Abstract
Here we report that Caenorhabditis elegans nematodes fed Listeria monocytogenes die over the course of several days, as a consequence of an accumulation of bacteria in the worm intestine. Mutant strains previously shown to be important for virulence in mammalian models were also found to be attenuated in their virulence in C. elegans. However, ActA, which is required for actin-based intracellular motility, appears to be dispensable during infection of C. elegans, indicating that L. monocytogenes remains extracellular in C. elegans.
There is a continuing need for the development of simple animal models for the study of host-pathogen interactions. The implementation of nonmammalian model organisms such as Drosophila melanogaster and Caenorhabditis elegans has recently proven successful, thereby reducing both cost and the ethical constraints associated with virulence studies using mammalian hosts. The realization that innate immunity plays a significant role in resistance to bacterial infections has further supported the use of nonmammalian model organisms which display an innate immune response but lack the adaptive immune system (6).
Listeria monocytogenes kills C. elegans.
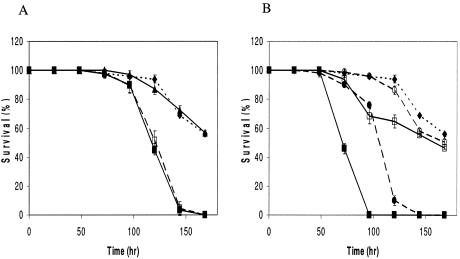
Listeria monocytogenes is a gram-positive facultative intracellular bacterial pathogen that gives rise to serious invasive infections in humans and animals. With the aim of investigating whether Listeria is also a pathogen of C. elegans, we examined the killing of the nematode when it was fed various L. monocytogenes strains. While the wild-type N2 worms were feeding on Listeria, locomotion was progressively decreased and the number of eggs laid was decreased when compared to OP50 (data not shown). In addition, we observed that the worms became laden with eggs, which in some cases hatched internally, a phenotype called “bag of worms” (data not shown). Bagging is suggested to be caused by a weakening of the infected worm, rendering it unable to lay eggs normally (12). Since killing assays extend over the generation time of C. elegans, new progeny may interfere with the counting of surviving nematodes. Therefore, to avoid new progeny and the bagging phenotype, we used the temperature-sensitive sterile C. elegans pha-1(e2123ts), which grows normally at 15°C but at 25°C is embryonic lethal (11). Mutation in the pha-1 gene does not affect the immune function of C. elegans (data not shown). Individual bacterial strains were grown overnight in either Luria broth (LB) or brain heart infusion broth at 37°C. Ten microliters of overnight culture was spread on 3.5-cm LB agar plates and grown for 16 h at 37°C. Thirty L4 hermaphrodites were transferred from nematode growth medium plates seeded with OP50 to each 3.5-cm LB agar plate seeded with the various strains, and plates were incubated at 25°C and scored for live or dead worms every 24 h. Each assay was carried out in triplicate and repeated three times. We examined the ability of several well-characterized L. monocytogenes strains to kill C. elegans. The wild-type strains EGDe, EGD (BUG600), and 10403S are known to be virulent in a mouse model (2, 3, 8), and we found that they also kill C. elegans much faster compared to the avirulent Escherichia coli OP50 and Listeria innocua (Fig. 1). Furthermore, when nematodes were fed heat-killed L. monocytogenes EGDe or BUG600, no killing was observed and we saw a life span comparable to that of worms fed OP50 (results not shown), indicating the need for direct association between viable L. monocytogenes and C. elegans for killing.
FIG. 1.
Survival of C. elegans pha-1(e2123ts) fed on E. coli and Listeria. (A) Worms fed E. coli OP50 (dotted lines, diamonds), Listeria innocua (triangles), L. monocytogenes 10403S (solid squares), or L. monocytogenes 10403S ΔactA (dashed lines, open squares). (B) Worms fed E. coli OP50 (dotted lines, diamonds), L. monocytogenes EGDe (dashed lines, solid circles), L. monocytogenes EGDe ΔprfA (dashed lines, open circles), L. monocytogenes EGD (BUG600) (solid squares), or L. monocytogenes EGD (BUG600) ΔdegU (open squares).
Worms fed L. monocytogenes accumulate bacteria within their digestive tracts.
Two distinct modes of bacterium-mediated killing of C. elegans have been identified. Certain Pseudomonas aeruginosa strains can produce low-weight molecular toxins that can kill the nematode within hours, known as “fast killing” (13). In contrast, when the killing occurs over the course of several days, it is referred to as “slow killing” and correlates with the accumulation of bacteria in the nematode gut (13). To follow the fate of L. monocytogenes, we fed C. elegans with L. monocytogenes EGDe that expresses green fluorescent protein (GFP) (5), and, after 72 h, we observed a distension of the intestinal lumen with a large number of intact bacteria (data not shown). This distension was not observed when worms were fed OP50 expressing GFP (data not shown). Similar results have previously been found when C. elegans was fed GFP-expressing Salmonella enterica serovar Typhimurium for 72 h (1).
C. elegans can be used to identify L. monocytogenes virulence factors.
Several bacterial pathogens, both gram positive and gram negative, kill C. elegans when supplied as a food source, and a variety of bacterial virulence factors, identified in mammalian models, have also been shown to be required for nematode pathogenesis (9, 12, 13). To evaluate whether the C. elegans model will be useful for studying and identifying Listeria virulence factors, mutant strains that exhibit a reduced virulence in mammals were examined. PrfA, a regulator of virulence gene expression, and DegU, a two-component regulator involved in motility, are both important for virulence in mice (3, 8). We investigated the effect of prfA and degU mutants and found both mutants to have reduced nematocidal activity (Fig. 1B), demonstrating that the virulence factors PrfA and DegU are required for L. monocytogenes virulence in C. elegans as well as in vertebrate hosts. In addition to PrfA L. monocytogenes utilizes several factors during its intracellular life. One of these is ActA, which mediates actin-based motility in the host cytosol and is required for the bacterium to spread to adjacent cells (4). actA mutants are known to be highly attenuated in a murine model (2). However, we found that an actA mutant had the same ability to kill the nematode as the wild type (Fig. 1A), indicating that intracellular survival and spread are not important for killing C. elegans. This observation is consistent with our observation that Listeria expressing GFP was restricted to the luminal space (data not shown). In addition, extracellular replication of L. monocytogenes is not restricted to C. elegans but has also recently been demonstrated in the murine model (7).
In conclusion, we report here the establishment of the nematode Caenorhabditis elegans as a model for analyzing the virulence of L. monocytogenes. Our results show that several genes important for full virulence in vertebrates are also required for killing C. elegans, suggesting that the nematode could be an attractive model for identifying new virulence factors in L. monocytogenes. In D. melanogaster, another invertebrate model used for L. monocytogenes (10), an actA mutant is able to replicate within the insect cells but is unable to spread to adjacent cells (10). However, ActA appears to be dispensable during the infection of the worm, and future work will reveal the genetic repertoire required for infection of C. elegans by L. monocytogenes.
Acknowledgments
We thank D. A. Portnoy, G. Knudsen, L. Dons, N. Fortinea, and W. Goebel for generous gifts of strains and plasmids. The E. coli and C. elegans strains used in this work were provided by the Caenorhabditis Genetics Center (University of Minnesota, Minneapolis).
This work was supported by a grant from the Danish Technical Research Council and a fellowship from the Howard Hughes Medical Institute.
REFERENCES
- 1.Aballay, A., P. Yorgey, and F. M. Ausubel. 2000. Salmonella typhimurium proliferates and establishes a persistent infection in the intestine of Caenorhabditis elegans. Curr. Biol. 10:1539-1542. [DOI] [PubMed] [Google Scholar]
- 2.Brundage, R. A., G. A. Smith, A. Camilli, J. A. Theriot, and D. A. Portnoy. 1993. Expression and phosphorylation of the Listeria monocytogenes ActA protein in mammalian cells. Proc. Natl. Acad. Sci. USA 90:11890-11894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chakraborty, T., M. Leimeister-Wachter, E. Domann, M. Hartl, W. Goebel, T. Nichterlein, and S. Notermans. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cossart, P., J. Pizarro-Cerda, and M. Lecuit. 2003. Invasion of mammalian cells by Listeria monocytogenes: functional mimicry to subvert cellular functions. Trends Cell Biol. 13:23-31. [DOI] [PubMed] [Google Scholar]
- 5.Fortinea, N., P. Trieu-Cuot, O. Gaillot, E. Pellegrini, P. Berche, and J. L. Gaillard. 2000. Optimization of green fluorescent protein expression vectors for in vitro and in vivo detection of Listeria monocytogenes. Res. Microbiol. 151:353-360. [DOI] [PubMed] [Google Scholar]
- 6.Gravato-Nobre, M. J., and J. Hodgkin. 2005. Caenorhabditis elegans as a model for innate immunity to pathogens. Cell. Microbiol. 7:741-751. [DOI] [PubMed] [Google Scholar]
- 7.Hardy, J., K. P. Francis, M. DeBoer, P. Chu, K. Gibbs, and C. H. Contag. 2004. Extracellular replication of Listeria monocytogenes in the murine gall bladder. Science 303:851-853. [DOI] [PubMed] [Google Scholar]
- 8.Knudsen, G. M., J. E. Olsen, and L. Dons. 2004. Characterization of DegU, a response regulator in Listeria monocytogenes, involved in regulation of motility and contributes to virulence. FEMS Microbiol. Lett. 240:171-179. [DOI] [PubMed] [Google Scholar]
- 9.Labrousse, A., S. Chauvet, C. Couillault, C. L. Kurz, and J. J. Ewbank. 2000. Caenorhabditis elegans is a model host for Salmonella typhimurium. Curr. Biol. 10:1543-1545. [DOI] [PubMed] [Google Scholar]
- 10.Mansfield, B. E., M. S. Dionne, D. S. Schneider, and N. E. Freitag. 2003. Exploration of host-pathogen interactions using Listeria monocytogenes and Drosophila melanogaster. Cell. Microbiol. 5:901-911. [DOI] [PubMed] [Google Scholar]
- 11.Schnabel, H., and R. Schnabel. 1990. An organ-specific differentiation gene, pha-1, from Caenorhabditis elegans. Science 250:686-688. [DOI] [PubMed] [Google Scholar]
- 12.Sifri, C. D., J. Begun, F. M. Ausubel, and S. B. Calderwood. 2003. Caenorhabditis elegans as a model host for Staphylococcus aureus pathogenesis. Infect. Immun. 71:2208-2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan, M. W., S. Mahajan-Miklos, and F. M. Ausubel. 1999. Killing of Caenorhabditis elegans by Pseudomonas aeruginosa used to model mammalian bacterial pathogenesis. Proc. Natl. Acad. Sci. USA 96:715-720. [DOI] [PMC free article] [PubMed] [Google Scholar]