S. L., N. J. Fuller, K. M. Orcutt, O. Bridle, and D. J. Scanlan. 2004. PCR analysis of the distribution of unicellular cyanobacterial diazotrophs in the Arabian Sea. Appl. Environ. Microbiol. 70:7355-7364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Montoya, J. P., C. M. Holl, J. P. Zher, A. Hansen, T. A. Villareal, and D. G. Capone. 2004. High rates of N2 fixation by unicellular diazotrophs in the oligotrophic Pacific Ocean. Nature 430:1027-1031. [DOI] [PubMed] [Google Scholar]
- 19.Moore, L. R., M. Ostrowski, D. J. Scanlan, K. Feren, and T. Sweetsir. 2005. Ecotypic variation in phosphorus-acquisition mechanisms within marine picocyanobacteria. Aquat. Microb. Ecol. 39:257-269. [Google Scholar]
- 20.Orchard, E., E. Webb, and S. T. Dyhrman. 2003. Characterization of phosphorus-regulated genes in Trichodesmium spp. Biol. Bull. 205:230-231. [DOI] [PubMed] [Google Scholar]
- 21.Palenik, B., B. Brahamsha, F. Larimer, M. Land, L. Hauser, P. Chain, J. Lamberdin, W. Regala, E. Allen, J. McCarren, I. Paulsen, A. Dufresne, F. Partensky, E. Webb, and J. Waterbury. 2003. The genome of a motile marine Synechococcus. Nature 424:1037-1042. [DOI] [PubMed] [Google Scholar]
- 22.Ray, J., D. Bhaya, M. A. Block, and A. R. Grossman. 1991. Isolation, transcription, and inactivation of the gene for an atypical alkaline phosphatase of Synechococcus sp. strain PCC 7942. J. Bacteriol. 173:4297-4309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sañudo-Wilhelmy, S. A., A. B. Kustka, C. J. Gobler, D. A. Hutchins, M. Yang, K. Lwiza, J. Burns, D. G. Capone, J. A. Raven, and E. J. Carpenter. 2001. Phosphorus limitation of nitrogen fixation by Trichodesmium in the central Atlantic Ocean. Nature 411:66-69. [DOI] [PubMed] [Google Scholar]
- 24.Scanlan, D. J., N. J. Silman, K. M. Donald, W. H. Wilson, N. G. Carr, I. Joint, and N. Mann. 1997. An immunological approach to detect phosphate stress in populations and single cells of photosynthetic picoplankton. Appl. Environ. Microbiol. 63:2411-2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scanlan, D. J., and N. J. West. 2002. Molecular ecology of the marine cyanobacteria Prochlorococcus and Synechococcus. FEMS Microbiol. Ecol. 40:1-12. [DOI] [PubMed] [Google Scholar]
- 26.Su, Z., P. Dam, X. Chen, V. Olman, T. Jiang, B. Palenik, and Y. Xu. 2003. Computational inference of regulatory pathways in microbes: an application to phosphorus assimilation pathways in Synechococcus sp. WH8102. Genome Informatics 14:3-13. [PubMed] [Google Scholar]
- 27.Suzuki, S., A. Ferjani, I. Suzuki, and N. Murata. 2004. The SphS-SphR two component system is the exclusive sensor for the induction of gene expression in response to phosphate limitation in Synechocystis. J. Biol. Chem. 279:13234-13240. [DOI] [PubMed] [Google Scholar]
- 28.Waterbury, J. B., F. W. Valois, and D. G. Franks. 1986. Biological and ecological characterization of the marine unicellular cyanobacterium Synechococcus, p. 71-120. In T. Platt and W. K. W. Li (ed.), Photosynthetic picoplankton, vol. 214. Canadian Department of Fisheries and Oceans, Ottawa. [Google Scholar]
- 29.White, A. W., and W. M. Metcalf. 2004. Two C-P lyase operons in Pseudomonas stutzeri and their roles in the oxidation of phosphonates, phosphite, and hypophosphite. J. Bacteriol. 186:4730-4739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu, J., W. G. Sunda, E. A. Boyle, and D. M. Karl. 2000. Phosphate depletion in the Western North Atlantic Ocean. Science 289:759-762. [DOI] [PubMed] [Google Scholar]
- 31.Zher, J. P., J. Waterbury, P. J. Turner, J. P. Montoya, E. Omoregie, G. F. Steward, A. Hansen, and D. M. Karl. 2001. Unicellular cyanobacteria fix N2 in the subtropical North Pacific Ocean. Nature 412:635-638. [DOI] [PubMed] [Google Scholar]