Abstract
Mycobacterium vanbaalenii PYR-1 is able to metabolize a wide range of low- and high-molecular-weight (HMW) polycyclic aromatic hydrocarbons (PAHs). A 20-kDa protein was upregulated in PAH-metabolizing M. vanbaalenii PYR-1 cells compared to control cultures. The differentially expressed protein was identified as a β subunit of the terminal dioxygenase using mass spectrometry. PCR with degenerate primers designed based on de novo sequenced peptides and a series of plaque hybridizations were done to screen the M. vanbaalenii PYR-1 genomic library. The genes, designated nidA3B3, encoding the α and β subunits of terminal dioxygenase, were subsequently cloned and sequenced. The deduced enzyme revealed close similarities to the corresponding PAH ring-hydroxylating dioxygenases from Mycobacterium and Rhodococcus spp. but had the highest similarity, 61.9%, to the α subunit from Nocardioides sp. strain KP7. The α subunit also showed 52% sequence homology with the previously reported NidA from M. vanbaalenii PYR-1. The genes nidA3B3 were subcloned into the expression vector pET-17b, and the enzyme activity in Escherichia coli cells was reconstituted through coexpression with the ferredoxin (PhdC) and ferredoxin reductase (PhdD) genes of the phenanthrene dioxygenase from Nocardioides sp. strain KP7. The recombinant PAH dioxygenase appeared to favor the HMW PAH substrates fluoranthene, pyrene, and phenanthrene. Several other PAHs, including naphthalene, anthracene, and benz[a]anthracene, were also converted to their corresponding cis-dihydrodiols. The recombinant E. coli, however, did not show any dioxygenation activity for phthalate and biphenyl. The upregulation of nidA3B3 in M. vanbaalenii PYR-1 induced by PAHs was confirmed by reverse transcription-PCR analysis.
The presence and distribution of microorganisms that can degrade polycyclic aromatic hydrocarbons (PAHs) in soils have been well documented (16, 23). Of several types of bacteria with the ability to degrade PAHs, the nocardioform actinomycetes, including species of Mycobacterium (4, 8, 13, 21, 24, 56, 66), Nocardioides (53), and Rhodococcus (5, 9, 65), are of particular interest because of their marked ability to aerobically degrade high-molecular-weight (HMW) PAHs to ring isomeric dihydroxylated metabolites and in some cases to complete oxidation to carbon dioxide and water.
Mycobacterium vanbaalenii PYR-1 (29) was isolated from oil-contaminated polluted sediment based on its ability to degrade pyrene (21). This bacterium can mineralize various aromatic hydrocarbons, including biphenyl, naphthalene, anthracene, fluoranthene, pyrene, 1-nitropyrene, phenanthrene, benzo[a]pyrene, benz[a]anthracene, and 7,12-dimethylbenz[a]anthracene (18-21, 25-28, 45-49, 51). Analysis of metabolites indicates that the oxidation of PAHs in the strain PYR-1 is primarily initiated by dioxygenation (21, 30, 34, 46-49) with minor monooxygenation of the aromatic ring (7). The enzyme catalyzing dioxygenation belongs to a family of aromatic-ring-hydroxylating dioxygenases that introduce two atoms of oxygen into aromatic hydrocarbons to form a cis-dihydrodiol. They are multicomponent enzyme systems consisting of an electron transport chain and a terminal dioxygenase composed of large (α) and small (β) subunits (43). The enzyme system has been extensively studied in many microorganisms, since this initial reaction mostly determines aromatic substrate ranges that can be degraded. However, a significant amount of information comes exclusively from dioxygenases for monocyclic aromatics or low-molecular-weight PAHs. In M. vanbaalenii PYR-1, identification of the genes encoding PAH degradation indicates that this bacterium has at least three copies of the terminal dioxygenase gene (6, 30, 59). Among them, nidAB and phtAaAb were shown to transform the PAHs phenanthrene and pyrene into phenanthrene and pyrene cis-dihydrodiols (30, 59) and phthalate into phthalate cis-3,4-dihydrodiol (58), respectively. In recent years, copies of terminal dioxygenase genes have been reported from HMW-PAH-degrading bacteria (6, 10, 14, 38, 44, 56). However, detailed information in regard to the actual role and properties of each gene product is limited, and no gene product has been assigned as a dioxygenase that significantly oxidizes HMW PAHs to cis-dihydrodiols. Previously, a 20-kDa protein which showed upregulated expression in response to the exposure of M. vanbaalenii PYR-1 to several PAHs was identified by two-dimensional gel electrophoresis (2-DE) and protein mass spectrometry (MS) as a β subunit of terminal dioxygenase (32). In this study, a gene cluster encoding the 20-kDa polypeptide was isolated and characterized. We cloned the fourth copy of the terminal dioxygenase gene with the aid of PCR and PYR-1 genome library screening. We then examined the substrate specificities and transformation rates of the enzyme that was expressed in Escherichia coli.
MATERIALS AND METHODS
Chemicals.
Pyrene and phenanthrene were purchased from Chem Service (Media, PA). Phthalate, naphthalene, biphenyl, anthracene, benz[a]anthracene, and pyrene were obtained from Sigma-Aldrich (St. Louis, MO). Fluoranthene was purchased from Fluka (Buchs, Switzerland).
Bacterial strains, plasmids, reagents, and growth conditions.
The bacterial strains, vectors, and plasmids used in this study are listed in Table 1. E. coli strains were grown at 30°C or 37°C on Luria-Bertani (LB) medium supplemented with 100, 15, and 10 μg/ml of ampicillin, tetracycline, and gentamicin, respectively (54). M. vanbaalenii PYR-1 was grown at 30°C in Middlebrook 7H10 (Remel, Lenexa, KS) or in phosphate-based minimal medium (33) with 2% sorbitol and PAHs as described in a previous report (32). Antibiotics and all other chemicals were purchased from Sigma.
TABLE 1.
Bacterial strains, phage, and plasmids used in this study
| Strain, phage, or plasmid | Relevant characteristicsa | Reference(s) or source |
|---|---|---|
| Strains | ||
| M. vanbaalenii PYR-1 | Mineralizes PAHs, such as fluoranthene, pyrene, and phenanthrene | 21, 29 |
| E. coli XL1-Blue MRF′ | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 supE44 thi-1 recA1 gyrA96 relA1 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] | Stratagene |
| E. coli XLOLR | Δ(mcrA)183 Δ(mcrCB-hsdSMR-mrr)173 endA1 thi-1 recA1 gyrA96 lac [F′ proAB lacIqZΔM15 Tn10 (Tetr)] Su (nonsuppressing) λR | Stratagene |
| E. coli DH5α | F− Φ80dlacZΔM15 Δ(lacZYA-argF) U169 deoR recA1 endA1 hsdR17 (rK− mK+) phoA supE44 λ−thi-1 gyrA96 relA1 | 17 |
| E. coli TOP10 | F−mcrA Δ(mrr-hsdRMS-mcrBC) υ80lacZΔM15 ΔlacX74 recA1 araD139 galU galK Δ(ara-leu)7697 rpsL (StrR) endA1 nupG | Invitrogen |
| E. coli BL21(DE3) | F−ompT hsdSB (rB− mB−) gal dcm (DE3) | Novagen |
| ExAssist helper phage | For in vivo excision of the pBK-CMV phagemid from ZAP Express vector with E. coli XLOLR | Stratagene |
| Plasmids | ||
| pGEM-T Easy | Apr, TA cloning vector | Promega |
| pCR2.1-TOPO | Apr, TA cloning vector | Invitrogen |
| pET-17b | Expression vector; Apr T7 promoter | 38 |
| pBRCD | Gmr, pBBR1MCS-5 containing phdCD | 60 |
| pNCK30 | pGEM-T Easy with a 109-bp PCR fragment | This study |
| pNCK31 | 3.7-kb BamHI fragment including nidA3B3 genes of M. vanbaalenii PYR-1 cloned into pBK-CMV phagemid | This study |
| pNCK32 | 1.87-kb PCR fragment including nidA3B3 genes from pNCK31 cloned into pCR2.1-TOPO | This study |
| pNCK33 | 1.87-kb NdeI-HindIII fragment from pNCK32 cloned into NdeI-HindIII of pET-17b | This study |
Apr, ampicillin resistance; Gmr, gentamicin resistance.
DNA manipulations, peptide de novo sequencing, cloning, and sequence analysis.
DNA manipulations were performed by standard procedures (54). Restriction enzymes, T4 DNA ligase, and Taq DNA polymerase were purchased from Promega (Madison, WI), Roche Applied Science (Indianapolis, IN), or New England Biolabs (Beverly, MA). Oligonucleotide primers were obtained from MWG Biotech, Inc. (High Point, NC). DNA fragments were purified from agarose gels using QIAquick spin columns (QIAGEN, Valencia, CA). Plasmid DNA was purified using a QIAprep plasmid mini kit (QIAGEN). M. vanbaalenii PYR-1 genomic DNA was isolated according to the protocol for gram-positive bacteria with a QIAGEN genomic DNA extraction kit. Tryptic peptides were de novo sequenced manually, and primary sequence tags were used for homology search as described previously (32). A set of degenerate primers, P18-F (5′-CANWSNGAYTGGGCNGARGAYCCN-3′) and P18-R (5′-GCRTTNSWNGTNACNCKRTAYTCRTCNCC-3′), was designed from peptide sequences (TSSDWAEDPP and SGEDGEDYRVTSNALLVR) of polypeptide P18. These were used to produce a probe against the gene for P18 and its neighboring region. A PCR with the primer pair (P18-F and P18-R) and PYR-1 total DNA as a template amplified a 0.1-kb fragment. This fragment was cloned in pGEM-T Easy vector (Promega), giving plasmid pNCK30, and its insert sequence was determined. The λ genomic library (67) was then screened using this 109-bp DNA fragment as a probe, which had been labeled with a digoxigenin oligonucleotide 3′ end labeling kit (Roche Applied Science). A plaque-lift hybridization procedure was used for the screening as recommended by the digoxigenin system user's guide (Roche Applied Science). Plaques showing a positive signal were in vivo excised to produce phagemid clones using ExAssist Helper Phage with the E. coli XLOLR strain (Stratagene, La Jolla, CA). The DNA sequence was determined on both strands using a model 377 DNA sequencer (Applied Biosystems, Foster City, CA).
Homology search was performed using the BLASTX program (1), and DNA sequences were processed using Lasergene (DNASTAR) software. Protein sequences were aligned with the CLUSTAL X program, version 1.81 (64), and phylogenetic trees were inferred from the alignments by the PHYLIP package version 3.63 (12, 31). The graphic for the phylogenetic tree was visualized by the TreeView program (50).
Construction of nidA3B3 dioxygenase expression system.
For gene expression in E. coli, the pET-17b expression system (Novagen, Madison, WI) was used. A DNA fragment containing nidA3B3 was amplified from M. vanbaalenii PYR-1 genomic DNA with primers EnidA3-F (5′-ACATATGGCGCCTGATGCGACGACAATG-3′) and EnidB3-R (5′-CAAGCTTTTAGATCCAGAATGACAGGTT-3′). The underlined sequences are the NdeI and HindIII sites, respectively. The 1.87-kb PCR product was initially cloned into pCR2.1-TOPO (Invitrogen, Carlsbad, CA) to give pNCK32 and subjected to DNA sequencing to confirm that PCR amplification did not introduce mutations. The insert of plasmid pNCK32 was isolated by digestion with NdeI and HindIII. This fragment was ligated between the NdeI and HindIII site of pET-17b, resulting in plasmid pNCK33. Both plasmids pNCK33, containing the dioxygenase genes (nidA3B3), and pBRCD, expressing PhdCD of KP7 (38), were transformed into E. coli strain BL21(DE3).
Analysis and identification of PAH metabolites.
The transformed E. coli BL21(DE3)(pNCK33)(pBRCD) and BL21(DE3)(pNCK33) cultures and the control E. coli BL21(DE3)(pET-17b) culture were incubated overnight by inoculating a colony in 50 ml of LB medium at 30°C. A total of 5 ml of the overnight culture was transferred to 100 ml of LB medium and incubated under the same conditions. When the bacterial cells reached an optical density at 600 nm of 0.5 to 0.7, the culture was induced by adding IPTG (isopropyl-β-d-thiogalactopyranoside) at a final concentration of 1 mM and further incubated for 2 h. The culture was harvested by centrifugation, washed with M9-glucose medium, and resuspended in 40 ml of the same solution supplemented with either ampicillin or gentamicin. PAHs were added to a final concentration of 200 μM, and the cells were further incubated overnight at 30°C. The supernatant was extracted with ethyl acetate, which was dried by a rotary evaporator, and the dried residues were dissolved in methanol for high-performance liquid chromatography (HPLC) analysis. Metabolites were resolved, isolated, and purified with an HPLC system (Hewlett-Packard 1100 series). A C18 column (Prodigy 5 μm ODS-3, 260 by 40 mm; Phenomenex, Torrance, CA) was used with a mobile phase of a 30-min linear gradient of methanol-water (from 45:55 to 95:5 [vol/vol]) at a flow rate of 1.0 ml/min.
Gas chromatography-mass spectrometry (GC-MS) was performed on either a Finnigan TSQ 700 or a 7000 triple quadrupole mass spectrometer (Thermo Finnigan, San Jose, CA) with separation of the metabolites using a J&W DB-5ms capillary column (30 m by 0.25 mm, 0.25-μm film thickness). GC-MS analyses were performed with a capillary column temperature rate of 10°C/min and a total analysis time of 30 min or less, depending upon the PAH analyzed. Retention times between the two instrument configurations were slightly different. Electron ionization (EI) was achieved at 70 eV electron energy and 150°C ion source temperature. Derivatization prior to GC/EI-MS analysis was performed by silylation with N,O-bis(trimethylsilyl)-trifluoroacetamide with 1% trimethylchlorosilane (Regis Technologies, Morton Grove, IL). The samples were dissolved in 400 μl of acetonitrile. Two hundred microliters of dissolved sample and 100 μl of silylation reagent were mixed in an Xpertek high-recovery 1.5-ml sample vial (P.J. Cobert, St. Louis, MO) sealed with a septum cap and allowed to react for 1 h at 68°C. The injection volumes were 0.5 μl. All mass spectrometric measurements were at low resolution, and no tandem mass spectrometry methods were employed. Therefore, all fragmentation losses are reported as assumptions based on the proposed structures and available moieties.
RT-PCR analysis.
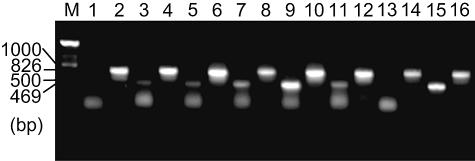
Levels of nidA3 RNA in induced M. vanbaalenii PYR-1 cultures and uninduced cultures were compared by reverse transcription (RT)-PCR analysis. The total RNA was extracted from cells grown in phosphate-based minimal medium supplemented with 100 μM naphthalene, anthracene, phenanthrene, fluoranthene, pyrene, benz[a]anthracene, or sorbitol (2%) using RNA-Bee (Tel-Test, Inc., Friendswood, TX). RNA was further extracted with phenol-chloroform. RNA was then recovered after precipitation with ethanol and treatment with RNase-free DNase I (Promega). PCR without reverse transcription was performed with the DNase-treated RNA to ensure the complete digestion of DNA. RT-PCR analyses were carried out using an Access RT-PCR system (Promega) according to the manufacturer's instructions. Two primer sets were used for RT-PCR. Primer set RnidA3-F3 (5′-GCCGAGATATTCCAGGCTATTA-3′) and RnidB3-R3 (5′-CGCATCTTCAGACTCGTGTAGT-3′) amplified the intergenic 469-bp sequence between nidA3 and nidB3, and the other primer set, R16S-F2 (5′-GAGAAGAAGGACCGGCCAACTACG-3′) and R16S-R2 (5′-AGACCCCGATCCGAACTGAGACC-3′), was used to produce an 826-bp fragment corresponding to 16S rRNA as an amplification control. The RT-PCR conditions were as follows: reverse transcription at 48°C for 45 min followed by 95°C for 15 min, 30 cycles each consisting of 94°C for 1 min, 53°C for 1 min, and 68°C for 1 min, with a final elongation step at 68°C for 7 min. As a positive control, genomic DNA from M. vanbaalenii PYR-1 was used as a template.
Nucleotide sequence accession number.
The gene sequences were deposited in the GenBank database under accession number DQ028634.
RESULTS
Cloning and sequence analysis of genes nidA3B3 encoding PAH dioxygenase.
We recently used 2-DE methods to simultaneously compare and quantitate more than 1,000 gel-separated proteins when M. vanbaalenii PYR-1 was grown in the presence of pyrene, phenanthrene, anthracene, fluoranthene, and sorbitol (32). From the protein profiles and liquid chromatography-MS/MS quadrupole time of flight de novo sequencing data, we detected a 20-kDa (P18) polypeptide (Fig. 1A), which showed identity to a β subunit of a ring-hydroxylating terminal dioxygenase (32). The results indicated that the 20-kDa protein was most highly expressed, showing a sevenfold increase when M. vanbaalenii PYR-1 was exposed to fluoranthene compared to the other PAH substrate inducers (Fig. 1B). To investigate the nucleotide sequence and expression of the gene encoding the PAH dioxygenase, two degenerate primers (P18-F and P18-R) were designed based on the amino acid sequences of the internal peptides and used to amplify a 109-bp fragment from the genomic DNA. The deduced amino acid sequence encoded by the amplified DNA fragment correctly contained the peptide sequence on which the degenerate primers were based. Using this DNA fragment as a probe, a series of plaque hybridizations were done to screen the M. vanbaalenii PYR-1 genomic library. Four different phagemids with a positive signal were isolated. Among the positive clones, a phagemid, designated pNCK31 (see the supplemental material), was shown to contain a 3,703-bp BamHI insert, whose nucleotide sequence was determined. As shown in the physical map in Fig. 2, four open reading frames (ORFs) were found in the sequence region. Two ORFs, designated nidA3 and nidB3, exhibited homology to the genes encoding the α and β subunits of the dioxygenase components of bacterial aromatic-ring-hydroxylating dioxygenases. The translated amino acid sequence of the β subunit was in perfect agreement with the MS-generated de novo peptide sequences (LRHYLTNVR, SGEDGDEYR, TSSDWAEDPPSR, VTSNALLVR, AETVSAER, AGGPGFSER, SMHLSDNYTSLK, and TTVPGADPP). The calculated molecular mass of NidB3 (21 kDa) is similar to the observed molecular mass of the P18 polypeptide, which was examined by 2-DE (32).
FIG. 1.
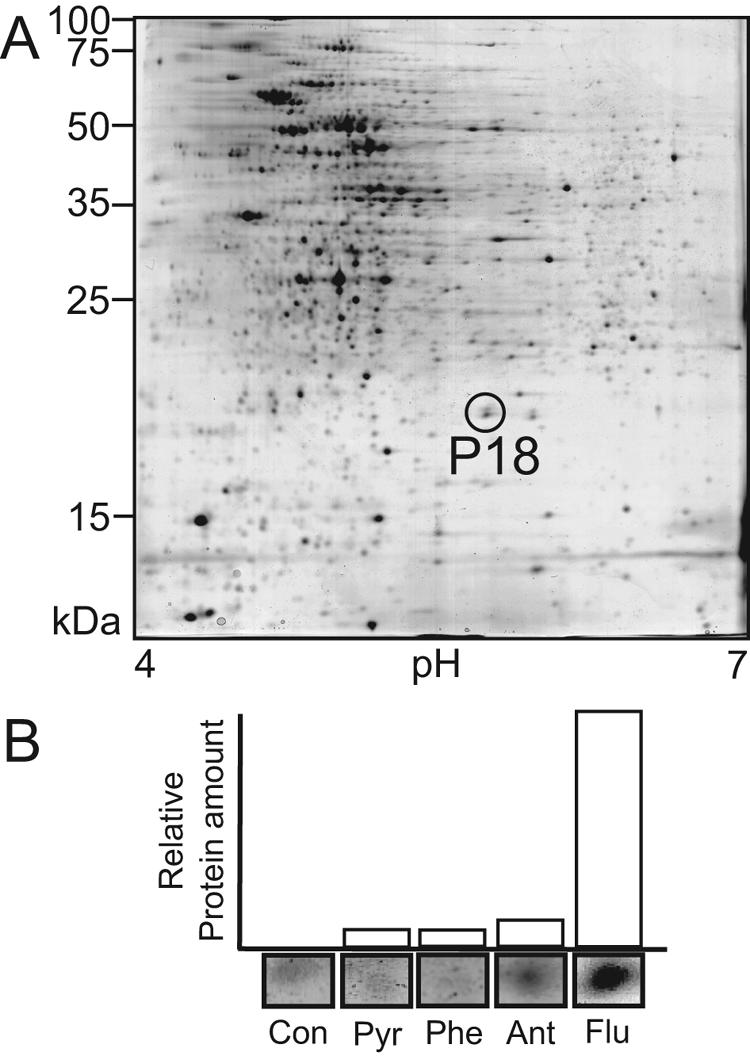
(A) 2-DE protein expression profile of M. vanbaalenii PYR-1 induced by PAH fluoranthene. (B) Relative synthesis rates of the 20-kDa polypeptide from 2-DE analysis of M. vanbaalenii PYR-1 during growth with glucose (Con), pyrene (Pyr), phenanthrene (Phe), anthracene (Ant), and fluoranthene (Flu). The inset below each bar shows a 2-DE spot image of the corresponding P18 polypeptide from each condition.
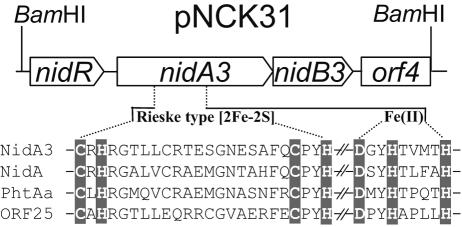
FIG. 2.
Physical map of the phagemid clone pNCK31 carrying nidA3B3 genes and conserved sequence alignment of the four α subunits of ring-hydroxylating dioxygenase discovered in M. vanbaalenii PYR-1. The amino acid residues involved in binding to the Rieske-type [2Fe-2S] cluster and to the mononuclear Fe(II) atom are indicated by highlighted characters.
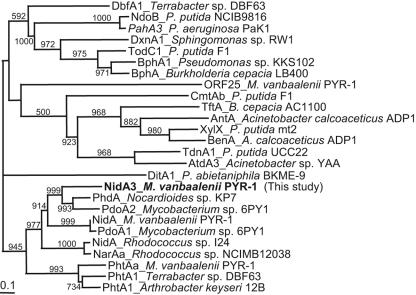
Analysis of NidA3 showed the consensus sequences for the Rieske-type [2Fe-2S] cluster binding region, CXHX17CX2H, and the catalytic nonheme iron, DX2HX3H (and Asp 372), which are well conserved among Rieske nonheme iron dioxygenases (43). Figure 2 presents a detailed amino acid sequence alignment for the conserved residues of NidA3 with the three other α subunits recently identified from M. vanbaalenii PYR-1. Although NidA3 and NidB3 share common sequence features with other terminal dioxygenases, the overall identity of the NidA3B3 sequence with those of other dioxygenases was moderate. The α subunit of a phenanthrene dioxygenase from Nocardioides sp. strain KP7 showed the highest degree of sequence similarity to NidA3 (61.7%). It also exhibited 52 to 60% identity with that of several Mycobacterium spp. (6, 14, 38, 44, 56) and 37 to 48% with Rhodococcus spp. (36, 40, 65), whereas other well-known α subunits from Pseudomonas and Sphingomonas spp. revealed levels of sequence identity lower than 32%. The enzyme showed 52% sequence homology with the previously reported NidA from M. vanbaalenii PYR-1. The NidB3 protein exhibited 46% identity at most to the β subunit from Mycobacterium spp. and about 42% to those from strain KP7 and other Rhodococcus spp. A sequence comparison of NidA3 with three other α subunits from M. vanbaalenii PYR-1 and other known α subunits of ring-hydroxylating dioxygenases is presented in Fig. 3. In this phylogenetic analysis, NidA3 clustered with groups of the α oxygenase subunits that catalyzed the hydroxylation of PAHs.
FIG. 3.
Phylogenetic tree of NidA3 obtained from alignment with related proteins. The protein sequences of the 26 α subunits of ring-hydroxylating dioxygenases, including NidA3, are classified. The multiple-alignment analysis was performed with the PHYLIP software package, and the phylogenetic unrooted tree was drawn by using TreeView. The numbers on branches refer to the percentage confidence, estimated by a bootstrap analysis with 1,000 replications. Scale bar, percentage divergence. GenBank accession numbers for the sequences are as follows: DbfA1, AB054975; NdoB, M23914; PahA3, D84146; DxnA1, X72850; TodC1, J04996; BphA1, D17319; BphA, M86348; ORF25, AY365117; CmtAb, U24215; TftA, U11420; AntA, AF071556; XylX, M64747; BenA, AF009224; TdnA1, D85415; AtdA3, D86080; DitA1, AF119621; PhdA, AB017794; PdoA2, AJ494743; NidA, AY365117; PdoA1, AJ494745; NidA, AF121905; NarAa, AF082663; PhtAa, AY365117; PhtA1, AB084235; PhtA1, AF331043.
Two additional ORFs were found adjacent to nidA3B3. In the 291-bp upstream region of nidA3B3, an ORF (orf1) designated nidR, coding for a 17.5-kDa protein, was detected. The orf1 sequence exhibits some similarities to certain bacterial putative MarR-family transcriptional regulators. The MarR-like proteins are a diverse group of transcriptional regulators, some of which are known to respond to aromatic compounds or are involved in the gene regulation for metabolism of aromatic compounds (11). An additional partial ORF (orf4) was also found to be located immediately following nidA3B3 with the same transcriptional direction. orf4 seemed to encode a protein similar to alcohol dehydrogenase, which is sometimes clustered with dioxygenase genes (15, 40, 42, 56, 63, 65). The organization of the genes nidA3 and nidB3, encoding terminal dioxygenase α and β subunits, respectively, was different from the previously identified dioxygenase nidAB of M. vanbaalenii PYR-1, where the gene order was found to be the reverse (30). The presence of the putative MarR-family regulator (nidR) has not been reported for other Mycobacterium spp., such as strain 6PY1 (38) or S65 (56), or for Nocardioides sp. KP7 (53) or Rhodococcus sp. I24 (65).
Functional analysis of nidA3B3.
To test whether the NidA3B3 proteins conferred any aromatic-ring-hydroxylating abilities to E. coli, the genes were cloned into pET-17b to construct the expression vector pNCK33. The E. coli strain transformed with pNCK33 seemed to actively express the NidA3B3 enzyme; however, no PAH degradation activity was detected. It appeared that the enzyme system was inactive, probably due to the lack of an electron carrier system, which often consists of reductase and ferredoxin, two components normally needed for electron transfer to the terminal dioxygenase. Since none of these genes were found in the 3.7-kb clone, the second plasmid, providing the electron transfer components from another system, was introduced into E. coli BL21(DE3)(pNCK33). For this purpose, the plasmid pBRCD, containing the cistrons encoding the ferredoxin (phdC) and ferredoxin reductase (phdD) of the phenanthrene dioxygenase from Nocardioides sp. strain KP7 (38), was used. The plasmid pBRCD is a derivative of pBBR11MCS-5 (37) and compatible with pET-17b. The enzyme activity was then achieved by coexpressing these two plasmids, which resulted in the hydroxylation of aromatic substrates.
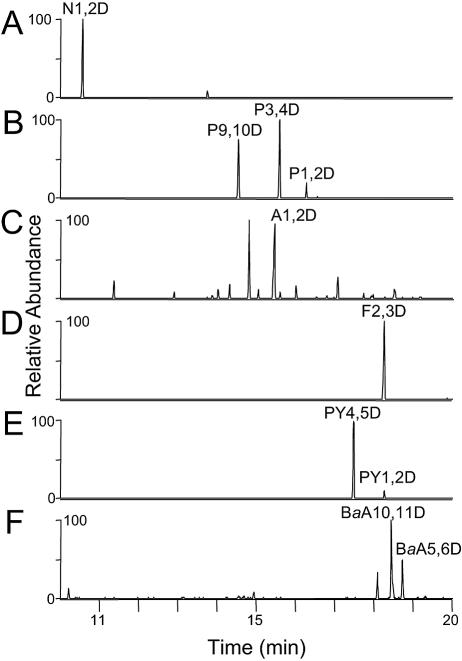
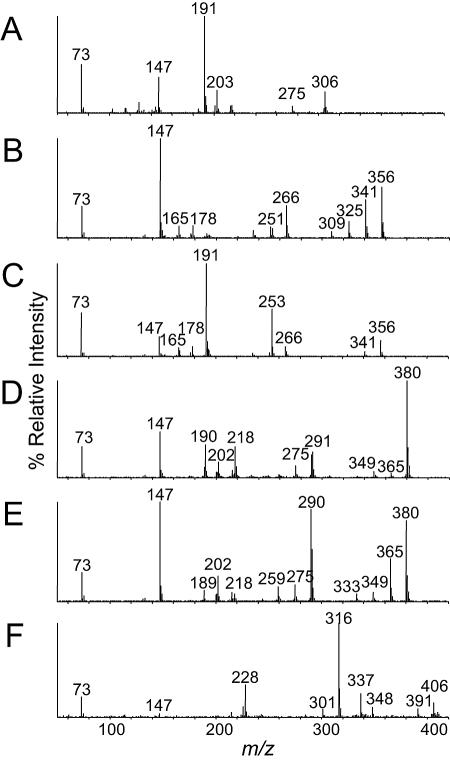
To determine the specificity of the NidA3B3 enzyme, phthalate, biphenyl, naphthalene, phenanthrene, anthracene, pyrene, fluoranthene, and benz[a]anthracene were tested (Table 2). The accumulated products formed from aromatic substrates were analyzed by HPLC and GC-MS (Fig. 4). The observed PAH intermediates in this study were compared to authentic compounds and to the previously identified initial PAH catabolic intermediates proposed from the biotransformation experiments using the wild-type M. vanbaalenii PYR-1 whole cells. The ethyl acetate-extractable metabolites were analyzed by GC-MS as trimethylsilylated derivatives. In many cases, since cis-PAH dihydrodiol metabolites easily undergo dehydration during analysis, the phenolic compounds detected are interpreted as the thermal dehydration products. In addition, dihydroxylated PAH (diols) compounds were also detected as PAH metabolic products, which are considered to be catalyzed by a nonspecific dehydrogenase(s) from the host E. coli. As shown in Table 2, the relative conversion percentages for initial oxidation of PAHs showed differences in substrate preference. Dihydroxylated metabolites of phthalate and biphenyl were not detected in the GC-MS and HPLC assays. Figure 4 shows GC-extracted ion chromatograms of naphthalene, phenanthrene, anthracene, fluoranthene, pyrene, and benz[a]anthracene degradation products from E. coli (DE3)(pNCK33)(pBRCD). In the analysis for naphthalene biotransformation (Fig. 4A), the detected metabolite had the same GC retention time (10.58 min), GC-MS properties (molecular ion at m/z 306) (Fig. 5A), and UV characteristics (λmax, 262 nm) as authentic cis-1,2-dihydroxy-1,2-dihydronaphthalene (26). Small amounts of 1- and 2-naphthol (molecular ion at m/z 216) gave an identical GC retention time and mass spectral fragment ions identical to authentic compounds (Table 1). GC-MS analysis of the neutral extracts of phenanthrene metabolites produced by the recombinant E. coli cells gave two major (14.56 min and 15.61 min) and one minor (16.29 min) compound (Fig. 4B) that give a molecular ion at m/z 356 [M+·] (Fig. 5B) that is consistent with trimethylsilylated derivative of phenanthrene cis-dihydrodiol (34). These results are consistent with the previous reports in which M. vanbaalenii PYR-1 was proposed to generate three cis-dihydrodiols bearing hydroxyl groups at the 1,2-, 3,4-, and 9,10- positions (34, 35, 47). The UV absorption spectra of the two major compounds (44% and 21%) corresponded to those of phenanthrene 3,4- and 9,10-dihydrodiol, respectively (3, 47, 61). The minor component (17%) was tentatively identified as phenanthrene cis-1,2-dihydrodiol. Small amounts of 2- and 3-hydroxyphenanthrene and two phenanthrenediols were also detected and considered to be formed by the dehydration and dehydrogenation, respectively, of the dihydrodiols (Table 1). Approximately 85% of the phenanthrene added was converted to hydroxylated metabolites (Table 1). For anthracene bioconversion studies (Fig. 4C), GC-MS analysis of the derivatized ethyl acetate neutral extract showed a major metabolite (22%) eluting at 15.5 min with a molecular ion at m/z 356 and fragment ions (Fig. 5C) identical to authentic anthracene cis-1,2-dihydrodiol. HPLC analysis indicated that the metabolite has the same UV absorption properties as those reported for anthracene cis-1,2-dihydrodiol (47). Two other minor metabolites with molecular ions at m/z 356 were also detected at 14.02 min and 15.05 min in the GC-MS analysis (Fig. 4C). These compounds could be other dihydrodiol isomers; however, the position of substitution could not be determined due to limited material and the lack of authentic standards. Two additional metabolites with apparent molecular ions at m/z 354 in the GC-MS analysis were presumed to be anthracenediols formed from dehydrogenation of anthracene cis-dihydrodiols. Anthraquinone ([M+·] at m/z 208) was also detected as an autoxidation product. In the fluoranthene experiment, the GC-MS analysis of the trimethylsilylated product showed a major compound (61%) at 18.27 min (Fig. 4D), which is consistent with fluoranthene cis-dihydrodiol (molecular ion at m/z 380 in Fig. 5D). HPLC and UV spectral analysis indicated that this metabolite had a UV absorption spectrum similar to that reported for fluoranthene cis-2,3-dihydrodiol (52). As shown in Table 2, we also detected compounds whose mass spectra were similar to those of monohydroxyfluoranthene ([M+·] at m/z 290) and fluoranthenediols ([M+·] at m/z 378) which we considered to be derived from dehydration of fluoranthene cis-dihydrodiol. Thus, with fluoranthene as the substrate, the sum (>99) of the areas of all these metabolite peaks was the highest of the studied aromatic compounds. Fluoranthene was not detected in the culture medium after overnight incubation. In the pyrene analysis, two metabolites (17.49 min and 18.27 min) were determined by GC-MS to have the same molecular ion at m/z 380 (Fig. 4E and 5E), which is consistent with a pyrene cis-dihydrodiol (34). The compound eluting at 17.49 min had the same retention time as authentic pyrene cis-4,5-dihydrodiol. The predominant metabolite peak (53%) further detected by HPLC and had a UV spectrum identical to that of pyrene cis-4,5-dihydrodiol (2, 21). The minor metabolite at 18.27 min was tentatively identified as pyrene cis-1,2-dihydrodiol based on the symmetry of the pyrene structure and previous biotransformation studies in M. vanbaalenii PYR-1 (34). Two pyrenols ([M+·] at m/z 290) and two pyrenediols ([M+·] at m/z 378) were also detected, and the former compounds are believed to be dehydrated products from cis-pyrene dihydrodiols. GC-MS analysis of the benz[a]anthracene extract revealed two trimethylsilylated metabolites at 18.45 min and 18.70 min that gave molecular ions at m/z 406 (Fig. 4F and 5F). The mass spectra are consistent with benz[a]anthracene dihydrodiols (46). HPLC analysis indicated that the major compound eluting at 18.4 min had a UV absorption spectrum identical to that of benz[a]anthracene cis-10,11-dihydrodiol (46). The compound at 18.7 min (Fig. 4F) had a GC retention time identical to that of authentic benz[a]anthracene cis-5,6-dihydrodiol. In addition, we found benz[a]anthracene dione ([M+·] at m/z 258), which was due to autoxidation of benz[a]anthracene.
TABLE 2.
GC-MS and HPLC data for the oxidation products showing PAH selectivity of NidA3B3 as expressed in E. coli
| Compound | Product identificationa | Retention time (min) | M+b | Conversion %c |
|---|---|---|---|---|
| Naphthalene | Naphthalene cis-1,2-dihydrodiol | 10.58 | 306 | 16d |
| 1-Naphthol | 9.18 | 216 | ∼1 | |
| 2-Naphthol | 9.49 | 216 | ∼2 | |
| Phenanthrene | Phenanthrene cis-3,4-dihydrodiol | 14.56 | 356 | 44 |
| Phenanthrene cis-9,10-dihydrodiol | 15.61 | 356 | 21 | |
| Phenanthrene cis-1,2-dihydrodiol | 16.29 | 356 | 17 | |
| 3-Hydroxyphenanthrene | 15.66 | 266 | 2.6 | |
| 2-Hydroxyphenanthrene | 16.03 | 266 | 0.7 | |
| Phenanthrene-9,10-diol | 16.62 | 354 | 0.3 | |
| Phenanthrenediol | 17.93 | 354 | 0.2 | |
| Anthracene | Anthracene dihydrodiol | 14.02 | 356 | 1 |
| Anthracene dihydrodiol | 15.05 | 356 | 1 | |
| Anthracene cis-1,2-dihydrodiol | 15.50 | 356 | 22 | |
| Monohydroxyanthracene | 13.75 | 266 | 1 | |
| Anthracenediol | 16.02 | 354 | <1 | |
| Anthracenediol | 17.08 | 354 | 5 | |
| Fluoranthene | Fluoranthene cis-2,3-dihydrodiol | 18.27 | 380 | 61 |
| Monohydroxyfluoranthene | 18.18 | 290 | <1 | |
| Monohydroxyfluoranthene | 18.39 | 290 | ∼1 | |
| Monohydroxyfluoranthene | 18.49 | 290 | 33 | |
| Fluoranthenediol | 19.89 | 378 | ∼4 | |
| Fluoranthenediol | 20.38 | 378 | <1 | |
| Pyrene | Pyrene cis-4,5-dihydrodiol | 17.49 | 380 | 53 |
| Pyrene cis-1,2-dihydrodiol | 18.27 | 380 | 4 | |
| Monohydroxypyrene | 15.10 | 290 | ∼1 | |
| Monohydroxypyrene | 15.20 | 290 | ∼2 | |
| Pyrenediol | 19.41 | 378 | ∼4 | |
| Pyrenediol | 19.90 | 378 | <1 | |
| Benz[a]anthracene | Benz[a]anthracene cis-10,11-dihydrodiol | 18.45 | 406 | ∼1 |
| Benz[a]anthracene cis-5,6-dihydrodiol | 18.70 | 406 | Trace |
Product identity based on GC-MS analysis of trimethylsilyl derivatives.
Molecular ion, mass to charge ratio (m/z).
Based on the GC-MS peak area of the detected metabolites.
Percentage of the total HPLC peak area.
FIG. 4.
Extracted ion chromatograms of the trimethylsilylated metabolites formed from transformed E. coli BL21(DE3)(pNCK33)(pBRCD) cells incubated with naphthalene (A), phenanthrene (B), anthracene (C), fluoranthene (D), pyrene (E), and benz[a]anthracene (F). Metabolite abbreviations are as follows: naphthalene cis-1,2-dihydrodiol, N1,2D; phenanthrene cis-1,2-dihydrodiol, P1,2D; phenanthrene cis-3,4-dihydrodiol, P3,4D; phenanthrene cis-9,10-dihydrodiol, P9,10D; anthracene cis-1,2-dihydrodiol, A1,2D; fluoranthene cis-2,3-dihydrodiol, F2,3D; pyrene cis-1,2-dihydrodiol, PY1,2D; pyrene cis-4,5-dihydrodiol, PY4,5D; benz[a]anthracene cis-5,6-dihydrodiol, BaA5,6D; benz[a]anthracene cis-10,11-dihydrodiol, BaA10,11D.
FIG. 5.
EI mass spectra of the dihydrodiol trimethylsilyl ethers: naphthalene cis-dihydrodiol (A), phenanthrene cis-dihydrodiol (B), anthracene cis-dihydrodiol (C), fluoranthene cis-dihydrodiol (D), pyrene cis-dihydrodiol (E), and benz[a]anthracene cis-dihydrodiol (F).
RT-PCR assay of the dioxygenase gene.
To determine whether the dioxygenase genes nidA3B3 are actually transcribed in response to PAHs, RT-PCR experiments were conducted with total RNA isolated from induced (grown in the presence of naphthalene, anthracene, phenanthrene, pyrene, fluoranthene, and benz[a]anthracene) and uninduced (grown in the presence of sorbitol) M. vanbaalenii PYR-1 (Fig. 6). With the use of primer set RnidA3-F3 and RnidB3-R3, an intercistronic fragment of 469 bp was amplified from the total RNA extracted from all cells grown with PAHs except for benz[a]anthracene, while the product was barely amplified from cells grown with sorbitol. The primer set for the amplification of 16S rRNA (R16S-F2 and R16S-R2) used as a control gave the expected PCR results (826 bp) for each experiment. These results suggested that growth of M. vanbaalenii PYR-1 in the presence of pyrene or fluoranthene leads to an increase in nidA3B3 transcription.
FIG. 6.
Transcriptional analysis to compare levels of nidA3B3 mRNA in M. vanbaalenii PYR-1 cells grown with sorbitol (uninduced) or with PAHs (induced). For reference purposes, the PCR signal obtained with M. vanbaalenii PYR-1 genomic DNA is also shown. The length of DNA size standards (1,000 and 500 bp) in lane M and the sizes of transcript (826 and 469 bp) are indicated. Lanes 1 and 2, RT-PCR products from total RNA from PYR-1 cells grown with sorbitol; lanes 3 and 4, RT-PCR products from total RNA from cells grown with naphthalene; lanes 5 and 6, RT-PCR products from cells grown with anthracene; lanes 7 and 8, RT-PCR products from cells grown with phenanthrene; lanes 9 and 10, RT-PCR products from cells grown with fluoranthene; lanes 11 and 12, RT-PCR products from cells grown with pyrene; lanes 13 and 14, RT-PCR products from cells grown with benz[a]anthracene; lanes 15 and 16, PCR products from M. vanbaalenii PYR-1 genomic DNA. Samples in lanes 1, 3, 5, 7, 9, 11, 13, and 15 were with the primer set for nidA3B3 and lanes 2, 4, 6, 8, 10, 12, 14, and 16 were with the primer set for 16 rRNA.
DISCUSSION
It has been suggested that M. vanbaalenii PYR-1 may possess multiple copies of several different dioxygenase genes besides nidAB because of its ability to degrade a broad range of aromatic substrates (30, 47). This prediction was supported by the recent evidence for the existence of at least two more dioxygenase genes and the cloning of two copies of new dioxygenase genes (phtAaAb, Orf25, and Orf26) in M. vanbaalenii PYR-1 (6, 58, 59). Proteome approaches resulted in the presence of another β subunit of a terminal dioxygenase protein, which was upregulated upon exposure of M. vanbaalenii PYR-1 to several PAHs (Fig. 1) (32). In this study, we felt that the α subunit, which is typically coupled with the β subunit, would be discovered by gene walking methodologies.
As revealed in the sequence analysis, transcription of these genes was thought to be coupled, since the start codon of nidB3 overlaps with the stop codon of nidA3. This was further confirmed by an RT-PCR experiment, which showed that RNA extracts derived from cells grown with naphthalene, anthracene, phenanthrene, pyrene, and fluoranthene gave amplified products of the anticipated size, whereas RNA from cells grown with sorbitol barely amplified the gene product. From these results, we concluded that nidA3B3 is induced in response to PAHs other than benz[a]anthracene but not sorbitol, and these genes are therefore regulated at the transcriptional level. The results were consistent with the previous 2-DE investigation (32), in which the upregulation of NidB3 after exposure to PAHs in M. vanbaalenii PYR-1 was demonstrated at the protein level.
In the segment of cloned DNA flanking nidA3B3, we found two additional ORFs, designated orf1 and orf4, which are oriented in the same direction. The product of orf1 (NidR) showed similarity to various members of the putative MarR-family transcriptional regulators. Because many proteins belonging to this family have been known to control a variety of microbial functions, including antibiotic resistance and catabolism of various substrates (11, 22, 55), the gene product might be involved in the transcriptional control of nidA3B3. The deduced amino acid sequence of orf4 immediately downstream of nidA3B3 was similar to those of short-chain alcohol dehydrogenases, to which most cis-dihydrodiol dehydrogenase genes involved in the initial dehydrogenation of PAH cis-dihydrodiols normally belong. Since the dehydrogenase genes are often located in the neighboring region of dioxygenase genes (15, 40, 42, 56, 63, 65), the ORF4 protein may be the cis-dihydrodiol dehydrogenase enzyme that converts the products of NidA3B3 to their dihydroxy compounds.
Recombinant dioxygenase NidA3B3 produced in E. coli required coexpression of a [3Fe-4S] type ferredoxin and reductase gene for activity. Previously, the nidAB dioxygenase genes of M. vanbaalenii PYR-1 were reported to be functional in an E. coli system, which was not provided with ferredoxin and reductase components (30). At the time, as has been demonstrated by other investigations (39, 41, 57), it was speculated that the NidAB dioxygenase subunits might borrow the ferredoxin and reductase components of another electron transport system in the recombinant E. coli cells. In the current study, despite the resting cells of recombinant E. coli BL21(DE3)(pNCK33) containing nidA3B3 being assumed to actively express the nidA3B3 genes, no enzyme activity was detected with PAH substrates, as determined by HPLC analysis, which indicates that an additional supplemental electron transport system must be required for enzyme activity. As revealed in the sequence comparison, NidA3B3 is similar to those Rieske nonheme iron dioxygenases from strains of Nocardioides, Mycobacterium, Rhodococcus, Terrabacter, and Arthrobacter spp. Interestingly, without exception, these dioxygenase genes are always found with genes for [3Fe-4S] type iron-sulfur ferredoxin, if there are any genes for ferredoxin components in the nearby region (GenBank accession numbers AB031319, AB048709, AY502076, AF331043, and AB084235 for Nocardioides sp. KP7, Rhodococcus sp. RHA1, Rhodococcus sp. DK17, Arthrobacter keyseri 12B, and Terrabacter sp. DBF63, respectively). Among them, phdCD, encoding a [3Fe-4S] type ferredoxin and ferredoxin reductase component from Nocardioides sp. KP7 (53), was previously used with the PdoA2B2 dioxygenase from Mycobacterium sp. 6PY1 (38) as well as with the respective dioxygenase PhdAB from Nocardioides sp. KP7 for the recovery of enzyme activity. It is also worth noting that surrogate [2Fe-2S] type ferredoxins did not result in functional enzymes when coupled to dioxygenases requiring [3Fe-4S] ferredoxin as an electron carrier protein (42, 62). NidA3B3 dioxygenase activity was reconstituted when coexpressed with pBRCD carrying phdCD from Nocardioides sp. KP7.
In the biotransformation test using E. coli, a range of PAHs was converted by the action of NidA3B3 dioxygenase to their corresponding cis-dihydrodiol compounds (Fig. 4). Monohydroxylated compounds were also identified as oxidized metabolites produced during biotransformation analysis. The compounds were presumed to be derived by nonenzymatic loss of water of the corresponding cis-dihydrodiols produced by NidA3B3. Small amounts of dihydroxy compounds were also detected as PAH metabolic products. In fact, we previously reported a similar experimental result with M. vanbaalenii PYR-1's first dioxygenase, NidAB, which showed the biotransformation of pyrene into 4,5-dihydroxypyrene (pyrene-4,5-diol) (30). Thus, we speculated that these dihydroxy compounds in the present study might be the product of a nonspecific dehydrogenation reaction catalyzed by an E. coli enzyme. As shown in Table 2, fluoranthene was the most preferred substrate followed by phenanthrene, but pyrene, anthracene, and naphthalene were also oxidized significantly. The results showing the NidA3B3 enzyme's highest substrate preference for fluoranthene support our previous 2-DE study (32), in which the NidB (P18) polypeptide showed the highest upregulation when PYR-1 was incubated with fluoranthene compared to other PAHs. It was sevenfold higher than the next highest inducer, pyrene.
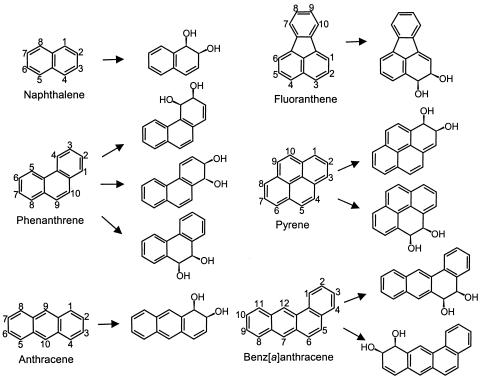
Based on the identification of isomeric cis-dihydrodiols formed during the degradation of phenanthrene, anthracene, benz[a]anthracene, pyrene, and benzo[a]pyrene by M. vanbaalenii PYR-1, it was proposed that this strain might have a dioxygenase with a relaxed substrate specificity for degrading a broad range of PAHs (34, 46-48). The results in this study indicate that this hypothesis has merit because the fourth dioxygenase, NidA3B3, extensively metabolized those representative (HMW) PAHs with positional hydroxylation, which resembled the pattern of PAH metabolism found in wild-type M. vanbaalenii PYR-1 (Fig. 7). Although the occurrence of bacterial species degrading HMW PAHs has been reported (6, 8, 10, 14, 38, 44, 56, 66), relatively little is known about HMW PAH dioxygenases. M. vanbaalenii PYR-1 was one of the first microorganisms found to utilize HMW PAHs (21). The nidA3B3 gene discussed in this investigation is the fourth copy of a terminal dioxygenase gene from M. vanbaalenii PYR-1, and the gene product NidA3B3 can oxidize PAHs, including HMW PAHs. To our knowledge, this is the first report to describe the dioxygenase which shows high enzyme activity for the four-ring HMW PAHs fluoranthene and pyrene.
FIG. 7.
Structures of products converted from aromatic substrates by recombinant E. coli cells.
Supplementary Material
Acknowledgments
We thank J. B. Sutherland, H. Chen, and F. Rafii for critical review of the manuscript and Y. Jouanneau, Unité Mixte de Recherche CEA-CNRS-Université Joseph Fourier, France, for providing plasmid pBRCD. We also thank R. E. Parales, University of California, Davis, for providing cis-1,2-dihydroxy-1,2-dihydronaphthalene, cis-1,2-dihydroxy-1,2-dihydroanthracene, and cis-2,3-dihydroxy-2,3-dihydrobiphenyl and Peter Fu, National Center for Toxicological Research, for cis-5,6-dihydroxy-5,6-dihydrobenz[a]anthracene for use as analytical standards.
This work was supported in part by an appointment to the Postgraduate Research Program at the National Center for Toxicological Research administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and the U.S. Food and Drug Administration.
Footnotes
Supplemental material for this article may be found at http://aem.asm.org/.
REFERENCES
- 1.Altschul, S. F., T. L. Madden, A. A. Schaffer, J. Zhang, Z. Zhang, W. Miller, and D. J. Lipman. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25:3389-3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bezalel, L., Y. Hadar, P. P. Fu, J. P. Freeman, and C. E. Cerniglia. 1996. Initial oxidation products in the metabolism of pyrene, anthracene, fluorene, and dibenzothiophene by the white rot fungus Pleurotus ostreatus. Appl. Environ. Microbiol. 62:2554-2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bezalel, L., Y. Hadar, P. P. Fu, J. P. Freeman, and C. E. Cerniglia. 1996. Metabolism of phenanthrene by the white rot fungus Pleurotus ostreatus. Appl. Environ. Microbiol. 62:2547-2553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boldrin, B., A. Tiehm, and C. Fritzsche. 1993. Degradation of phenanthrene, fluorene, fluoranthene, and pyrene by a Mycobacterium sp. Appl. Environ. Microbiol. 59:1927-1930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bouchez, M., D. Blanchet, and J. P. Vandecasteele. 1995. Degradation of polycyclic aromatic hydrocarbons by pure strains and by defined strain associations: inhibition phenomena and cometabolism. Appl. Microbiol. Biotechnol. 43:156-164. [DOI] [PubMed] [Google Scholar]
- 6.Brezna, B., A. A. Khan, and C. E. Cerniglia. 2003. Molecular characterization of dioxygenases from polycyclic aromatic hydrocarbon-degrading Mycobacterium spp. FEMS Microbiol. Lett. 223:177-183. [DOI] [PubMed] [Google Scholar]
- 7.Brezna, B., O. Kweon, R. L. Stingley, J. P. Freeman, A. A. Khan, B. Polek, R. C. Jones, and C. E. Cerniglia. Molecular characterization of cytochrome P450 genes in the polycyclic aromatic hydrocarbons degrading Mycobacterium vanbaalenii PYR-1. Appl. Microbiol. Biotechnol., in press. [DOI] [PubMed]
- 8.Churchill, S. A., J. P. Harper, and P. F. Churchill. 1999. Isolation and characterization of a Mycobacterium species capable of degrading three- and four-ring aromatic and aliphatic hydrocarbons. Appl. Environ. Microbiol. 65:549-552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean-Ross, D., J. D. Moody, J. P. Freeman, D. R. Doerge, and C. E. Cerniglia. 2001. Metabolism of anthracene by a Rhodococcus species. FEMS Microbiol. Lett. 204:205-211. [DOI] [PubMed] [Google Scholar]
- 10.Demaneche, S., C. Meyer, J. Micoud, M. Louwagie, J. C. Willison, and Y. Jouanneau. 2004. Identification and functional analysis of two aromatic-ring-hydroxylating dioxygenases from a Sphingomonas strain that degrades various polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 70:6714-6725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Egland, P. G., and C. S. Harwood. 1999. BadR, a new MarR family member, regulates anaerobic benzoate degradation by Rhodopseudomonas palustris in concert with AadR, an Fnr family member. J. Bacteriol. 181:2102-2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Felsenstein, J. 1993. PHYLIP (phylogenetic inference package) version 3.5c. Department of Genetics, University of Washington, Seattle, Wash. (Distributed by the author.)
- 13.Grosser, R. J., D. Warshawsky, and J. R. Vestal. 1991. Indigenous and enhanced mineralization of pyrene, benzo[a]pyrene, and carbazole in soils. Appl. Environ. Microbiol. 57:3462-3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Habe, H., M. Kanemitsu, M. Nomura, T. Takemura, K. Iwata, H. Nojiri, H. Yamane, and T. Omori. 2004. Isolation and characterization of an alkaliphilic bacterium utilizing pyrene as a carbon source. J. Biosci. Bioeng. 98:306-308. [DOI] [PubMed] [Google Scholar]
- 15.Habe, H., M. Miyakoshi, J. Chung, K. Kasuga, T. Yoshida, H. Nojiri, and T. Omori. 2003. Phthalate catabolic gene cluster is linked to the angular dioxygenase gene in Terrabacter sp. strain DBF63. Appl. Microbiol. Biotechnol. 61:44-54. [DOI] [PubMed] [Google Scholar]
- 16.Habe, H., and T. Omori. 2003. Genetics of polycyclic aromatic hydrocarbon metabolism in diverse aerobic bacteria. Biosci. Biotechnol. Biochem. 67:225-243. [DOI] [PubMed] [Google Scholar]
- 17.Hanahan, D. 1983. Studies on transformation of Escherichia coli with plasmids. J. Mol. Biol. 166:557-580. [DOI] [PubMed] [Google Scholar]
- 18.Heitkamp, M. A., and C. E. Cerniglia. 1988. Mineralization of polycyclic aromatic hydrocarbons by a bacterium isolated from sediment below an oil field. Appl. Environ. Microbiol. 54:1612-1614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heitkamp, M. A., and C. E. Cerniglia. 1989. Polycyclic aromatic hydrocarbon degradation by a Mycobacterium sp. in microcosms containing sediment and water from a pristine ecosystem. Appl. Environ. Microbiol. 55:1968-1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heitkamp, M. A., W. Franklin, and C. E. Cerniglia. 1988. Microbial metabolism of polycyclic aromatic hydrocarbons: isolation and characterization of a pyrene-degrading bacterium. Appl. Environ. Microbiol. 54:2549-2555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heitkamp, M. A., J. P. Freeman, D. W. Miller, and C. E. Cerniglia. 1988. Pyrene degradation by a Mycobacterium sp.: identification of ring oxidation and ring fission products. Appl. Environ. Microbiol. 54:2556-2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkins, J. R., and R. A. Cooper. 1988. Molecular cloning, expression, and analysis of the genes of the homoprotocatechuate catabolic pathway of Escherichia coli C. J. Bacteriol. 170:5317-5324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kanaly, R. A., and S. Harayama. 2000. Biodegradation of high-molecular-weight polycyclic aromatic hydrocarbons by bacteria. J. Bacteriol. 182:2059-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kelley, I., and C. E. Cerniglia. 1995. Degradation of a mixture of high-molecular-weight polycyclic aromatic hydrocarbons by a Mycobacterium strain, PYR-1. J. Soil Contam. 4:77-91. [Google Scholar]
- 25.Kelley, I., and C. E. Cerniglia. 1991. The metabolism of fluoranthene by a species of Mycobacterium. J. Ind. Microbiol. Biotechnol. 7:19-26. [Google Scholar]
- 26.Kelley, I., J. P. Freeman, and C. E. Cerniglia. 1990. Identification of metabolites from degradation of naphthalene by a Mycobacterium sp. Biodegradation 1:283-290. [DOI] [PubMed] [Google Scholar]
- 27.Kelley, I., J. P. Freeman, F. E. Evans, and C. E. Cerniglia. 1991. Identification of a carboxylic acid metabolite from the catabolism of fluoranthene by a Mycobacterium sp. Appl. Environ. Microbiol. 57:636-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kelley, I., J. P. Freeman, F. E. Evans, and C. E. Cerniglia. 1993. Identification of metabolites from the degradation of fluoranthene by Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 59:800-806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khan, A. A., S. J. Kim, D. D. Paine, and C. E. Cerniglia. 2002. Classification of a polycyclic aromatic hydrocarbon-metabolizing bacterium, Mycobacterium sp. strain PYR-1, as Mycobacterium vanbaalenii sp. nov. Int. J. Syst. Evol. Microbiol. 52:1997-2002. [DOI] [PubMed] [Google Scholar]
- 30.Khan, A. A., R. F. Wang, W. W. Cao, D. R. Doerge, D. Wennerstrom, and C. E. Cerniglia. 2001. Molecular cloning, nucleotide sequence, and expression of genes encoding a polycyclic aromatic ring dioxygenase from Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 67:3577-3585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim, S. J., J. Chun, K. S. Bae, and Y. C. Kim. 2000. Polyphasic assignment of an aromatic-degrading Pseudomonas sp., strain DJ77, in the genus Sphingomonas as Sphingomonas chungbukensis sp. nov. Int. J. Syst. Evol. Microbiol. 50:1641-1647. [DOI] [PubMed] [Google Scholar]
- 32.Kim, S. J., R. C. Jones, C. J. Cha, O. Kweon, R. D. Edmondson, and C. E. Cerniglia. 2004. Identification of proteins induced by polycyclic aromatic hydrocarbon in Mycobacterium vanbaalenii PYR-1 using two-dimensional polyacrylamide gel electrophoresis and de novo sequencing methods. Proteomics 4:3899-3908. [DOI] [PubMed] [Google Scholar]
- 33.Kim, Y. H., K. H. Engesser, and C. E. Cerniglia. 2003. Two polycyclic aromatic hydrocarbon o-quinone reductases from a pyrene-degrading Mycobacterium. Arch. Biochem. Biophys. 416:209-217. [DOI] [PubMed] [Google Scholar]
- 34.Kim, Y. H., J. P. Freeman, J. D. Moody, K. H. Engesser, and C. E. Cerniglia. 2005. Effects of pH on the degradation of phenanthrene and pyrene by Mycobacterium vanbaalenii PYR-1. Appl. Microbiol. Biotechnol. 67:275-285. [DOI] [PubMed] [Google Scholar]
- 35.Kim, Y. H., J. D. Moody, J. P. Freeman, K. H. Engesser, and C. E. Cerniglia. 2004. Evidence for the existence of PAH-quinone reductase and catechol-O-methyltransferase in Mycobacterium vanbaalenii PYR-1. J. Ind. Microbiol. Biotechnol. 31:507-516. [DOI] [PubMed] [Google Scholar]
- 36.Kitagawa, W., N. Kimura, and Y. Kamagata. 2004. A novel p-nitrophenol degradation gene cluster from a gram-positive bacterium, Rhodococcus opacus SAO101. J. Bacteriol. 186:4894-4902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kovach, M. E., P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop II, and K. M. Peterson. 1995. Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes. Gene 166:175-176. [DOI] [PubMed] [Google Scholar]
- 38.Krivobok, S., S. Kuony, C. Meyer, M. Louwagie, J. C. Willison, and Y. Jouanneau. 2003. Identification of pyrene-induced proteins in Mycobacterium sp. strain 6PY1: evidence for two ring-hydroxylating dioxygenases. J. Bacteriol. 185:3828-3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kurkela, S., H. Lehvaslaiho, E. T. Palva, and T. H. Teeri. 1988. Cloning, nucleotide sequence and characterization of genes encoding naphthalene dioxygenase of Pseudomonas putida strain NCIB9816. Gene 73:355-362. [DOI] [PubMed] [Google Scholar]
- 40.Larkin, M. J., C. C. Allen, L. A. Kulakov, and D. A. Lipscomb. 1999. Purification and characterization of a novel naphthalene dioxygenase from Rhodococcus sp. strain NCIMB12038. J. Bacteriol. 181:6200-6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Laurie, A. D., and G. Lloyd-Jones. 1999. The phn genes of Burkholderia sp. strain RP007 constitute a divergent gene cluster for polycyclic aromatic hydrocarbon catabolism. J. Bacteriol. 181:531-540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Martin, V. J., and W. W. Mohn. 1999. A novel aromatic-ring-hydroxylating dioxygenase from the diterpenoid-degrading bacterium Pseudomonas abietaniphila BKME-9. J. Bacteriol. 181:2675-2682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mason, J. R., and R. Cammack. 1992. The electron-transport proteins of hydroxylating bacterial dioxygenases. Annu. Rev. Microbiol. 46:277-305. [DOI] [PubMed] [Google Scholar]
- 44.Miller, C. D., K. Hall, Y. N. Liang, K. Nieman, D. Sorensen, B. Issa, A. J. Anderson, and R. C. Sims. 2004. Isolation and characterization of polycyclic aromatic hydrocarbon-degrading Mycobacterium isolates from soil. Microb. Ecol. 48:230-238. [DOI] [PubMed] [Google Scholar]
- 45.Moody, J. D., D. R. Doerge, J. P. Freeman, and C. E. Cerniglia. 2002. Degradation of biphenyl by Mycobacterium sp. strain PYR-1. Appl. Microbiol. Biotechnol. 58:364-369. [DOI] [PubMed] [Google Scholar]
- 46.Moody, J. D., J. P. Freeman, and C. E. Cerniglia. 2005. Degradation of benz[a]anthracene by Mycobacterium vanbaalenii strain PYR-1. Biodegradation 16:513-526. [DOI] [PubMed] [Google Scholar]
- 47.Moody, J. D., J. P. Freeman, D. R. Doerge, and C. E. Cerniglia. 2001. Degradation of phenanthrene and anthracene by cell suspensions of Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 67:1476-1483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moody, J. D., J. P. Freeman, P. P. Fu, and C. E. Cerniglia. 2004. Degradation of benzo[a]pyrene by Mycobacterium vanbaalenii PYR-1. Appl. Environ. Microbiol. 70:340-345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moody, J. D., P. P. Fu, J. P. Freeman, and C. E. Cerniglia. 2003. Regio- and stereoselective metabolism of 7,12-dimethylbenz[a]anthracene by Mycobacterium vanbaalenii PYR-1. Appl. Environ. Microbiol. 69:3924-3931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Page, R. D. 1996. TreeView: an application to display phylogenetic trees on personal computers. Comput. Appl. Biosci. 12:357-358. [DOI] [PubMed] [Google Scholar]
- 51.Rafii, F., A. L. Selby, R. K. Newton, and C. E. Cerniglia. 1994. Reduction and mutagenic activation of nitroaromatic compounds by a Mycobacterium sp. Appl. Environ. Microbiol. 60:4263-4267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rehmann, K., N. Hertkorn, and A. A. Kettrup. 2001. Fluoranthene metabolism in Mycobacterium sp. strain KR20: identity of pathway intermediates during degradation and growth. Microbiology 147:2783-2794. [DOI] [PubMed] [Google Scholar]
- 53.Saito, A., T. Iwabuchi, and S. Harayama. 2000. A novel phenanthrene dioxygenase from Nocardioides sp. strain KP7: expression in Escherichia coli. J. Bacteriol. 182:2134-2141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 55.Seoane, A. S., and S. B. Levy. 1995. Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J. Bacteriol. 177:3414-3419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sho, M., C. Hamel, and C. W. Greer. 2004. Two distinct gene clusters encode pyrene degradation in Mycobacterium sp. strain S65. FEMS Microbiol. Ecol. 48:209-220. [DOI] [PubMed] [Google Scholar]
- 57.Simon, M. J., T. D. Osslund, R. Saunders, B. D. Ensley, S. Suggs, A. Harcourt, W. C. Suen, D. L. Cruden, D. T. Gibson, and G. J. Zylstra. 1993. Sequences of genes encoding naphthalene dioxygenase in Pseudomonas putida strains G7 and NCIB 9816-4. Gene 127:31-37. [DOI] [PubMed] [Google Scholar]
- 58.Stingley, R. L., B. Brezna, A. A. Khan, and C. E. Cerniglia. 2004. Novel organization of genes in a phthalate degradation operon of Mycobacterium vanbaalenii PYR-1. Microbiology 150:3749-3761. [DOI] [PubMed] [Google Scholar]
- 59.Stingley, R. L., A. A. Khan, and C. E. Cerniglia. 2004. Molecular characterization of a phenanthrene degradation pathway in Mycobacterium vanbaalenii PYR-1. Biochem. Biophys. Res. Commun. 322:133-146. [DOI] [PubMed] [Google Scholar]
- 60.Studier, F. W., A. H. Rosenberg, J. J. Dunn, and J. W. Dubendorff. 1990. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 185:60-89. [DOI] [PubMed] [Google Scholar]
- 61.Sutherland, J. B., A. L. Selby, J. P. Freeman, F. E. Evans, and C. E. Cerniglia. 1991. Metabolism of phenanthrene by Phanerochaete chrysosporium. Appl. Environ. Microbiol. 57:3310-3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Takagi, T., H. Habe, T. Yoshida, H. Yamane, T. Omori, and H. Nojiri. 2005. Characterization of [3Fe-4S] ferredoxin DbfA3, which functions in the angular dioxygenase system of Terrabacter sp. strain DBF63. Appl. Microbiol. Biotechnol. 68:336-345. [DOI] [PubMed] [Google Scholar]
- 63.Takizawa, N., N. Kaida, S. Torigoe, T. Moritani, T. Sawada, S. Satoh, and H. Kiyohara. 1994. Identification and characterization of genes encoding polycyclic aromatic hydrocarbon dioxygenase and polycyclic aromatic hydrocarbon dihydrodiol dehydrogenase in Pseudomonas putida OUS82. J. Bacteriol. 176:2444-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Thompson, J. D., T. J. Gibson, F. Plewniak, F. Jeanmougin, and D. G. Higgins. 1997. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25:4876-4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Treadway, S. L., K. S. Yanagimachi, E. Lankenau, P. A. Lessard, G. Stephanopoulos, and A. J. Sinskey. 1999. Isolation and characterization of indene bioconversion genes from Rhodococcus strain I24. Appl. Microbiol. Biotechnol. 51:786-793. [DOI] [PubMed] [Google Scholar]
- 66.Vila, J., Z. Lopez, J. Sabate, C. Minguillon, A. M. Solanas, and M. Grifoll. 2001. Identification of a novel metabolite in the degradation of pyrene by Mycobacterium sp. strain AP1: actions of the isolate on two- and three-ring polycyclic aromatic hydrocarbons. Appl. Environ. Microbiol. 67:5497-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang, R. F., D. Wennerstrom, W. W. Cao, A. A. Khan, and C. E. Cerniglia. 2000. Cloning, expression, and characterization of the katG gene, encoding catalase-peroxidase, from the polycyclic aromatic hydrocarbon-degrading bacterium Mycobacterium sp. strain PYR-1. Appl. Environ. Microbiol. 66:4300-4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.