Abstract
The cascade of reactive nitrogen species generated from nitric oxide causes modification of proteins, lipids, and nucleic acids in a wide range of organisms. 3-Nitrotyrosine is one of the most common products of the action of reactive nitrogen species on proteins. Although a great deal is known about the formation of 3-nitrotyrosine, the subsequent metabolism of this compound is a mystery. Variovorax paradoxus JS171 and Burkholderia sp. strain JS165 were isolated from soil slurries when 3-nitrotyrosine was provided as the sole carbon, nitrogen, and energy source. During growth on 3-nitrotyrosine stoichiometric amounts of nitrite were released along with approximately one-half of the theoretically available ammonia. The catabolic pathway involving oxidative denitration is distinct from the pathway for tyrosine metabolism. The facile isolation and the specific, regulated pathway for 3-nitrotyrosine degradation in natural ecosystems suggest that there is a significant flux of 3-nitrotyrosine in such environments.
Nitric oxide (NO) is produced by a wide range of organisms (31) via a pathway having an ancient evolutionary origin (14). The cascade of reactive nitrogen species generated from NO causes modification of proteins, lipids, and nucleic acids (6, 11, 17, 22), including the nitration of a variety of catecholamines (21, 23). The nitration of aromatic amino acids, principally tyrosine (Tyr) and tryptophan, in proteins results in alteration of the function of the proteins (22). It is not clear whether the alterations are part of a signaling system (8, 16) or a by-product of the process that produces NO (22).
Numerous studies on the formation of 3-nitrotyrosine (3NTyr) and related molecules have yielded little information about the subsequent behavior and disposition of this compound. There is some evidence of enzymatic denitration of modified proteins (10) in animals, but the mechanism of the reaction is a mystery. The reported activity seems to interact only with the nitrated protein and not with free 3NTyr. It requires no cofactors, and the products of the reaction are unknown. The diversity of sources of NO and the resultant nitration of Tyr suggested the potential for metabolism of 3NTyr by bacteria, either in the infection-inflammation process or as part of the recycling involved in the carbon cycle.
A substantial number of bacteria that are able to degrade synthetic nitroaromatic compounds have been isolated, and the degradation mechanisms have been established (19). No information is available on the biodegradation of natural nitroaromatic compounds, such as 3NTyr. We report here the isolation of bacteria that are able to degrade 3NTyr via an oxidative catabolic pathway.
(A preliminary report of this work has been presented previously [Nishino and Spain, Abstr. 104th Gen. Meet. Am. Soc. Microbiol., abstr. Q-299, 2004].)
MATERIALS AND METHODS
Isolation and growth of bacteria.
A sandy loam topsoil from Cape Cod, MA, was inoculated (10%, wt/vol) into nitrogen-free minimal medium (4) (BLK) at pH 7.2 containing 3NTyr (50 μM), and the preparations were incubated with shaking at room temperature. After 3NTyr disappeared from the slurries, samples (10%, vol/vol) were transferred into fresh BLK containing 3NTyr (100 μM). Subsequent cultures were incubated at 30°C with shaking, and 3NTyr concentrations were monitored by high-performance liquid chromatography (HPLC) (see below). After several transfers into fresh media, samples were spread on 3NTyr plates (500 μM 3NTyr in BLK solidified with 1.8% agar). Individual colonies that were able to grow with 3NTyr as the sole carbon and nitrogen source were sent to MIDI Labs (Newark, DE) for identification by 16S rRNA gene sequence analysis.
Cultures were routinely maintained on 3NTyr plates. Liquid cultures were initially grown in 1 liter of BLK containing 3NTyr (500 μM). When the 3NTyr disappeared from the cultures, additional 3NTyr was added (at final concentrations up to 2 mM) until the optical density at 600 nm was>1.0. In later experiments cultures were grown on either glucose (5 mM), pyruvate (10 mM), or succinate (7.5 mM) provided as the carbon source with 3NTyr (500 μM) as the nitrogen source. When the 3NTyr disappeared, the cells were centrifuged, and each pellet was suspended in fresh BLK with 3NTyr (0.5 to 2 mM) without the additional carbon source and incubated for 2 to 4 h until the 3NTyr again disappeared. Control cultures were grown overnight in mineral medium (25) with succinate or glucose (both at a concentration of 5 mM) as the carbon source and NH4Cl (9.4 mM) as the nitrogen source. Tyrosine-grown cultures were started in 100 ml of BLK containing 500 μM Tyr as the sole carbon and nitrogen source. Variovorax paradoxus strain JS171 was transferred to 2-liter shake flasks containing 1 liter of BLK supplemented with 5 mM Tyr. Burkholderia sp. strain JS165 was grown in a 2-liter bioreactor (Applikon, Dover, NJ) with an initial Tyr concentration of 500 μM. When the Tyr in the culture medium disappeared, additional substrate (500 mM) was added via a syringe pump at rates (0.1 to 0.6 ml h−1) that kept the concentration of Tyr in the culture fluid below 100 μM. The temperature was maintained at 30°C, the O2 concentration was maintained at >30%, and the culture was stirred at 1,000 rpm. Cells were harvested by centrifugation and washed twice with phosphate buffer (0.02 M, pH 7.0) before use in experiments.
Respirometry.
Oxygen uptake was measured polarographically with a Clark-type oxygen electrode connected to a YSI model 5300 biological oxygen monitor. The reaction mixtures contained cells (0.05 to 0.2 mg protein ml−1) plus substrates (10 μM) in air-saturated phosphate buffer (0.02 M, pH 7.0) at 30°C.
Enzyme assays.
Cell extracts were prepared as previously described (18). 3NTyr deaminase reaction mixtures (final volume, 1 ml) contained 0.05 to 1 μmol of 3NTyr, 0.1 μmol of α-ketoglutarate (α-KG), 9.8 μmol of sodium phosphate (pH 7.0), and cell extract (0.1 to 0.5 mg of protein). 4-Hydroxy-3-nitrophenylacetic acid (HNPA) denitrase reaction mixtures (final volume, 1 ml) contained 0.05 to 1 μmol HNPA, 0.1 μmol NADH, 9.8 μmol of sodium phosphate (pH 7.0), and cell extract (0.1 to 0.5 mg of protein). Suboptimal concentrations of NADH were used to avoid interference with the nitrite assay (30); therefore, only initial rates were used to calculate specific activities. Some cell extracts were dialyzed for 1 h against phosphate buffer (0.02 M, pH 7.0) before use. Substrates and products in enzyme assays were assayed by HPLC. Rates were determined at 30°C. Reactions were stopped by addition of the reaction mixture (100 μl) to trichloroacetic acid (1 μl). The acidified reaction mixture was clarified by centrifugation before analysis.
Analytical methods.
Nitrite and ammonia concentrations were measured by standard methods (24). Protein was measured with a Pierce (Rockford, IL) BCA protein assay reagent kit. HPLC analyses were performed with a Merck Chromolith RP18e column (4.6 mm [inside diameter] by 50 mm) attached to an Agilent 1100 HPLC with a diode array detector. The mobile phase consisted of a mixture of part A (13.5 mM trifluoroacetic acid in water) and part B (6.75 mM trifluoroacetic acid in acetonitrile). The initial conditions were 98% part A and 2% part B at a flow rate of 1 ml/min for 1 min. Between 1 and 7 min a linear gradient changed the proportion to 50% part A and 50% part B and the flow rate to 1.5 ml/min. The final conditions were held for 1 min. The column and autosampler temperatures were 35 and 5°C, respectively. 3NTyr, HNPA, and homoprotocatechuate were monitored at 220 nm and eluted at 3.57, 4.73, and 2.14 min, respectively. Potential metabolites of 3NTyr were identified by HPLC comparison with authentic standards.
Chemicals.
3NTyr (purity, >99%) was obtained from Cayman Chemical (AnnArbor, MI), and HNPA (purity, 99%) was obtained from Sigma-Aldrich (Milwaukee, WI). 3-Chlorocatechol was obtained from Helix Biotech.
Nucleotide sequence accession numbers.
The 16S rRNA gene sequences of Burkholderia sp. strain JS165 and V. paradoxus JS171 have been deposited in the GenBank database under accession numbers DQ285572 and DQ285573.
RESULTS
Isolation and identification of 3NTyr-degrading bacteria.
3NTyr degradation began in soil enrichment cultures after 1 to 2 weeks when 3NTyr was provided as the sole growth substrate. 3NTyr disappearance was accompanied by the accumulation of nitrite and ammonia in the culture medium. Pure cultures able to grow on 3NTyr as the sole carbon, nitrogen, and energy source were isolated after several transfers with increasing concentrations of 3NTyr. Thirteen strains that represented a variety of colony morphologies were selected for identification by 16S rRNA gene analysis. Nine of the isolates could not be matched to any organism in the GenBank or MicroSeq databases. In preliminary experiments dense suspensions of 3NTyr-grown cells all degraded 200 μM 3NTyr with the release of nitrite and ammonia. The various isolates could grow on different maximum concentrations of 3NTyr in the range from 0.5 to 5 mM. Burkholderia sp. strain JS165 and V. paradoxus strain JS171 were selected for detailed studies because they grew on higher concentrations of 3NTyr than the other isolates (5 and 2 mM, respectively).
Growth of bacteria.
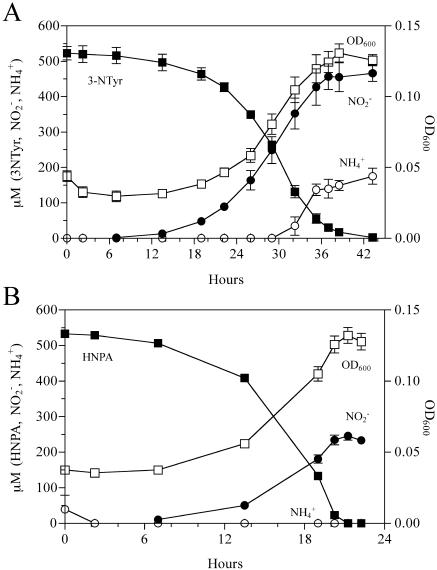
Burkholderia sp. strain JS165 grew on 3NTyr with stoichiometric accumulation of nitrite and accumulation of about one-half of the theoretical ammonia (Fig. 1A). HNPA, a potential metabolic intermediate of 3NTyr degradation, was an even better growth substrate (Fig. 1B). The growth yields from the two substrates were similar, but only one-half the nitrite accumulated in the growth medium when HNPA was the growth substrate. Similar results were obtained for V. paradoxus JS171, but the growth yields with the two substrates were about 60% of those of strain JS165 (data not shown).
FIG. 1.
Conversion and accumulation of 3NTyr metabolites. (A) Growth of Burkholderia sp. strain JS165 on 3NTyr with stoichiometric accumulation of nitrite. Cultures were grown in duplicate, and the error bars represent one standard deviation. (B) Growth of Burkholderia sp. strain JS165 on HNPA. OD600, optical density at 600 nm.
Both strains grew rapidly when 3NTyr or HNPA was supplied as the nitrogen source and glucose (for JS165), succinate, or pyruvate was the carbon source. Burkholderia sp. strain JS165 grew poorly on 1 mM or less Tyr; higher Tyr concentrations were toxic. V. paradoxus JS171 grew well on 5 mM Tyr and poorly on glucose.
Resting cell assays.
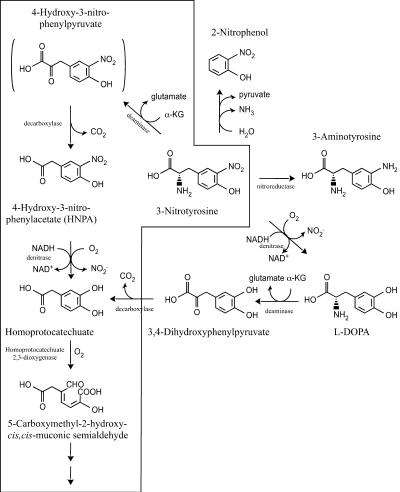
The preliminary results suggested that HNPA was an intermediate in the 3NTyr degradation pathway and that the nitro group was released as nitrite. Plausible initial transformations of 3NTyr (Fig. 2) could involve elimination of either nitrite or ammonia. The predominant bacterial pathways for degradation of Tyr involve α-KG-dependent deamination of Tyr to 4-hydroxyphenylpyruvate and then further degradation through either homogentisate (2) or homoprotocatechuate (3). Potential intermediates of the 3NTyr catabolic pathways, based on analogy to the most common transformations of Tyr, were tested for the ability to stimulate oxygen uptake in resting cells. 3NTyr, HNPA, homoprotocatechuate, and 3,4-dihydroxyphenylalanine (L-DOPA) stimulated oxygen uptake by washed cells grown on 3NTyr or HNPA (Table 1) but not by Tyr-, glucose-, or succinate-grown cells. Lack of stimulation by 2-nitrophenol suggested that an initial lyase reaction was unlikely. The failure of 3-aminotyrosine to stimulate oxygen uptake by 3NTyr-grown cells argues strongly against initial reduction of the nitro group. Tyr-grown cells of both strains oxidized homogentisate more rapidly than homoprotocatechuate, whereas the reverse was true of 3NTyr-grown cells. We concluded that the 3NTyr and HNPA degradation pathways in the two strains are tightly regulated, similar, and distinct from the Tyr degradation pathway.
FIG. 2.
Potential pathways for 3NTyr degradation based on analogy to Tyr degradation pathways. The reactions in the box represent the proposed degradation pathway.
TABLE 1.
Oxygen uptake by resting cellsa
| Test substrate | Oxygen uptake (nmol min−1 mg protein−1) (mean ± SD)
|
|||||
|---|---|---|---|---|---|---|
|
Burkholderia sp. strain JS165 with the following growth substrates:
|
V. pardoxus JS171 with the following growth substrates:
|
|||||
| 3NTyr | HNPA | Tyr | 3NTyr | HNPA | Tyr | |
| 3NTyr | 120 ± 40 (9)b | 58 (1) | NDc | 110 ± 29 (5) | 160 (1) | 6 ± 12 (4) |
| HNPA | 260 ± 150 (6)d | 170 (1) | ND | 160 ± 22 (3) | 300 (1) | 12 ± 16 (2) |
| Tyrosine | 38 ± 15 (5) | NTe | 23 (1) | 53 ± 14 (4) | 83 (1) | 200 ± 160 (4) |
| Homoprotocatechuate | 280 ± 140 (5)f | NT | ND | 55 ± 17 (4) | 130 (1) | 8 ± 14 (4) |
| Homogentisate | ND | NT | 13 (1) | 41 ± 22 (4) | 69 (1) | 64 ± 73 (4) |
| L-DOPA | 120 ± 64 (5) | 160 (1) | ND | 190 ± 63 (4) | 300 (1) | 41 ± 29 (4) |
| Hydroxyphenylpyruvate | 58 ± 40 (4) | NT | NT | 46 ± 25 (4) | NT | 80 ± 29 (4) |
| Dopamine | ND | NT | NT | ND | NT | NT |
| 2-Nitrophenol | ND | NT | NT | ND | NT | NT |
Substrates were provided at a concentration 100 μM for rate determinations and at a concentration of 25 μM for stoichiometry determinations. Succinate- and glucose-grown control cultures had no activity with 3NTyr, HNPA, or homoprotocatechuate. Succinate-grown JS165 had no activity with Tyr; succinate-grown JS171 had low activity(36% of succinate) with Tyr.
The numbers in parentheses are numbers of experiments(two to four assays per experiment).
ND, not detected.
The stoichiometry was 2.9 nmol O2/nmol substrate.
NT, not tested.
The stoichiometry was 1.8 nmol O2/nmol substrate.
Deamination of 3NTyr.
Growth studies indicated that both an amino group and a nitro group are released from 3NTyr during degradation of the compound, but time courses did not reveal which group was released first. HNPA, a nitrated analog of 4-hydroxyphenylacetate, has been suggested as a marker for nitrosation and nitration of Tyr (20), with possible involvement of a decarboxylase in the formation of HNPA from 3NTyr (15). Such formation would also require deamination either before or after decarboxylation. Because 3NTyr-grown cultures readily oxidized HNPA, cell extracts were tested for an initial α-KG-dependent deamination of 3NTyr (Table 2). When extracts of 3NTyr-grown JS165 were provided with 3NTyr and α-KG, 3NTyr disappearance was accompanied by stoichiometric accumulation of a metabolite that had a UV-visible light spectrum and retention time identical to those of authentic HNPA. No 3NTyr disappeared and no HNPA accumulated in controls that lacked α-KG or when extracts of Tyr- or succinate-grown cells were substituted in the reaction mixtures. The enzyme activity was not stable in extracts of JS171 cells. Neither tetrahydrobiopterin nor pyridoxal phosphate in any combination with α-KG or NAD(P)H increased the rate or extent of deamination.
TABLE 2.
Enzyme activities in cell extracts of cultures grown on 3NTyra
| Strain | Enzyme assayed | Assay substrateb | Sp act (nmol min−1 mg protein−1)c |
|---|---|---|---|
| Burkholderia sp. strain JS165 | 3-Nitrotyrosine deaminase | 3NTyr, α-KG | 88 |
| 3-Nitrotyrosine deaminase | 3NTyr, no α-KG | NDd | |
| HNPA denitrase | HNPA, NADH | 40 | |
| HNPA denitrase | HNPA, no added NADH | 14 | |
| HNPA denitrase | HNPA, NAD+ recyclee | ND | |
| 3NTyr denitrase | 3NTyr, NADH | ND | |
| V. paradoxus JS171 | HNPA denitrase | HNPA, NADH | 26 |
| HNPA denitrase | HNPA, no NADH | ND | |
| 3NTyr denitrase | 3NTyr, NADH | ND |
No activity was detected in extracts of cells grown on Tyr or succinate.
Substrates were provided at a concentration of 200 μM.
The values are averages from duplicate reactions.
ND, not detected.
Preincubation of cell extract with lactate dehydrogenase and pyruvate.
Denitration of HNPA.
Ultracentrifuged extracts of 3NTyr-grown cells catalyzed the NADH-dependent stoichiometric release of nitrite from HNPA but not from 3NTyr (Table 2). The initial rates were consistent with oxygen uptake rates determined in resting cell assays (Table 1), as well as with the maximum rate of substrate disappearance during growth on 3NTyr and HNPA (8 and 11 nmol min−1 mg protein−1, respectively, for JS165). NADPH could not replace NADH. Dialysis abolished the denitration activity, which was only partially restored by incubation of the extract with ferrous iron (50 μM), but not with ferric iron, prior to the assay. Addition of flavin adenine dinucleotide and flavin mononucleotide had no effect on the reaction. Cell extracts without addition of NADH catalyzed a low level of nitrite release from HNPA, but extracts that were preincubated with lactate dehydrogenase and pyruvate to remove native NADH prior to the HNPA-nitrite release assay catalyzed no nitrite release. No nitrite release was detected in the presence of extracts of Tyr- or succinate-grown cells, and HPLC analysis revealed no transformation of HNPA in any of the reaction mixtures containing extracts from Tyr- or succinate-grown cells. The results clearly indicate that the NADH-dependent denitrating enzyme is expressed during growth on 3NTyr.
Ring cleavage of homoprotocatechuate.
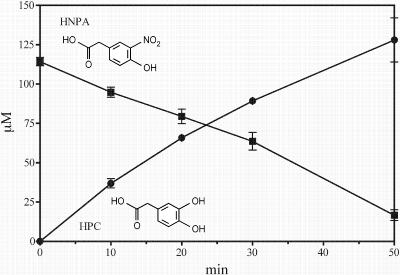
Extracts from 3NTyr-grown cells converted homoprotocatechuate to a yellow product with absorbance maxima at 380 and 315 nm at pH 7.0, consistent with 5-carboxymethyl-2-hydroxy-cis,cis-muconic semialdehyde, the product of cleavage of homoprotocatechuate by homoprotocatechuate 2,3-dioxygenase (1, 13). 3-Chlorocatechol is a powerful inhibitor of extradiol ring cleavage enzymes (27), andwhen 3NTyr-grown cells were preincubated with 3-chlorocatechol (750 μM), upon addition of HNPA homoprotocatechuate accumulated stoichiometrically (Fig. 3). The stoichiometry of oxygen consumption versus substrate for HNPA was 1 mol greater than that for homoprotocatechuate (Table 1), which clearly indicates that one mole of oxygen is required for the denitration of HNPA and suggests that the enzyme is a monooxygenase.
FIG. 3.
Accumulation of homoprotocatechuate (HPC) from HNPA when 3NTyr-grown cells were incubated with 3-chlorocatechol.
DISCUSSION
We found that 3NTyr is degraded by a pathway that is inducible and distinct from the Tyr degradation pathways found in Burkholderia sp. strain JS165 and V. paradoxus JS171. The entire pathway is expressed after growth on HNPA or 3NTyr but not after growth on tyrosine, glucose, or succinate. The activities of the key enzymes were demonstrated in intact cells or cell extracts, and the rates were sufficient to account for the growth of the bacteria. The proposed pathway (Fig. 2) is initiated by an α-KG-dependent deamination analogous to the initial deamination catalyzed by the broadly distributed tyrosine transaminase (EC 2.6.1.5) (5). The proposed intermediate, 4-hydroxy-3-nitrophenylpyruvate, was not confirmed in the pathway due to the lack of an authentic standard and the fact that it did not accumulate during the conversion of 3NTyr to HNPA. Decarboxylation of 4-hydroxy-3-nitrophenylpyruvate would produce HNPA, which was detected with a stoichiometric yield from 3NTyr incubated with cell extracts in the presence of α-KG. The requirement for α-KG for catabolism of 3NTyr rules out initial decarboxylation followed by deamination. Monooxygenase-catalyzed denitration of HNPA yieldshomoprotocatechuate, which is cleaved by homoprotocatechuate-2,3-dioxygenase, yielding 5-carboxymethyl-2-hydroxy-cis,cis-muconic semialdehyde. Subsequent metabolism of the ring fission product has been well established in a variety of bacteria (28).
L-DOPA stimulation of oxygen uptake by 3NTyr- and HNPA-grown cells is probably due to the broad substrate ranges of enzymes expressed after growth on 3NTyr and HNPA. The stoichiometric accumulation of HNPA in enzyme assay mixtures provided with 3NTyr and α-KG strongly argues against the hypothesis that L-DOPA is in the 3NTyr degradation pathway. HNPA production from 3NTyr requires loss of the amino group and retention of the nitro group, whereas L-DOPA production would require loss of the nitro group and retention of the amino group. There was no transformation of 3NTyr in the presence of NADH and the absence of α-KG, as would be expected for conversion to L-DOPA. Enzymes of the 3NTyr degradation pathway might catalyze deamination of L-DOPA to dihydroxyphenylpyruvate, followed by decarboxylation to homoprotocatechuate, thus accounting for the strong stimulation of oxygen uptake. L-DOPA can also be degraded via decarboxylation to dopamine in some systems, but we detected no activity towards dopamine by resting cells. L-DOPA did not stimulate any oxygen uptake by Tyr-grown cells of JS165, which suggests that L-DOPA is a better substrate for 3NTyr deaminase than for Tyr deaminase.
The key reaction in the 3NTyr degradation pathway is the denitration of HNPA. This reaction is reminiscent of the reaction catalyzed by the flavoprotein monooxygenase of the 2-nitrophenol degradation pathway (32). Work is currently under way to purify and characterize the enzyme responsible for the denitration activity. It is possible that the 3NTyr degradation pathway is derived from an HNPA degradation pathway. HNPA is excreted into the environment at concentrations that are an order of magnitude greater than the 3NTyr concentration (15, 26) and so might be more available as a growth substrate than 3NTyr. Other studies have reported that mammalian peroxidases, including enzymes in human saliva, catalyze the nitration of 4-hydroxyphenylacetate to HNPA (9, 29). On the other hand, the environmental flux of 3NTyr may be much higher than previously thought. When urban pollution coincides with high pollen loads, up to 10% of the airborne pollen protein can be nitrated (7). Estimates of airborne particulates of biological origin, principally cellular materials and pollen, have recently been revised upward 200-fold to 1,000 Tg year−1 (12).
The pathway for 3NTyr catabolism is closely analogous to thetyrosine degradation pathway that goes through homoprotocatechuate, and it might easily have been derived from the latter pathway by small changes in the substrate specificity of the key enzymes. Thus, the biochemistry of the 3NTyr degradation pathway is not surprising. The important question is what the presence of the pathway reveals about the flux of 3NTyr in natural ecosystems. It is clear that there was a substantial community of bacteria in our modest array of soil samples that could grow on 3NTyr. The ubiquity of 3NTyr in biological systems and the facile isolation of bacteria able to grow on 3NTyr suggest that considerable quantities of this compound might be released in the biosphere, which has led to the evolution and wide distribution of the pathway for degradation of 3NTyr. We are currently exploring the distribution of the ability to degrade 3NTyr among bacteria in a variety of ecosystems. The discovery of a biochemical mechanism for degradation of 3NTyr by bacteria provides the basis for examination of other systems for the existence of similar enzymes and may help to clarify the role of 3NTyr in mammalian systems.
Acknowledgments
We thank Stephen Spiro, Allen Orville, and Andreas Bommarius for helpful discussions and Rayford Payne for reviewing the manuscript.
Support for this project was provided by the Air Force Office of Scientific Research and the Defense Environmental Restoration Account.
REFERENCES
- 1.Adachi, K., Y. Takeda, S. Senoh, and H. Kita. 1964. Metabolism of p-hydroxyphenylacetic acid in Pseudomonas ovalis. Biochim. Biophys. Acta 93:483-493. [DOI] [PubMed] [Google Scholar]
- 2.Arias-Barrau, E., E. R. Olivera, J. M. Luengo, C. Fernandez, B. Galan, J. L. Garcia, E. Diaz, and B. Minambres. 2004. The homogentisate pathway: a central catabolic pathway involved in the degradation of l-phenylalanine, l-tyrosine, and 3-hydroxyphenylacetate in Pseudomonas putida. J. Bacteriol. 186:5062-5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arunachalam, U., V. Massey, and C. Vaidyanathan. 1992. p-Hydroxyphenylacetate-3-hydroxylase. A two-protein component enzyme. J. Biol. Chem. 267:25848-25855. [PubMed] [Google Scholar]
- 4.Bruhn, C., H. Lenke, and H.-J. Knackmuss. 1987. Nitrosubstituted aromatic compounds as nitrogen source for bacteria. Appl. Environ. Microbiol. 53:208-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Canellakis, Z. N., and P. P. Cohen. 1956. Purification studies of tyrosine-alpha-ketoglutaric acid transaminase. J. Biol. Chem. 222:53-62. [PubMed] [Google Scholar]
- 6.Davis, K. L., E. Martin, I. V. Turko, and F. Murad. 2001. Novel effects of nitric oxide. Annu. Rev. Pharmacol. Toxicol. 41:203-236. [DOI] [PubMed] [Google Scholar]
- 7.Franze, T., M. G. Weller, R. Niessner, and U. Pöschl. 2005. Protein nitration by polluted air. Environ. Sci. Technol. 39:1673-1678. [DOI] [PubMed] [Google Scholar]
- 8.Hanafy, K. A., J. A. Krumenacker, and F. Murad. 2001. NO, nitrotyrosine, and cyclic GMP in signal transduction. Med. Sci. Monit. 7:801-819. [PubMed] [Google Scholar]
- 9.Hirota, S., U. Takahama, T. N. Ly, and R. Yamauchi. 2005. Quercetin-dependent inhibition of nitration induced by peroxidase/H2O2/nitrite systems in human saliva and characterization of an oxidation product of quercetin formed during the inhibition. J. Agric. Food Chem. 53:3265-3272. [DOI] [PubMed] [Google Scholar]
- 10.Irie, Y., M. Saeki, Y. Kamisaki, E. Martin, and F. Murad. 2003. Histone H1.2 is a substrate for denitrase, an activity that reduces nitrotyrosine immunoreactivity in proteins. Proc. Natl. Acad. Sci. USA 100:5634-5639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ischiropoulos, H. 2003. Biological selectivity and functional aspects of protein tyrosine nitration. Biochem. Biophys. Res. Commun. 305:776-783. [DOI] [PubMed] [Google Scholar]
- 12.Jaenicke, R. 2005. Abundance of cellular material and proteins in the atmosphere. Science 308:73. [DOI] [PubMed] [Google Scholar]
- 13.Jamaluddin, M. P. 1977. Purification and properties of homoprotocatechuate 2,3-dioxygenase from Bacillus stearothermophilus. J. Bacteriol. 129:690-697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kers, J. A., M. J. Wach, S. B. Krasnoff, J. Widom, K. D. Cameron, R. A. Bukhalid, D. M. Gibson, B. R. Crane, and R. Loria. 2004. Nitration of a peptide phytotoxin by bacterial nitric oxide synthase. Nature 429:79-82. [DOI] [PubMed] [Google Scholar]
- 15.Mani, A. R., A. S. Pannala, N. N. Orie, R. Ollosson, D. Harry, C. A. Rice-Evans, and K. P. Moore. 2003. Nitration of endogenous para-hydroxyphenylacetic acid and the metabolism of nitrotyrosine. Biochem. J. 374:521-527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monteiro, H. P. 2002. Signal transduction by protein tyrosine nitration: competition or cooperation with tyrosine phosphorylation-dependent signaling events? Free Radic. Biol. Med. 33:765-773. [DOI] [PubMed] [Google Scholar]
- 17.Nathan, C. 2004. The moving frontier in nitric oxide-dependent signaling. Sci. STKE 2004:pe52. [DOI] [PubMed] [Google Scholar]
- 18.Nishino, S. F., G. Paoli, and J. C. Spain. 2000. Aerobic degradation of dinitrotoluenes and pathway for bacterial degradation of 2,6-dinitrotoluene. Appl. Environ. Microbiol. 66:2139-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Nishino, S. F., and J. C. Spain. 2004. Catabolism of nitroaromatic compounds, p. 575-608. In J.-L. Ramos (ed.), Pseudomonas, vol. 3. Kluwer Academic/Plenum Publishers, New York, N.Y. [Google Scholar]
- 20.Ohshima, H., M. Friesen, I. Brouet, and H. Bartsch. 1990. Nitrotyrosine as a new marker for endogenous nitrosation and nitration of proteins. Food Chem. Toxicol. 28:647-652. [DOI] [PubMed] [Google Scholar]
- 21.Palumbo, A., A. Napolitano, P. Barone, and M. d'Ischia. 1999. Nitrite- and peroxide-dependent oxidation pathways of dopamine: 6-nitrodopamine and 6-hydroxydopamine formation as potential contributory mechanisms of oxidative stress- and nitric oxide-induced neurotoxicity in neuronal degeneration. Chem. Res. Toxicol. 12:1213-1222. [DOI] [PubMed] [Google Scholar]
- 22.Radi, R. 2004. Nitric oxide, oxidants, and protein tyrosine nitration. Proc. Natl. Acad. Sci. USA 101:4003-4008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shintani, F., T. Kinoshita, S. Kanba, T. Ishikawa, E. Suzuki, N. Sasakawa, R. Kato, M. Asai, and T. Nakaki. 1996. Bioactive 6-nitronorepinephrine identified in mammalian brain. J. Biol. Chem. 271:13561-13565. [DOI] [PubMed] [Google Scholar]
- 24.Smibert, R. M., and N. R. Krieg. 1994. Phenotypic characterization, p.607-654. In P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg(ed.), Methods for general and molecular bacteriology. ASM Press, Washington, D.C.
- 25.Stanier, R. Y., N. J. Palleroni, and M. Doudoroff. 1966. The aerobic pseudomonads: a taxonomic study. J. Gen. Microbiol. 43:159-271. [DOI] [PubMed] [Google Scholar]
- 26.Tabrizi-Fard, M. A., T. S. Maurer, and H.-L. Fung. 1999. In vivo disposition of 3-nitro-l-tyrosine in rats: implications on tracking systemic peroxynitrite exposure. Drug Metab. Dispos. 27:429-431. [PubMed] [Google Scholar]
- 27.Vaillancourt, F. H., G. Labbe, N. M. Drouin, P. D. Fortin, and L. D. Eltis. 2002. The mechanism-based inactivation of 2,3-dihydroxybiphenyl 1,2-dioxygenase by catecholic substrates. J. Biol. Chem. 277:2019-2027. [DOI] [PubMed] [Google Scholar]
- 28.Vaillancourt, F. H., J. T. Bolin, and L. D. Eltis. 2004. Ring-cleavage dioxygenases, p. 359-395. In J.-L. Ramos (ed.), Pseudomonas, vol. 3. Kluwer Academic/Plenum Publishers, New York, N.Y. [Google Scholar]
- 29.van der Vliet, A., J. P. Eiserich, B. Halliwell, and C. E. Cross. 1997. Formation of reactive nitrogen species during peroxidase-catalyzed oxidation of nitrite. A potential additional mechanism of nitric oxide-dependent toxicity. J. Biol. Chem. 272:7617-7625. [DOI] [PubMed] [Google Scholar]
- 30.Verdon, C. P., B. A. Burton, and R. L. Prior. 1995. Sample pretreatment with nitrate reductase and glucose-6-phosphate dehydrogenase quantitatively reduces nitrate while avoiding interference by NADP+ when the Griess reaction is used to assay for nitrite. Anal. Biochem. 224:502-508. [DOI] [PubMed] [Google Scholar]
- 31.Wendehenne, D., A. Pugin, D. F. Klessig, and J. Durner. 2001. Nitric oxide: comparative synthesis and signaling in animal and plant cells. Trends Plant Sci. 6:177-183. [DOI] [PubMed] [Google Scholar]
- 32.Zeyer, J., and H. P. Kocher. 1988. Purification and characterization of a bacterial nitrophenol oxygenase which converts ortho-nitrophenol to catechol and nitrite. J. Bacteriol. 170:1789-1794. [DOI] [PMC free article] [PubMed] [Google Scholar]