Abstract
In this study we evaluated the short-term effects of copper, cadmium, and mercury, added singly or in combination at different doses, on soil bacterial community structure using the bacterial automated ribosomal intergenic spacer analysis (B-ARISA) fingerprinting technique. Principal-component analysis of B-ARISA profiles allowed us to deduce the following order of impact: (Cu + Cd + Hg) >> Hg ≥ Cd > Cu. These results demonstrated that there was a cumulative effect of metal toxicity. Furthermore, the trend of modifications was consistent with the “hump-backed” relationships between biological diversity and disturbance described by Giller et al. (K. E. Giller, E. Witler, and S. P. McGrath, Soil Biol. Biochem. 30:1389-1414, 1998).
Among the heavy metals, cadmium (Cd), mercury (Hg), and copper (Cu) are known to have deleterious effects on the density, activity, and diversity of soil microflora (8, 11, 16). However, most of the experiments dealing with the impact of metals on soil microorganisms have been field experiments in which there has been polymetal contamination, generally associated with inorganic nutrients and organic matter, which has made it difficult to distinguish the effect of one metal from the effect of another (3, 7, 14, 17). In this context, Renella et al. (12, 13) performed a short-term laboratory experiment to evaluate the impact on soil microorganisms of different metals (Cu, Zn, and Cd) added singly or in combination. They found that soil respiration and microbial biomass were significantly affected by Zn and Cu, respectively (12). In all cases, polymetal contamination had a greater impact than single-metal contamination, leading the workers to conclude that there were additive effects of metal toxicity (13). Here, we conducted a complementary study by characterizing modifications in the genetic structures of soil bacterial communities induced by different doses of Cu, Cd, and Hg added independently or in combination (Cu + Cd + Hg).
The soil used for this study was a calcareous sandy luvisol under a garden market (southwest of Paris, France). The pH was 7.9 and the soil contained 83.5% sand, 7.9% silt, 8.6% clay, 2.5% CaCO3, 0.97% organic C, and 0.76% organic N. The soil was not affected by any known metal contamination and had a classical soil background (8 mg Cu · kg−1, 0.25 mg Cd · kg−1, 0.07 mg Hg · kg−1) (1). It was collected from the plowed layer (depth, 0 to 20 cm) and was sieved with a 4-mm sieve.
Microcosms were set up by placing 10 g (dry weight) of soil into 125-ml plasma flasks. Cu, Hg, and Cd were added as aqueous chloride solutions at a level that was 80% of the water-holding capacity of the soil. The low level of contamination (LC) corresponded to final total concentrations of Cu, Hg, and Cd of 50, 2, and 2 mg · kg soil−1, respectively. The medium level (MC) corresponded to 150, 10, and 10 mg · kg soil−1, respectively, and the high level (HC) corresponded to 300, 20, and 40 mg · kg soil−1, respectively. These concentrations were chosen by compiling data from the literature dealing with soil contamination and metal effects on the density and diversity of soil microbial communities (5, 7, 8, 11, 12). In parallel, control incubation mixtures were supplemented independently with sterile water and with KCl at concentrations of 150 mg and 300 mg · kg soil−1 to simulate addition of chloride anions and with HCl at concentrations of 0.05 N and 0.1 N to simulate soil acidification due to MC and HC metal spiking, respectively. Metals were applied singly or in combination (Cu + Cd + Hg) for each concentration. Soil was incubated at 20°C for 60 days, and triplicate microcosms were killed at zero time and on days 10 and 60 for analyses.
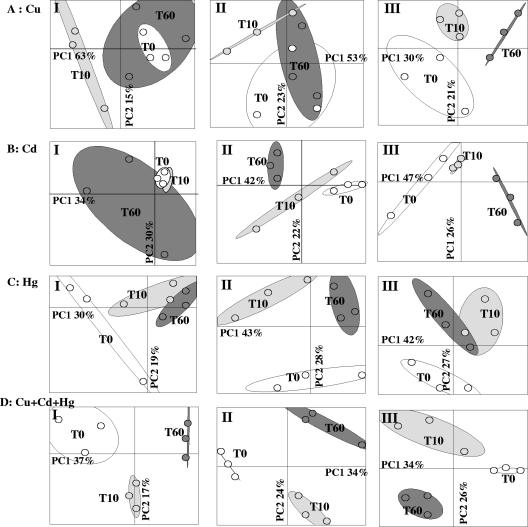
The genetic structure of the bacterial community was characterized by using the bacterial automated ribosomal intergenic spacer analysis (B-ARISA) fingerprinting approach for soil DNA extract (10). This approach provided complex profiles with peaks ranging from 250 bp to more than 1,000 bp for each sample studied (Fig. 1). B-ARISA profiles were analyzed by principal-component analysis (PCA), which resulted in plotting, on a factorial map, of the genetic structure of the bacterial community based on similarities (Fig. 2 to 4). In Fig. 2 to 4, statistical ellipses indicate (with 90% confidence) the replicates of microcosms subjected to similar treatments. Overlapping of statistical ellipses means that the genetic structures were not significantly discriminated by PCA (10). The PCA of control microcosms (supplemented with water, HCl, and KCl) demonstrated that the genetic structure of the bacterial community was not modified by these treatments during the incubation period (data not shown). Except for Cd at LC, the three metals added singly resulted in significant modifications of the bacterial genetic structure at each level of contamination (Fig. 2A to C). The changes induced by Cd increased with time during incubation, whereas for Hg the modifications of the genetic structure occurred between zero time and day 10 and after day 10 remained stable. For Cu, shifts were observed between zero time and day 10 for LC and MC, but the modifications seemed to be resilient since the genetic structure at day 60 was similar to that observed at zero time. For HC (300 mg Cu · kg soil−1), the changes in the genetic structure gradually increased with the incubation period. Other authors have observed similar resilience of microbial communities (defined as the speed with which a system returns to its predisturbance level following a disturbance) with copper concentrations close to LC and MC (16). Furthermore, our results were in agreement with those of Frostegard and Bååth (7), who found that modifications in the microbial composition became irreversible at Cu concentrations greater than 130 mg · kg soil−1. The trimetal treatments resulted in modifications which were more marked than the modifications observed with the corresponding single-metal treatments and were irreversible (Fig. 2D). These findings were consistent with the findings of Renella et al. (13), who demonstrated the additive effect on soil enzymatic activities of polymetal contamination (Cu + Cd + Zn) compared to the effect of each metal added singly.
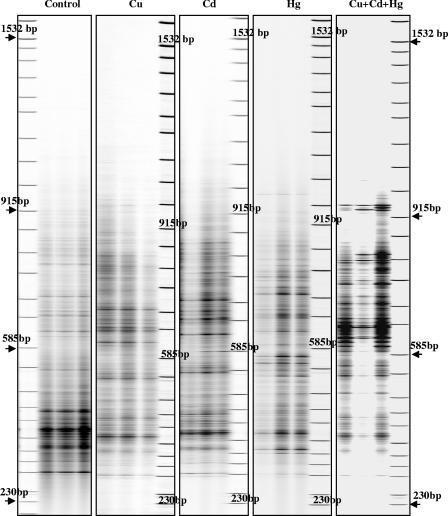
FIG. 1.
B-ARISA profiles obtained at day 60 with DNA extracted from triplicate microcosms treated with water (control) and with copper (Cu), cadmium (Cd), mercury (Hg), and all three metals (Cu + Hg + Cd) at HC.
FIG. 2.
PCA ordination of the bacterial genetic structure for microcosms contaminated with copper (A), cadmium (B), mercury (C), and all three metals (Cu + Hg + Cd) (D) at LC (panels I), MC (panels II), and HC (panels III). The statistical ellipses indicate 90% confidence for replicates. Open ellipses, zero time (T0); light gray ellipses, day 10 (T10); dark gray ellipses, day 60 (T60).
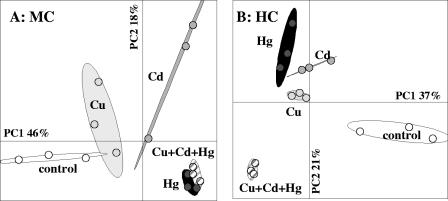
FIG. 4.
PCA ordination of the bacterial genetic structure for control microcosms (open ellipses) and microcosms contaminated with copper (light gray ellipses), cadmium (dark gray ellipses), mercury (black ellipses), and all three metals (striped ellipses) at MC (A) and HC (B) at day 60. The statistical ellipses indicate 90% confidence.
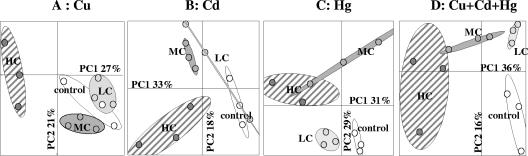
At day 60 the magnitude of shifts was positively correlated with the level of contamination for each metal added singly or the metals added simultaneously (Fig. 3). This observation could suggest that there is stronger selection of adapted populations with higher levels of metal (6). When we compared the effects of Cd, Cu, and Hg added singly and in combination, we observed distinct population structures depending on the metal treatment (Fig. 4). Bacterial resistance to Cd, bacterial resistance to Hg, and bacterial resistance to Cu involved different physiological pathways and genetic determinants (for example, czc, mer, and the cop operon, respectively) (4). Consequently, our data are not consistent with the studies which suggested that these genes were harbored by the same populations, which could be enriched by different metal treatments (9, 15). However, such discrepancy could be partly explained by the short-time of spiking of soils that were not previously exposed to metal contamination and also by the fact that metal treatments could select opportunist populations. For MC, the trimetal and mercury treatments at day 60 selected similar bacterial genetic structures (Fig. 4A), suggesting that Hg was the most stressful element in the metal combination. Hg contamination seemed to select adapted populations that were also able to survive in the presence of Cd and Cu (Fig. 4A). However, these populations were not stimulated by the Cu and Cd treatments applied independently, since in these cases other populations were selected. The HC trimetal treatment induced a genetic structure which was easily distinguished from the genetic structures resulting from independent Cu, Cd, and Hg treatments (Fig. 4B). This observation confirmed the cumulative effects of metal toxicity when metals are applied simultaneously and could be partially explained by the loss of dominant populations, since in this case a dramatic reduction in the B-ARISA profile complexity, in terms of the number of bands, was observed (Fig. 1). Such drastic modification of the bacterial community genetic structure has been observed previously in other cases of deleterious chemical stress (2).
FIG. 3.
PCA ordination of the bacterial genetic structure for the different levels of copper (A), cadmium (B), mercury (C), and trimetal (D) contamination at day 60. The statistical ellipses indicate 90% confidence. Open ellipses, control microcosms (H2O); light gray ellipses, LC; dark gray ellipses, MC; striped ellipses, HC.
Altogether, we deduced from the various observations using (i) the magnitude of modifications and irreversible effects observed for each metal applied singly or simultaneously at the three doses (as shown in Fig. 2) and (ii) the similarity of the genetic structures induced by the treatments (as shown in Fig. 4) the following general order of impact: (Cu + Cd + Hg) > Hg ≥ Cd > Cu. Furthermore, most of the profile modifications resulted in the appearance of new bands and/or an increase in the intensity of preexisting bands, except for HC trimetal contamination. Although B-ARISA did not allow estimates of bacterial diversity, the modifications seemed to be consistent with the “hump-backed” relationships between biological diversity and metal stress (8). This model hypothesized that for the relationship between pollution stresses and genetic diversity in microbial communities there was not a simple negative correlation. It assumed that the increase in stress (from LC to HC single-metal treatments and from LC to MC trimetal treatments in our study) reduced the innate competitive exclusion between populations and induced the enrichment and predominance of other types of populations (as illustrated by increasing band intensity), leading to possible higher diversity, followed, when stress reached a critical high level (HC trimetal treatment), by progressive extinction of organisms, leading to a loss of diversity.
Acknowledgments
We thank P. A. Maron, J. C. Lata, C. Mougel, and D. P. H. Lejon for their comments on the manuscript.
REFERENCES
- 1.Alloway, B. J. 1995. Soil processes and the behaviour of metals, p. 9-38. In B. J. Alloway (ed.), Heavy metals in soils. Chapman and Hall, London, United Kingdom.
- 2.Atlas, R. M., A. Horwitz, M. Krichevsky, and A. K. Bej. 1991. Response of microbial populations to environmental disturbance. Microb. Ecol. 22:249-256. [DOI] [PubMed] [Google Scholar]
- 3.Bååth, E., M. Diaz-Raviña, Å. Frostegard, and C. D. Campbell. 1998. Effects of metal-rich sludge amendments on the soil microbial community. Appl. Environ. Microbiol. 64:238-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bruins, M. R., S. Kapil, and W. Oehme. 2000. Microbial resistance to metals in the environment. Ecotoxicol. Environ. Saf. 45:198-207. [DOI] [PubMed] [Google Scholar]
- 5.Diaz-Raviña, M., and E. Bååth. 1996. The development of metal tolerance of soil bacterial communities and its measurement by a thymidine incorporation technique. Appl. Environ. Microbiol. 60:2238-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duxburry, T., and B. Bicknell. 1983. Metal-tolerant bacterial populations from natural and metal-polluted soils. Soil Biol. Biochem. 15:243-250. [Google Scholar]
- 7.Frostegard, Å., A. Tunlid, and E. Bååth. 1993. Phospholipid fatty acid composition, biomass, and activity of microbial communities from two soil types experimentally exposed to different heavy metals. Appl. Environ. Microbiol. 59:3605-3617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giller, K. E., E. Witler, and S. P. McGrath. 1998. Toxicity of heavy metals to microorganisms and microbial processes in agricultural soils: a review. Soil Biol. Biochem. 30:1389-1414. [Google Scholar]
- 9.Huysman, F., W. Verstraete, and P. C. Brookes. 1994. Effect of manuring practices and increased copper concentrations on soil microbial populations. Soil Biol. Biochem. 26:103-110. [Google Scholar]
- 10.Ranjard, L., D. P. H. Lejon, C. Mougel, L. Schehrer, D. Merdinoglu, and R. Chaussod. 2003. Sampling strategy in molecular microbial ecology: influence of soil sample size on DNA fingerprinting analysis of fungal and bacterial communities. Environ. Microbiol. 5:1111-1120. [DOI] [PubMed] [Google Scholar]
- 11.Ranjard, L., S. Nazaret, F. Gourbière, J. Thioulouse, P. Linet, and A. Richaume. 2000. A soil microscale study to reveal the heterogeneity of Hg(II) impact on indigenous bacteria by quantification of adapted phenotypes and analysis of DNA community fingerprints. FEMS Microbiol. Ecol. 31:107-115. [DOI] [PubMed] [Google Scholar]
- 12.Renella, G., A. M. Chaudri, and P. C. Brookes. 2002. Fresh additions of heavy metals do not model long-term effects on microbial biomass and activity. Soil Biol. Biochem. 34:121-124. [Google Scholar]
- 13.Renella, G., A. L. R. Ortigoza, L. Landi, and P. Nannipieri. 2003. Additive effects of copper and zinc on cadmium toxicity on phosphatase activities and ATP content of soil as estimated by the ecological dose (ED50). Soil Biol. Biochem. 35:1203-1210. [Google Scholar]
- 14.Sandaa, R. A., V. Torsvik, and Ø. Enger. 2001. Influence of long-term heavy-metal contamination on microbial communities in soil. Soil Biol. Biochem. 33:287-295. [Google Scholar]
- 15.Silver, S., B. T. O. Lee, N. L. Brown, and D. A. Cooksey. 1993. Bacterial plasmid resistances to copper, cadmium and zinc, p. 38-53. In A. J. Welch and K. Chapman (ed.), The chemistry of the copper and zinc triads. The Royal Society of Chemistry, Cambridge, United Kingdom.
- 16.Tom-Petersen, A., T. Leser, T. L. Marsh, and O. Nybroe. 2003. Effects of copper amendment on the bacterial community in agricultural soil analysed by the T-RFLP technique. FEMS Microbiol. Ecol. 46:53-62. [DOI] [PubMed] [Google Scholar]
- 17.Turpeinen, R., T. Kairesalo, and M. M. Haggblom. 2004. Microbial community structure and activity in arsenic-, chromium- and copper-contaminated soils. FEMS Microbiol. Ecol. 47:39-50. [DOI] [PubMed] [Google Scholar]