Abstract
Freezing and lyophilization are common methods used for preservation and storage of microorganisms during the production of concentrated starter cultures destined for industrial fermentations or product formulations. The compatible solute trehalose has been widely reported to protect bacterial, yeast and animal cells against a variety of environmental stresses, particularly freezing and dehydration. Analysis of the Lactobacillus acidophilus NCFM genome revealed a putative trehalose utilization locus consisting of a transcriptional regulator, treR; a trehalose phosphoenolpyruvate transferase system (PTS) transporter, treB; and a trehalose-6-phosphate hydrolase, treC. The objective of this study was to characterize the tre locus in L. acidophilus and determine whether or not intracellular uptake of trehalose contributes to cryoprotection. Cells subjected to repeated freezing and thawing cycles were monitored for survival in the presence of various concentrations of trehalose. At 20% trehalose a 2-log increase in survival was observed. The trehalose PTS transporter and trehalose hydrolase were disrupted by targeted plasmid insertions. The resulting mutants were unable to grow on trehalose, indicating that both trehalose transport into the cell via a PTS and hydrolysis via a trehalose-6-phosphate hydrolase were necessary for trehalose fermentation. Trehalose uptake was found to be significantly reduced in the transporter mutant but unaffected in the hydrolase mutant. Additionally, the cryoprotective effect of trehalose was reduced in these mutants, suggesting that intracellular transport and hydrolysis contribute significantly to cryoprotection.
Freezing and freeze-drying are methods commonly used for preservation and storage of microorganisms and for production of concentrated starter cultures used in industrial fermentations. The sensitivity of microorganisms to cryogenic stress results in significant cell injury and death, ultimately reducing productivity or activity. The mechanisms by which freezing causes cell damage include efflux of water from the cell, resulting in precipitation of cytoplasmic solutes and components (25), mechanical stress to cellular components, and rupture of cell membranes due to ice crystal formation (26, 36, 37). Cryoprotectants are protective compounds added to cell suspensions, prior to low-temperature exposure, to reduce these deleterious effects.
Trehalose (α-d-glucopyranosyl-α-d-glycopyranoside) is a disaccharide found naturally in a variety of organisms including both prokaryotes and eukaryotes (3). Trehalose has been shown to act in a number of important physiological roles including serving as a primary carbon source, and as a compatible solute accumulated in response to heat shock (4) and osmotic stress (11). Trehalose is an effective cryoprotectant that has been demonstrated to protect different cell types from freezing and desiccation, including bacteria (23), yeasts (10), and human cells (14, 27). Trehalose is thought to confer cryoprotection by mimicking the hydrogen-bonding properties of water (32) and preventing the formation of large crystals and reducing or delaying the onset of a phase shift of the plasma membrane from a liquid-crystal state to a gel state (9, 30). As a result, trehalose promotes stress tolerance, in particular conferring cryoprotection to cells upon freezing. Despite widespread interest in cryoprotection, there is limited information available regarding the molecular mechanisms responsible for cryoprotection by the compatible solute trehalose.
The lactic acid bacteria are important organisms widely used as starter cultures in the food and dairy industries. One particular class of lactic acid bacteria, probiotics, are microbes used as health-promoting functional food ingredients thought to have a beneficial effect on health via interactions in the gastrointestinal tract. One such organism, Lactobacillus acidophilus, has been shown to benefit from cryoprotection by trehalose (8). The genome sequence of L. acidophilus NCFM was recently determined (1). The genome sequence provided insights as to its probiotic functionalities and revealed a region that was implicated in the potential utilization of trehalose.
This study investigated the genetic and biochemical basis for cryoprotection by trehalose in L. acidophilus NCFM. The tre locus was characterized in detail and the involvement of the trehalose phosphoenolpyruvate transferase system (PTS) transporter and hydrolase in cryoprotection was investigated.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. L. acidophilus strains were propagated statically at 37°C in MRS broth (Difco, Detroit, MI), or on MRS supplemented with 1.5% agar (Difco). For growth and cryoprotection studies, lactobacilli were grown in a semisynthetic medium (SSM) described previously (7). The carbohydrates added to the SSM included glucose (Fisher, Fair Lawn, NJ), fructose (Sigma, St. Louis, MO), sucrose (Sigma), and trehalose (Sigma). When appropriate, chloramphenicol (Fisher) and/or erythromycin (Sigma) was added at a concentration of 5 μg/ml.
TABLE 1.
Bacterial strains and plasmids
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| L. acidophilus | ||
| NCK56 | NCFM, human intestinal isolate | 6 |
| NCK1392 | NCFM with pTRK669 | 31 |
| NCK1398 | NCFM lacL::pTRK670, Lac− | 31 |
| NCK1624 | NCFM treB::pTRK801, Tre− | This study |
| NCK1626 | NCFM msmE::pTRK802, FOS | 7 |
| NCK1725 | NCFM treC::pTRK836, Tre− | This study |
| E. coli | ||
| EC 1000 | RepA+ MC1000, Kmr, carrying a single copy of the pWVO1 repA gene in the glgB gene; host for pORI28-based plasmids | 31 |
| NCK1609 | EC1000(pORI28) | 31 |
| NCK1623 | EC1000(pTRK801) | This study |
| NCK1724 | EC1000(pTRK836) | This study |
| Plasmids | ||
| pORI28 | Emr, ori (pWV01), replicates only with repA provided in trans | 31 |
| pTRK669 | ori (pWV01), Cmr, temperature sensitive, provides repA in trans | 31 |
| pTRK801 | 2.4 kb, pORI28 with 776-bp internal L. acidophilus NCFM treB fragment | This study |
| pTRK836 | 2.4 kb, pORI28 with 739-bp internal L. acidophilus NCFM treC fragment | This study |
Escherichia coli cells were propagated at 37°C in Luria-Bertani (LB) broth (Difco) with shaking or on 1.5% LB agar plates. When appropriate, kanamycin (Sigma) was added at a concentration of 40 μg/ml and erythromycin was added at a concentration of 150 μg/ml. E. coli transformants were selected on brain heart infusion agar (Difco) supplemented with 150 μg/ml erythromycin and maintained afterwards in LB broth supplemented with 150 μg/ml erythromycin.
Comparative genomics.
Several microbial gene clusters containing EC 3.2.1.93 orthologs were selected from public databases with the gene clusters aligned and centered on EC 3.2.1.93.
DNA isolation, manipulations, and transformations.
L. acidophilus genomic DNA was isolated according to the method of Walker and Klaenhammer (39). E. coli plasmid DNA was isolated using the QIAprep Spin Miniprep (QIAGEN Inc., Valencia, CA) kit.
All DNA manipulations were performed according to standard procedures. Restriction enzymes, T4 ligase, and Taq DNA polymerase were purchased from Roche Molecular Biochemicals (Indianapolis, IN). DNA fragments were purified from agarose gels using the GeneClean kit (Qbiogene Inc, Carlsbad, CA). All PCRs were performed according to standard procedures using Taq polymerase. PCR primers were synthesized by IDT (Coralville, IA) with restriction sites designed at the 5′ end of the primers to facilitate cloning. PCR products were purified using the QIAquick PCR purification kit (QIAGEN).
Chemically competent E. coli EC1000 cells were prepared using the Z-competent E. coli transformation buffer set (Zymo Research, Orange, CA). Electrocompetent L. acidophilus NCFM cells were prepared using 3.5× sucrose MgCl electroporation buffer as described by Luchansky et al. (24). Electrotransformation was performed using a Gene-Pulser (Bio-Rad, Hercules, CA).
Survival analysis.
An 18-hour culture of L. acidophilus was inoculated at 1% (vol/vol) into 10 ml of SSM with 1% fructose and grown to an optical density at 600 nm (OD600) of 0.5 to 0.6, corresponding to the mid-exponential growth phase. Cells were harvested by centrifugation (3,220 × g, 15 min, 4°C) and resuspended in various concentrations of trehalose (0 to 30%, wt/vol) in Butterfield's buffer yielding a fourfold concentration of cells. The cells were incubated at 37°C for 1 hour, and subsequently subjected to repeated cycles of freezing at −70°C in a dry ice and ethanol bath for 5 min and thawing at 20°C in a water bath for 10 min. Cell survival was monitored by CFU enumerations plated on MRS agar with a Whitley automatic spiral plater (Don Whitley Scientific Limited, West Yorkshire, England) and counted using a ProtoCOL colony counter (Synbiosis, Frederick, MD).
Gene inactivations.
The inactivation of treB and treC was carried out by targeted insertion of an erythromycin resistance cassette via homologous recombination according to the method of Russell and Klaenhammer (31). Internal fragments for each gene targeted for inactivation were amplified by PCR using L. acidophilus NCFM chromosomal DNA as the template. The primers were designed using Clone Manager 7 (Scientific and Educational Software, Cary, NC) with restriction enzyme sites to facilitate cloning and are listed in Table 2.
TABLE 2.
Primers
| ORF | Gene | Primers (orientation) | Sequence (5′→3′) |
|---|---|---|---|
| La1012 | treB | LA1012Eco(F) | GATCGAATTCCGTTATAGCTCATGCATACC |
| LA1012Bam(R) | GATCGGATCCTTAAGCCAATCTTCGCATTT | ||
| La1014 | treC | La1014Eco(F) | GATCGAATTCCGGTGGTACTGCTTGG |
| La1014Bam(R) | GATCGGATCCGGCCTCAACATCTACG |
Plasmids pTRK801 and pTRK836 were created by ligating these products into pORI28 using the BamHI and EcoRI sites and transformed into E. coli EC1000. Clones containing pTRK801 and pTRK836 were identified as E. coli NCK1623 and E. coli NCK1725, respectively. Plasmids pTRK801 and pTRK836 were propagated in E. coli and transformed by electroporation into L. acidophilus NCK1392, harboring the temperature-sensitive helper plasmid pTRK669. Selection for clones harboring an insertion of pTRK801 and pTRK836 into the chromosome was performed at 43°C, a temperature nonpermissive to the pORI28 replicon and under selection by erythromycin.
Southern hybridization.
Southern hybridization of genomic DNA was performed according to standard protocols (5). EcoRI-digested chromosomal DNA was blotted onto a MagnaGraph nylon membrane (Osmonics, Inc., Westborough, MA) and probed using the treB and treC internal fragments labeled with [α-32P]ATP using the Strip-EZ DNA kit (Ambion, Inc., Austin, TX).
Trehalose uptake.
Cultures of L. acidophilus were grown to OD600 of 0.5 to 0.6 in SSM plus 1% fructose and harvested by centrifugation (3,220 × g, 15 min, 4°C). Pellets were washed three times with 50 mM potassium phosphate buffer containing 5 mM MgCl2 (pH 6.5) and resuspended in the same buffer yielding a fourfold concentration in cells. Cells were incubated for 1 h at 37°C and transport assays were started by the addition of [14C]trehalose (0.600 mCi/mmol) (American Radiolabeled Chemicals, Inc., St. Louis, MO) to cell suspensions to give a final trehalose concentration of 150 μM. After a 1-h incubation at 37°C, cells were centrifuged through silicon oil (12,000 × g, 15 min) (17). The radioactivity in the supernatant and in the cell pellets was determined using a Beckman LS 3801 liquid scintillation counter (Beckman Instruments, Fullerton, CA).
Nucleotide sequence accession number.
The DNA sequences discussed are included in the complete genome sequence of L. acidophilus submitted to GenBank and assigned accession number CP000033 (1).
RESULTS
Trehalose cryoprotection of L. acidophilus.
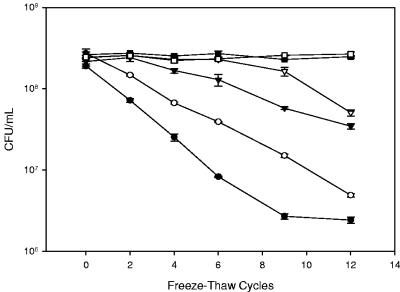
When subjected to successive cycles of freezing and thawing, L. acidophilus cells frozen in various concentrations of trehalose had significantly higher survival rates than cells frozen in Butterfield's buffer alone (Fig. 1). Over the course of 12 successive freeze-thaw cycles, cells frozen in buffer alone experienced a 99% reduction in CFU. In contrast, cells frozen in the presence of trehalose displayed increased survival rates with increasing amounts of trehalose, ranging from 2.5 to 30%. The increase in survival was incremental over the range of 2.5 to 20% trehalose. However, addition of trehalose beyond 20% did not significantly improve survival further. Cells frozen in 20 and 30% trehalose displayed survival rates of greater than 94% after 12 cycles of freezing and thawing.
FIG. 1.
Dose response to trehalose cryoprotection in L. acidophilus NCFM. Cells were grown to an OD600 of 0.5 to 0.6 in SSM plus 1% fructose. Cells were harvested by centrifugation and resuspended in various concentrations of trehalose in Butterfield's buffer and subjected to repeated freeze-thaw cycles (−70°C, 5 min; 20°C, 10 min). Cell survival was monitored by CFU enumeration on MRS agar. The error bars represent 1 standard deviation of duplicate measurements. Symbols represent trehalose concentrations of (•) 0%, (○) 2.5%, (▾) 5%, (▿) 10%, (▪) 20%, and (□) 30%.
Analysis of the tre locus.
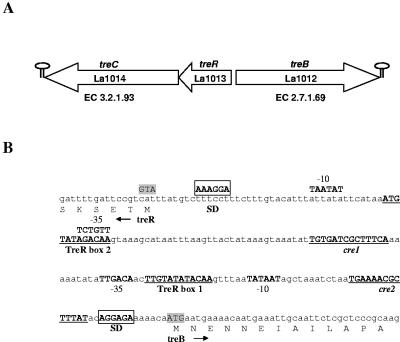
Analysis of the L. acidophilus NCFM genome sequence revealed the presence of a tre locus (1) encoding three putative open reading frames, La1012, La1013, and La1014 (Fig. 2A). Because the encoded proteins were homologous to those present in E. coli (15, 22, 29), a similar nomenclature was used. Analysis of the predicted open reading frames (ORFs) suggested the presence of the following: a trehalose-specific EIIABC (EIItre) of the phosphoenolpyruvate transferase system family of transporters EC 2.7.1.69, treB (La1012); a transcriptional repressor from the GntR family, treR (La1013); and a trehalose-6-phosphate hydrolase, EC 3.2.1.93, treC (La1014).
FIG. 2.
L. acidophilus tre locus. (A) White arrows indicate the treC, treR, and treB genes. (B) Nucleotide sequence of treR-treB intergenic region. Putative −35 and −10 regions are in bold, putative cre element and TreR boxes are underlined, the putative ribosome binding sites (SD) are boxed, and putative translation start sites are in shaded text.
The transcriptional regulator contained two distinct domains: an N-terminal helix-turn-helix containing the DNA binding domain of the GntR family (PFam 00392) and a UTRA ligand-binding domain (PFam 07702) at the C terminus. The transcriptional regulator was found to share 47% identity with a previously described repressor of the trehalose operon in Bacillus subtilis (35). The transcriptional regulator was also found to have high homology to the transcriptional regulators in tre loci from Lactobacillus johnsonii (28) and Lactobacillus plantarum (21). The putative trehalose transporter was found to consist of components typical of PTS transporters, namely the EIIA (PFam 00358), EIIB (PFam 00367), and EIIC (PFam 00367) domains. The putative trehalose hydrolase was found to contain an alpha-amylase catalytic domain (Pfam 00128) from family 13 of the glycosylhydrolases at the N terminus and to share 47% identity with a trehalose-6-phosphate hydrolase described previously in B. subtilis (34).
Analysis of the treB-treR intergenic region revealed the presence of putative promoter elements for both parts of the divergent operon, namely treB and treRC. Putative Shine-Dalgarno sequences were identified 12 bp upstream of treB (5′AGGAGA3′) and 13 bp upstream of treR (5′AGGAAA3′). Analysis of the intergenic region also revealed the presence of two cre-like sequences: cre1 (5′TGAAAACGCTTTAT3′) and cre2 (5′TGTGATCGCTTTCA3′) (7). Additionally two putative binding sites for TreR were also identified, treR1 (5′TTGTATATACAA3′) and treR2 (5′ATGTATAGACAA3′) (7, 35). Dyad symmetry analysis using Clone Manager suggested the presence of two stem loop structures that could act as rho-independent transcriptional terminators: one 24 bp downstream of treB with a free energy of −17.8 kcal mol−1, and the other 124 bp downstream of treC with a free energy of −16.3 kcal mol−1 (Fig. 2B).
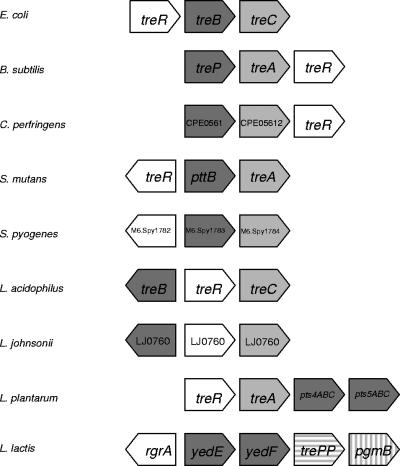
Comparative genomic analysis of the tre clusters from the selected genomes shows consistent cooccurrence of genes (Fig. 3). Primarily, this locus consisted of three or four elements: a transcriptional regulator, a trehalose-6-phosphate hydrolase (EC 3.2.1.93) and PTS transporter elements (EC 2.7.1.69). Alternatively, in addition to the regulator and PTS transporter, the Lactococcus lactis trehalose operon was found to encode a trehalose phosphate phosphorylase (EC 2.4.1.216) and a phosphoglucomutase (EC 5.4.2.6).
FIG. 3.
Comparative operon alignment. Alignment of the tre loci from Streptococcus mutans, Streptococcus pyogenes, L. acidophilus, L. johnsonii, L. plantarum, E. coli, B. subtilis, Clostridium perfringens, and Lactococcus lactis. Transcriptional regulators, white; PTS transporters, dark gray; trehalose-6-phosphate hydrolase, light gray; trehalose phosphate phosphorylase, horizontal hatching; and phosphoglucomutase, vertical hatching.
Insertion into treB and treC.
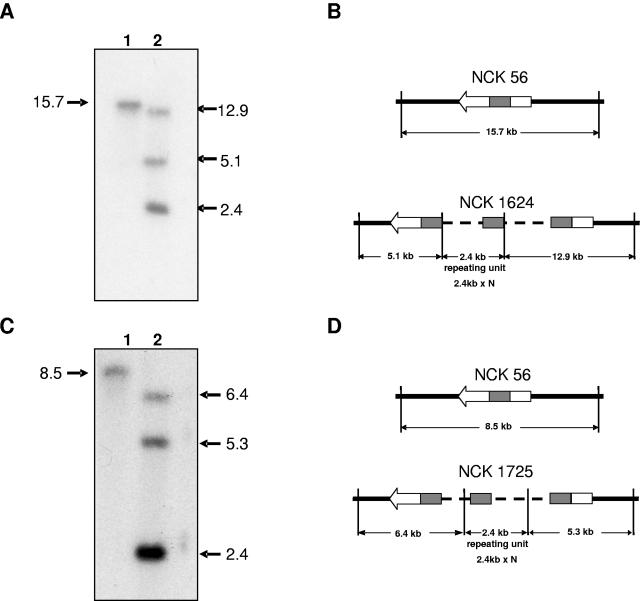
Insertional inactivation of treB and treC was performed and confirmed by Southern hybridization (Fig. 4) and PCR (data not shown). The treB probe hybridized to an EcoRI fragment of 15.7 kb in the wild-type L. acidophilus NCFM. In contrast, in the mutant strain L. acidophilus TreB− (NCK1624), this band was replaced by two junction fragments of 12.9 kb and 5.1 kb, indicating the integration of pTRK801 in treB. Similarly, the treC probe hybridized to an 8.5-kb EcoRV fragment in the wild type, whereas in the treC mutant (NCK1725), this band was replaced by two junction fragments of 6.5 and 5.3 kb. Also, an additional 2.4-kb band was observed in both knockouts, indicating amplification of the plasmid within the targeted loci of each mutant.
FIG. 4.
Confirmation of integrants. Southern hybridization analysis of NCK56, NCK1624, and NCK1725. (A) Genomic DNA was digested with EcoRI and probed with the internal treB fragment. Lane 1, NCK56; lane 2 NCK1624. (B) Schematic of treB locus in NCK56 and NCK1624. (C) Genomic DNA was digested with EcoRV and probed with treC internal fragment. Lane 1, NCK56; lane 2, NCK1725. (D) Schematic of treC locus in NCK56 and NCK1624. Chromosomal DNA is marked by a thick line, plasmid DNA by a dotted line, the tre genes by an arrow with internal tre fragments marked by a shaded box, and vertical lines indicate restriction sites.
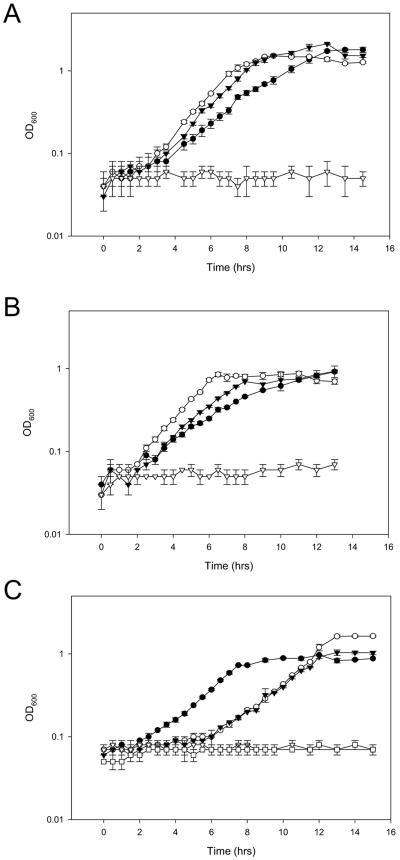
The tre mutants were evaluated for their ability to grow on various carbohydrates (Fig. 5A, B and C). Although the mutants retained the ability to grow on fructose, glucose and sucrose, their ability to grow on trehalose was completely abolished. Additionally, the inability of the mutants to grow on trehalose compared to other insertional mutants, namely lac (β-galactosidase) and msmE (FOS ABC transporter) mutants, indicated that inactivation of either TreB and TreC is responsible for the loss of the ability to ferment trehalose (Fig. 4C). The other mutants retained the ability to grow on trehalose, reaching a final OD600 similar to that of the wild type. A period of lag was observed in the growth of the lacL and msmE mutants compared to the wild type when grown on trehalose (Fig. 5). This lag was attributed to the presence of antibiotic to the medium in order to maintain the insertion within those respective loci. Inactivation of either the trehalose transporter or the hydrolase resulted in the inability to grow on trehalose.
FIG. 5.
Growth on trehalose and other carbohydrates. Cultures were propagated overnight in MRS broth. Cells were washed with SSM and inoculated at 1% into SSM plus 1% carbohydrate. Error bars represent 1 standard deviation in triplicate measurements. (A) treB and (B) treC knockouts were grown on (•) fructose, (○) glucose, (▾) sucrose, and (▿) trehalose. (C) Wild-type (•) NCFM, (○) lacL, (▾) msmE, (▿) treB, and (□) treC strains were grown on trehalose. Error bars represent 1 standard deviation of triplicate measurements.
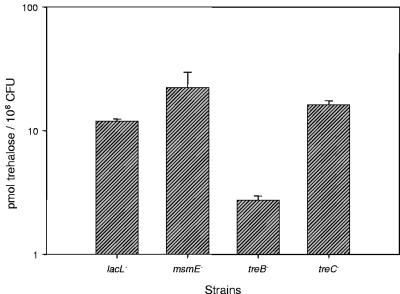
The transport of [14C]trehalose was measured to determine the ability of the L. acidophilus NCFM mutants to internalize trehalose (Fig. 6). The treB mutant was found to have a significantly diminished capacity for trehalose transport (2.7 pmol trehalose/108 CFU) in comparison to the lacL (11.9 pmol trehalose/108 CFU), msmE (22.4 pmol trehalose/108 CFU), and treC (16.2 pmol trehalose/108 CFU) mutants. The treC mutant transported trehalose at levels comparable to those of the msmE and lacL control strains.
FIG. 6.
Trehalose transport. Cells were grown to an OD600 of 0.5 to 0.6 in SSM plus 1% fructose and were harvested by centrifugation. Cells were washed three times, concentrated fourfold in 50 mM potassium phosphate buffer with 5 mM MgCl2, and incubated for 1 h at 37°C. [14C]trehalose was added at 150 uM and incubated for 1 h. Cells were centrifuged through silicon oil (12,000 × g, 15 min). Radioactivity in the cell pellet was measured by liquid scintillation. Error bars represent 1 standard deviation of triplicate measurements.
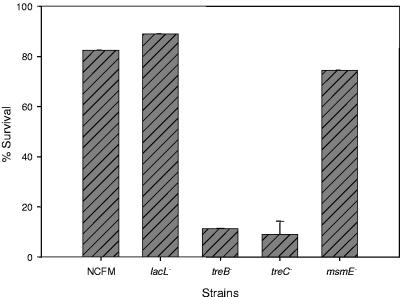
With the treB and treC mutants we were able to investigate whether intracellular trehalose contributed to cryoprotection of L. acidophilus (Fig. 7). In particular, with the treB mutant, trehalose transport was significantly lowered. Presumably, with the treC mutant intracellular hydrolysis of trehalose would be halted. Because survival of the wild-type parent was optimized in 20% trehalose (Fig. 1), survival of the tre mutants was evaluated at this concentration. The results showed that the survival was 74, 82, and 89% for msmE, NCFM, and lacL, respectively, after 12 freeze-thaw cycles. In contrast, survival of the treB mutant was greatly reduced at 11%. Surprisingly, survival of the treC mutant was also greatly reduced at 9%. Therefore, these data indicate that the cryoprotective effect results primarily from internalization and the subsequent hydrolysis of trehalose. Additionally, we observed a 2-log reduction in CFU/ml after 12 cycles in the wild-type strain in the absence of trehalose. However, only a 1-log reduction in CFU/ml was observed for the treB mutant in the presence of 20% trehalose. These data indicate extracellular trehalose also contributes to cryoprotection. However, the protective effects of external trehalose were less than the protection provided when trehalose was also transported into the cell.
FIG. 7.
Control and mutant survival in 20% trehalose. Cells were grown to an OD600 of 0.5 to 0.6 in SSM plus 1% fructose. Cells were harvested by centrifugation and resuspended in 20% trehalose in Butterfield's buffer and subjected to repeated freeze-thaw cycles (−70°C, 5 min; 20°C, 10 min). Cell survival was monitored by CFU enumeration on MRS agar. The error bars represent 1 standard deviation of triplicate measurements.
We were concerned that carbohydrate exhaustion in the L. acidophilus tre mutants might contribute to cell death during freeze-thaw cycling. The survival of the lacL, treB, and treC knockout mutants after freeze-thaw cycling in 20% trehalose was compared in the presence and absence of 1% fructose (Table 3). The results showed only a marginal change in survival when fructose was present. Therefore it is unlikely that carbohydrate exhaustion in the trehalose mutants contributed significantly to cell death.
TABLE 3.
Survival of L. acidophilus mutants frozen in trehalose with or without fructose
| Mutation | % Survivala
|
|
|---|---|---|
| Without 1% fructose | With 1% fructose | |
| lacL | 100 (3) | 94 (11) |
| treB | 24 (4) | 32 (2) |
| treC | 17 (2) | 30 (1) |
The data are means for triplicate measurements. The values in parentheses are errors represented as the relative standard deviation.
DISCUSSION
Trehalose is widely used by microorganisms as a compatible solute for the regulation of osmotic pressure. For example, E. coli has been shown to synthesize trehalose de novo in response to osmotic stress (11). Although this enzymatic machinery is not encoded within the L. acidophilus NCFM genome (1), this organism may use trehalose as a compatible solute by alternative means. The presence of a PTS transporter in the tre locus indicates that osmotic internalization of trehalose may occur through translocation, possibly allowing L. acidophilus to accumulate intracellular trehalose.
L. acidophilus NCFM has the ability to utilize trehalose as a primary carbohydrate source using a PTS transporter and carbohydrate hydrolase encoded within the genome. In E. coli and B. subtilis, trehalose has been shown to be transported by a PTS, which translocates and phosphorylates trehalose via a specific EIICBTre and the EIIAGlc of the glucose-PTS releasing trehalose-6-phosphate into the cell (22, 34). The resulting trehalose-6-phosphate is then subsequently hydrolyzed into glucose and glucose-6-phosphate by a trehalose-6-phosphate hydrolase (EC 3.2.1.93); these products can be used as substrates for glycolysis (13, 29). Alternatively, in the hyperthermophilic archaeon Thermococcus litoralis and the gram-positive soil bacterium Streptomyces reticuli, trehalose transport has been shown to occur via an ATP-binding cassette (ABC) transporter (16, 33).
The presence of the trehalose PTS and trehalose-6-phosphate hydrolase in the genome of L. acidophilus NCFM suggested that trehalose is transported and metabolized in a manner similar to that of E. coli and B. subtilis. However, in L. acidophilus NCFM, trehalose appears to be transported by an EIIABCTre rather than by EIICBTre and EIIAGlc. Based on genomic content, all of the trehalose-fermenting lactic acid bacteria available in the public database appear to utilize trehalose in a similar manner. In contrast, while L. lactis internalizes trehalose via a PTS as trehalose-6-phosphate, it is hydrolyzed by trehalose phosphate phosphorylase (EC 2.4.1.216) into glucose-6-phosphate and glucose-1-phosphate (2). Glucose-1-phosphate is isomerized to glucose-6-phosphate by β-phosphoglucomutase (EC 5.4.2.6) (2).
The transcriptional regulator encoded in the tre locus of L. acidophilus NCFM shares strong identity with a repressor of the trehalose operon in B. subtilis, suggesting that the regulator represses transcription by binding to the promoter-operator region of the tre locus, likely at the putative TreR binding sites. Additionally, identification of catabolite responsive element-like sequences within the promoter-operator region suggests that transcription from the operon is also likely under control of carbon catabolite repression (7).
Compared to the wild type, the ability of the organism to grow on trehalose was abolished when the PTS transporter was inactivated. However, the ability of the mutant to grow on other carbohydrates was unaffected. The ability of the transporter mutant to accumulate trehalose was also significantly diminished when compared to the other mutants. The inability of the treB knockout to grow on trehalose and its inability to accumulate trehalose confirm the involvement of this locus in trehalose metabolism and suggest the EIITre of the PTS is the sole path of trehalose transport. Additionally, the ability of theorganism to grow on trehalose was also abolished when the trehalose-6-phosphate hydrolase is inactivated. This indicates that TreC provides the only means for trehalose hydrolysis encoded in the L. acidophilus genome.
Previous work studying cryoprotection in L. acidophilus found the organism to be robust to freezing and found trehalose contributed to the survival and long-term stability of the organism after freeze-drying (8). In the current study, subjecting cells to repeated cycles of freezing and thawing provided a reproducible system to quickly study cryoprotection by trehalose and investigate the genetic and biochemical basis for utilization of trehalose in L. acidophilus.
The survival of L. acidophilus NCFM increased concomitantly with increasing concentration of trehalose in a dose-dependent manner with survival appearing to be optimal in 20% trehalose (Fig. 1). The mechanism by which trehalose confers cryoprotection is unknown. Specifically, our objective was to determine whether internalization of trehalose was important to confer cryoprotection. Inactivation of the trehalose PTS transporter significantly reduced the ability of the organism to survive freeze and thaw stress compared to that of the wild-type strain and other mutants. These results strongly suggest that the PTS transporter is needed for trehalose to confer cryoprotection via internalization. Moreover, extracellular trehalose also contributes to cryoprotection, likely by stabilizing the cellular membrane (36, 37).
Previously Leslie et al. (23) showed that trehalose can gain access to the cytoplasm and that trehalose did confer cryoprotection. While the implications are clear, this work did not definitively show that trehalose must be internalized to confer cryoprotection. Additionally, Leslie et al. (23) stated that it is unlikely that an organism could actively transport enough trehalose for it to be an effective cryoprotectant, and that accumulation of trehalose is likely due to the membrane becoming leaky during freezing and drying. By disruption of the trehalose transporter, we were able to definitively show that trehalose must be internalized to confer cryoprotection and that trehalose can be actively transported in sufficient concentration to be effective. This is the first study in prokaryotes to show that internalization of trehalose plays an important role in cryoprotection.
Lactic acid bacteria have been shown to accumulate compatible solutes in response to osmotic stress (12). In particular, accumulation of the compatible solute betaine in response to salt stress has been shown to enhance survivability of several Lactobacillus species to drying (18). Additionally, the adaptive response of L. acidophilus after exposure to sublethal levels of osmotic stress has been shown to confer increased resistance or cross-protection to other stresses such as heat and bile (20). Preconditioning L. acidophilus using osmotic stress may increase compatible solute accumulation and promote further cryoprotection to the organism. Similar genetic organization of tre loci in other lactic acid bacteria suggests that these organisms may also likely utilize trehalose in a similar manner.
Alternatively, work in Saccharomyces cerevisiae has shown that when the activity of the trehalose-hydrolyzing enzyme acid trehalase is disrupted, intracellular trehalose concentrations increase and the ability of the organism to survive dehydration and freezing is increased (19, 20). In contrast, in L. acidophilus, when the trehalose hydrolase was inactivated, but trehalose continued to be transported, cryoprotection is lost. This surprising result suggests that while trehalose may be able to confer cryoprotection through its action as a compatible solute, the hydrolytic products of trehalose may also contribute significantly to this process. Hibernating frogs have been shown to synthesize and accumulate low-molecular-weight carbohydrates in response to ice formation in peripheral tissues. In particular, Rana sylvatica and Hyla crucifer have been shown to accumulate glucose, while Hyla versicolor has been shown to use glycerol as a cryoprotectant (38). While this has not been shown in bacteria, it may be possible that the products of trehalose-6-phosphate hydrolysis, glucose and glucose-6-phosphate, may act as cryoprotectants in a similar manner. It is more likely that the accumulation of the glycolytic intermediates glyceraldehyde-3-phosphate and dihydroxyacetone phosphate may act in a manner similar to that of glycerol, which is commonly used as a cryoprotectant.
Based on these observations, we propose that the tre locus of L. acidophilus NCFM encodes a PTS transporter and a hydrolase involved in the transport and catabolism of trehalose. Disruption of both the trehalose transporter and hydrolase abolished the ability of L. acidophilus NCFM to grow on trehalose and reduced the survival of L. acidophilus NCFM when subjected to repeated cycles of freezing and thawing in the presence of trehalose. This indicates not only that the internalization of trehalose is important but that its subsequent hydrolysis also contributes. Extracellular trehalose also provided limited cryoprotection possibly by stabilizing the external membrane. A mechanistic understanding of how L. acidophilus can better survive exposure to freezing or freeze-drying is expected to enhance the production, distribution, and long-term storage of viable and active probiotic cultures.
Acknowledgments
We acknowledge Evelyn Durmaz, Michael J. Miller, Joseph M. Sturino, and Robert W. Hutkins for insightful discussions and technical help.
This work was partially sponsored by Dairy Management, Inc., the Southeast Dairy Foods Research Center, and Danisco USA, Inc. Tri Duong and Rodolphe Barrangou were recipients of National Science Foundation IGERT fellowships in functional genomics.
REFERENCES
- 1.Altermann, E., W. M. Russell, M. A. Azcarate-Peril, R. Barrangou, B. L. Buck, O. McAuliffe, N. Souther, A. Dobson, T. Duong, M. Callanan, S. Lick, A. Hamrick, R. Cano, and T. R. Klaenhammer. 2005. Complete genome sequence of the probiotic lactic acid bacterium Lactobacillus acidophilus NCFM. Proc. Natl. Acad. Sci. USA 102:3906-3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson, U., F. Levander, and P. Radstrom. 2001. Trehalose-6-phosphate phosphorylase is part of a novel metabolic pathway for trehalose utilization in Lactococcus lactis. J. Biol. Chem. 276:42707-42713. [DOI] [PubMed] [Google Scholar]
- 3.Arguelles, J. C. 2000. Physiological roles of trehalose in bacteria and yeasts: a comparative analysis. Arch. Microbiol. 174:217-224. [DOI] [PubMed] [Google Scholar]
- 4.Attfield, P. V. 1987. Trehalose accumulates in Saccharomyces cerevisiae during exposure to agents that induce heat shock response. FEBS Lett. 225:259-263. [DOI] [PubMed] [Google Scholar]
- 5.Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2001. Current protocols in molecular biology. John Wiley and Sons, New York, N.Y.
- 6.Barefoot, S. F., and T. R. Klaenhammer. 1983. Detection and activity of lactacin B, a bacteriocin produced by Lactobacillus acidophilus. Appl. Environ. Microbiol. 45:1808-1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrangou, R., E. Altermann, R. Hutkins, R. Cano, and T. R. Klaenhammer. 2003. Functional and comparative genomic analyses of an operon involved in fructooligosaccharide utilization by Lactobacillus acidophilus. Proc. Natl. Acad. Sci. USA 100:8957-8962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Conrad, P. B., D. P. Miller, P. R. Cielenski, and J. J. de Pablo. 2000. Stabilization and preservation of Lactobacillus acidophilus in saccharide matrices. Cryobiology 41:17-24. [DOI] [PubMed] [Google Scholar]
- 9.Crowe, J. H., L. M. Crowe, A. E. Oliver, N. Tsvetkova, W. Wolkers, and F. Tablin. 2001. The trehalose myth revisited: introduction to a symposium on stabilization of cells in the dry state. Cryobiology 43:89-105. [DOI] [PubMed] [Google Scholar]
- 10.Diniz-Mendes, L., E. Bernardes, P. S. de Araujo, A. D. Panek, and V. M. Paschoalin. 1999. Preservation of frozen yeast cells by trehalose. Biotechnol. Bioeng. 65:572-578. [DOI] [PubMed] [Google Scholar]
- 11.Giaever, H. M., O. B. Styrvold, I. Kaasen, and A. R. Strom. 1988. Biochemical and genetic characterization of osmoregulatory trehalose synthesis in Escherichia coli. J. Bacteriol. 170:2841-2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glaasker, E., W. N. Konings, and B. Poolman. 1996. Osmotic regulation of intracellular solute pools in Lactobacillus plantarum. J. Bacteriol. 178:575-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gotsche, S., and M. K. Dahl. 1995. Purification and characterization of the phospho-α(1,1)glucosidase (TreA) of Bacillus subtilis 168. J. Bacteriol. 177:2721-2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guo, N., I. Puhlev, D. R. Brown, J. Mansbridge, and F. Levine. 2000. Trehalose expression confers desiccation tolerance on human cells. Nat. Biotechnol. 18:168-171. [DOI] [PubMed] [Google Scholar]
- 15.Horlacher, R., W. Boos, K. Uhland, W. Klein, M. Ehrmann, and M. Rimmele. 1997. Characterization of TreR, the major regulator of the Escherichia coli trehalose system. J. Biol. Chem. 272:13026-13032. [DOI] [PubMed] [Google Scholar]
- 16.Horlacher, R., K. B. Xavier, H. Santos, J. DiRuggiero, M. Kossmann, and W. Boos. 1998. Archaeal binding protein-dependent ABC transporter: molecular and biochemical analysis of the trehalose/maltose transport system of the hyperthermophilic archaeon Thermococcus litoralis. J. Bacteriol. 180:680-689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaplan, H., and R. W. Hutkins. 2003. Metabolism of fructooligosaccharides by Lactobacillus paracasei 1195. Appl. Environ Microbiol. 69:2217-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kets, E. P. W., P. J. M. Teunissen, and J. A. M. de Bont. 1996. Effect of compatible solutes on survival of lactic acid bacteria subjected to drying. Appl. Environ. Microbiol. 62:259-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, J., P. Alizadeh, T. Harding, A. Hefner-Gravink, and D. J. Klionsky. 1996. Disruption of the yeast ATH1 gene confers better survival after dehydration, freezing, and ethanol shock: potential commercial applications. Appl. Environ. Microbiol. 62:1563-1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kim, W. S., L. Perl, J. H. Park, J. E. Tandianus, and N. W. Dunn. 2001. Assessment of stress response of the probiotic Lactobacillus acidophilus. Curr. Microbiol. 43:346-350. [DOI] [PubMed] [Google Scholar]
- 21.Kleerebezem, M., J. Boekhorst, R. van Kranenburg, D. Molenaar, O. P. Kuipers, R. Leer, R. Tarchini, S. A. Peters, H. M. Sandbrink, M. W. Fiers, W. Stiekema, R. M. Lankhorst, P. A. Bron, S. M. Hoffer, M. N. Groot, R. Kerkhoven, M. de Vries, B. Ursing, W. M. de Vos, and R. J. Siezen. 2003. Complete genome sequence of Lactobacillus plantarum WCFS1. Proc. Natl. Acad. Sci. USA 100:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Klein, W., R. Horlacher, and W. Boos. 1995. Molecular analysis of treB encoding the Escherichia coli enzyme II specific for trehalose. J. Bacteriol. 177:4043-4052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Leslie, S. B., E. Israeli, B. Lighthart, J. H. Crowe, and L. M. Crowe. 1995. Trehalose and sucrose protect both membranes and proteins in intact bacteria during drying. Appl. Environ. Microbiol. 61:3592-3597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luchansky, J. B., M. C. Tennant, and T. R. Klaenhammer. 1991. Molecular cloning and deoxyribonucleic acid polymorphisms in Lactobacillus acidophilus and Lactobacillus gasseri. J. Dairy Sci. 74:3293-3302. [DOI] [PubMed] [Google Scholar]
- 25.Mazur, P. 1970. Cryobiology: the freezing of biological systems. Science 168:939-949. [DOI] [PubMed] [Google Scholar]
- 26.Mazur, P. 1977. The role of intracellular freezing in the death of cells cooled at supraoptimal rates. Cryobiology 14:251-272. [DOI] [PubMed] [Google Scholar]
- 27.Pellerin-Mendes, C., L. Million, M. Marchand-Arvier, P. Labrude, and C. Vigneron. 1997. In vitro study of the protective effect of trehalose and dextran during freezing of human red blood cells in liquid nitrogen. Cryobiology 35:173-186. [DOI] [PubMed] [Google Scholar]
- 28.Pridmore, R. D., B. Berger, F. Desiere, D. Vilanova, C. Barretto, A. C. Pittet, M. C. Zwahlen, M. Rouvet, E. Altermann, R. Barrangou, B. Mollet, A. Mercenier, T. Klaenhammer, F. Arigoni, and M. A. Schell. 2004. The genome sequence of the probiotic intestinal bacterium Lactobacillus johnsonii NCC 533. Proc. Natl. Acad. Sci. USA 101:2512-2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rimmele, M., and W. Boos. 1994. Trehalose-6-phosphate hydrolase of Escherichia coli. J. Bacteriol. 176:5654-5664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rudolph, A. S., J. H. Crowe, and L. M. Crowe. 1986. Effects of three stabilizing agents—proline, betaine, and trehalose—on membrane phospholipids. Arch. Biochem. Biophys. 245:134-143. [DOI] [PubMed] [Google Scholar]
- 31.Russell, W. M., and T. R. Klaenhammer. 2001. Efficient system for directed integration into the Lactobacillus acidophilus and Lactobacillus gasseri chromosomes via homologous recombination. Appl. Environ. Microbiol. 67:4361-4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sano, F., N. Asakawa, Y. Inoue, and M. Sakurai. 1999. A dual role for intracellular trehalose in the resistance of yeast cells to water stress. Cryobiology 39:80-87. [DOI] [PubMed] [Google Scholar]
- 33.Schlosser, A. 2000. MsiK-dependent trehalose uptake in Streptomyces reticuli. FEMS Microbiol. Lett. 184:187-192. [DOI] [PubMed] [Google Scholar]
- 34.Schock, F., and M. K. Dahl. 1996. Analysis of DNA flanking the treA gene of Bacillus subtilis reveals genes encoding a putative specific enzyme IITre and a potential regulator of the trehalose operon. Gene 175:59-63. [DOI] [PubMed] [Google Scholar]
- 35.Schock, F., and M. K. Dahl. 1996. Expression of the tre operon of Bacillus subtilis 168 is regulated by the repressor TreR. J. Bacteriol. 178:4576-4581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Souzu, H. 1989. Changes in chemical structure and function in Escherichia coli cell membranes caused by freeze-thawing. I. Change of lipid state in bilayer vesicles and in the original membrane fragments depending on rate of freezing. Biochim. Biophys. Acta 978:105-111. [DOI] [PubMed] [Google Scholar]
- 37.Souzu, H., M. Sato, and T. Kojima. 1989. Changes in chemical structure and function in Escherichia coli cell membranes caused by freeze-thawing. II. Membrane lipid state and response of cells to dehydration. Biochim. Biophys. Acta 978:112-118. [DOI] [PubMed] [Google Scholar]
- 38.Storey, K. B. 1990. Life in a frozen state: adaptive strategies for natural freeze tolerance in amphibians and reptiles. Am. J. Physiol. 258:R559-568. [DOI] [PubMed] [Google Scholar]
- 39.Walker, D. C., and T. R. Klaenhammer. 1994. Isolation of a novel IS3 group insertion element and construction of an integration vector for Lactobacillus spp. J. Bacteriol. 176:5330-5340. [DOI] [PMC free article] [PubMed] [Google Scholar]