Abstract
Using the established commercial system Sherlock (MIDI, Inc.), cellular fatty acid methyl ester analysis for differentiation among Burkholderia cepacia complex species was proven. The identification key based on the diagnostic fatty acids is able to discern phenotypically related Ralstonia pickettii and Pandoraea spp. and further distinguish Burkholderia pyrrocinia, Burkholderia ambifaria, and Burkholderia vietnamiensis.
At present, nine Burkholderia species are combined into the Burkholderia cepacia complex (BCC): B. cepacia, B. multivorans, B. cenocepacia, B. stabilis, B. vietnamiensis, B. dolosa, B. ambifaria, B. anthina, and B. pyrrocinia (2, 12-14). In some cases, the severity of BCC infection is closely connected with one particular species. The proper identification of likely BCC isolates is crucial for patients suffering from cystic fibrosis (CF) (9).
Identification of BCC isolates to the species level based on the results obtained by genotypic methods, such as restriction fragment length polymorphism analysis of the recA gene or 16S rRNA genes (17, 18) and recA gene-based PCR (4), is mostly done with research tools provided in reference centers. Phenotypic testing is still frequently used in routine laboratories. Because of constantly changing taxonomy, such identification could be inaccurate. Databases of commonly used commercial kits are often incomplete and may even be insufficient to properly discern BCC isolates from other phylogenetically related isolates or from Ralstonia pickettii and Pandoraea spp. commonly isolated from clinical human sources.
Cellular fatty analysis using semiautomated gas chromatography by the microbial identification system (MIS) Sherlock (MIDI, Inc., Newark, Del.) is a relatively rapid and cost-effective method widely available and suitable for clinical laboratories. Identification based on the species-specific differences in fatty acid composition in cell lipidic structure was proved to be a good taxonomic marker (15). Contrary to phenotyping, previous studies have shown that analysis of cellular fatty acid components was able to distinguish between the genera Burkholderia, Ralstonia, and Pandoraea (1, 19). Fatty acids of the BCC are rather uniform; nevertheless, species-specific differences useful to separate B. anthina or B. ambifaria from B. cepacia and B. cenocepacia were observed (2, 13). So far there are no reports that include and compare the fatty acid compositions of all known BCC species and related taxa under standardized culture conditions. In this study, we tested whether comparing fatty acid profiles obtained on MIS Sherlock could yield the ability to sufficiently discriminate between BCC species and other closely related taxa.
Type strains (Tables 1 and 2) and a set including 47 well-characterized clinical isolates recovered during a half-year survey in 2002 in two town hospitals and several ambulatory medical practices in our region (Ostrava, Czech Republic) were studied. Isolates (one per patient) originated primarily from non-CF patients (sputum, blood cultures, wound and vaginal swabs, urine, catheter isolates, and isolates from the hospital environment; two B. cenocepacia strains were obtained from the sputum of CF patients). Tentative phenotypic identification as BCC was performed by commercially available biochemical tests (NEFERMtest24; Pliva-Lachema Diagnostika) and additional conventional testing (6; unpublished data). BCC species were determined by recA gene-based PCR assay as described previously (4). Differences in fatty acid compositions of other organisms (Inquilinus limosus and members of the genera Cupriavidus, Achromobacter, and Delftia) which were previously found in the respiratory tracts of cystic fibrosis patients or misidentified as BCC or Pandoraea spp. (1, 12) were also compared (Tables 1 and 2).
TABLE 1.
Fatty acid compositions of all nine BCC type strainsa
| Fatty acidb or comparison | Fatty acid composition (%)c or species identification of:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B. cepacia DSM 7288T |
B. multivorans
|
B. cenocepacia
|
B. stabilis
|
B. vietnamiensis DSM 11319T | B. dolosa LMG 18943T | B. ambifaria CCUG 44356T | B. anthina CCUG 46047T | B. pyrrocinia DSM 10685T | ||||
| DSM 13243T | 17 isolates | CCM 4899T | 15 isolates | CCM 4900T | 15 isolates | |||||||
| Saturated | ||||||||||||
| 12:0 | trd (1) | NDe | ND | ND | tr (2 CFi) | ND | ND | 0.1 ± 0.0 (2) | tr (2) | 0.9 ± 0.1 | ND | 2.8 ± 0.2 |
| 14:0 | 3.8 ± 0.1 | 5.2 ± 0.1 | 3.6 ± 0.2 | 3.4 ± 0.2 | 3.7 ± 0.2 | 4.1 ± 0.1 | 3.9 ± 0.2 | 4.0 ± 0.2 | 3.9 ± 0.2 | 2.9 ± 0.2 | 4.1 ± 0.2 | 0.5 ± 0.0 |
| 16:0 | 27.2 ± 2.1 | 30.2 ± 1.3 | 22.9 ± 1.1 | 23.4 ± 2.4 | 19.0 ± 1.0 | 19.7 ± 0.7 | 20.5 ± 0.9 | 21.2 ± 0.2 | 26.0 ± 6.6 | 17.4 ± 0.6 | 20.5 ± 1.2 | 18.1 ± 0.2 |
| 17:0 | 0.3 ± 0.0 (2) | ND | 0.2 (1) | ND | 0.2 ± 0.0 (3) | 0.3 ± 0.1 | 0.2 ± 0.0 (6) | 0.4 ± 0.1 | ND | 0.1 (1) | 0.2 ± 0.0 (2) | 0.4 ± 0.1 |
| 18:0 | 1.5 ± 0.1 | 0.1 ± 0.1 (2) | 1.4 ± 0.2 | 0.9 ± 0.2 | 0.9 ± 0.2 | 0.8 ± 0.1 | 0.9 ± 0.1 | 3.6 ± 0.5 | 0.6 ± 0.2 | 0.6 ± 0.1 | 0.6 ± 0.1 | 0.7 ± 0.0 |
| Unsaturated | ||||||||||||
| 13:1 AT 12-13 | 1.2 ± 1.0 | 1.0 ± 0.8 | 0.8 ± 0.7 (6) | 1.6 (1) | 0.7 ± 0.6 (9) | 1.0 ± 0.8 | 0.8 ± 0.8 (5) | 1.1 ± 0.9 | 1.0 ± 0.8 | 1.2 ± 1.0 | 1.2 ± 1.0 | 0.6 ± 0.6 |
| 16:1ω5c | 0.2 (1) | 0.7 ± 0.0 | 0.2 (1) | ND | 0.2 ± 0.0 (5) | 0.2 ± 0.1 (2) | 0.3 ± 0.3 (4) | ND | 0.4 ± 0.2 | 0.3 ± 0.1 | 0.4 ± 0.1 (2) | 0.3 ± 0.1 |
| 16:1ω7cf | 18.7 ± 5.2 | 27.6 ± 2.0 | 17.4 ± 1.4 | 12.6 ± 1.3 | 16.3 ± 2.9 | 18.6 ± 2.9 | 18.5 ± 2.5 | 23.2 ± 1.6 | 17.8 ± 1.7 | 13.5 ± 4.1 | 17.9 ± 6.7 | 19.1 ± 2.9 |
| 18:1ω7c | 26.9 ± 4.4 | 16.2 ± 1.1 | 40.8 ± 2.1 | 29.3 ± 2.3 | 37.4 ± 3.8 | 35.1 ± 1.1 | 33.0 ± 4.1 | 31.8 ± 0.6 | 27.9 ± 6.7 | 36.0 ± 1.6 | 32.6 ± 1.7 | 37.3 ± 1.2 |
| Hydroxy | ||||||||||||
| 10:0 3OH | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 12:0 2OH | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND | ND |
| 14:0 2OH | ND | 0.1 ± 0.0 (2) | 0.1 (1) | 0.5 ± 0.1 | 0.3 ± 0.1 (9) | ND | ND | ND | 0.2 ± 0.1 (2) | ND | ND | ND |
| 14:0 3OHg | 5.1 ± 0.1 | 5.8 ± 0.3 | 4.7 ± 0.4 | 5.0 ± 0.1 | 5.0 ± 0.4 | 5.5 ± 0.2 | 4.8 ± 0.4 | 5.0 ± 0.2 | 5.4 ± 0.0 | 4.8 ± 0.4 | 5.7 ± 0.4 | 4.9 ± 0.5 |
| 16:0 2OH | 0.4 ± 0.1 | 0.3 ± 0.0 | 0.5 ± 0.2 (12) | 0.6 ± 0.0 | 1.0 ± 0.3 | 1.0 ± 0.2 | 1.2 ± 0.2 | 0.7 ± 0.0 | 0.8 ± 0.4 | 0.8 ± 0.1 | 0.8 ± 0.2 | 1.4 ± 0.1 |
| 16:0 3OH | 3.2 ± 0.6 | 3.3 ± 0.7 | 3.8 ± 0.5 | 3.9 ± 0.8 | 3.8 ± 0.4 | 4.3 ± 1.7 | 3.9 ± 0.3 | 3.6 ± 0.8 | 3.4 ± 0.8 | 4.0 ± 0.7 | 3.5 ± 1.1 | 3.4 ± 0.7 |
| 16:1 2OH | 0.5 ± 0.0 | 1.7 ± 0.2 | 0.5 ± 0.2 (11) | 0.5 ± 0.1 | 1.1 ± 0.3 | 1.0 ± 0.1 | 1.6 ± 0.4 | 0.8 ± 0.1 | 1.2 ± 0.4 | 0.8 ± 0.1 | 0.7 ± 0.1 | 1.7 ± 0.2 |
| 18:1 2OH | 0.9 ± 0.3 | 0.6 ± 0.1 | 0.6 ± 0.3 (11) | 1.3 ± 0.2 | 2.3 ± 0.7 | 2.0 ± 0.2 | 2.2 ± 0.7 | 1.2 ± 0.1 | 1.5 ± 1.0 | 2.1 ± 0.1 | 2.2 ± 0.5 | 2.2 ± 0.1 |
| Cyclo | ||||||||||||
| 17:0 cyclo | 6.7 ± 4.2 | 5.1 ± 2.1 | 2.1 ± 1.2 | 11.1 ± 0.6 | 5.3 ± 3.1 | 3.9 ± 2.6 | 6.5 ± 3.2 | 2.6 ± 1.2 | 6.1 ± 2.0 | 10.2 ± 1.7 | 6.9 ± 4.3 | 4.9 ± 1.9 |
| 19:0 cyclo ω8c | 3.4 ± 2.4 | 1.3 ± 0.3 | 1.7 ± 0.9 | 6.6 ± 0.8 | 3.3 ± 2.2 | 2.1 ± 1.5 | 2.6 ± 1.3 | 0.7 ± 0.4 | 3.4 ± 1.3 | 5.1 ± 2.4 | 2.9 ± 2.4 | 1.2 ± 0.7 |
| Library comparison | ||||||||||||
| TSBA50h | B. cepacia/ B. gladioli | Serratia odorifera/Raoultella terrigena | B. cepacia/ B. caryophylli | B. cepacia | B. cepacia | B. cepacia | B. cepacia | B. cepacia | B. cepacia/ B. gladioli | B. cepacia | B. cepacia | B. pyrrocinia/ B. cepacia |
Fatty acid compositions of species related to the BCC are given in Table 2. Fatty acids present in all species in an amount lower than 0.5% were omitted. Cultivation conditions for the TSBA50 standard library were used.
In a designation, the first value is number of C atoms in the fatty acid. The number following the colon is the number of double-bound C atoms. The value following the letter ω signifies the position in the carbon chain (counted from opposite the carboxyl group). The number preceding OH indicates the number of OH groups. cyclo, cyclopropane circle; c, cis conformation.
Values are means ± standard deviations. Values in parentheses show how many times the fatty acid appeared. Mean values were calculated either for three independent analyses (type strains) or for several isolates (one isolate each for B. multivorans, B. cenocepacia, and B. stabilis) by using FAME data from the standard MIS Sherlock composition report. With the aid of MS Excel software, the standard deviations were calculated according to the formula [sgrt (Σ(x − x′)2/(n − 1))]. Values in italics are characteristic of B. multivorans, B. cenocepacia, or B. stabilis, but sometimes similar values are found with other strains. Values in boldface type clearly differentiate individual species or genera.
tr, trace amount (fatty acid was present in an amount lower than 0.1%).
ND, not detected.
Detected as Summed feature 3, which can comprise C15:0 iso 2OH, C16:1ω7c, or any combination of these fatty acids. Summed feature 3 probably corresponds to C16:1ω7c, as this fatty acid has been reported for Burkholderia species (2).
Detected as Summed feature 2, which can comprise C14:0 3OH, C16:1 iso I, an unidentified fatty acid with an equivalent chain length value of 10.928, C12:0 ALDE, or any combination of these fatty acids. Summed feature 2 probably corresponds to C14:0 3OH, as this fatty acid has been reported for Burkholderia species (2).
All similarity indexes were calculated as a good library match (0.7 or higher). The similarity index is a software-generated calculation of the distance, in multidimensional space, between the profile of the unknown and the mean profile of the closest library entry. Organisms given could come out as a good library match solely or together as the two closest matches. Apart from individual BCC species, Pandoraea norimbergensis, Pandoraea sputorum, and Inquilinus limosum are not included in the TSBA50 library.
The two isolates in which trace amounts of fatty acid were detected were both from patients with cystic fibrosis.
TABLE 2.
Fatty acid compositions of species related to the BCCa
| Fatty acidb or comparison | Fatty acid composition (%)c or species identification of:
|
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| B. gladioli DSM 4285T | P. norimberg. DSM 11628T | P. apista CCM 4976T | P. pnomenusa CCM 4978T | P. pulmonic. CCM 4979T | P. spurorum CCM 4980T | R. pickettii DSM 6297T | C. necatori | Achromobacter xylosoxidans xylosoxidansi | Delftia acidovoransi | I. limosum DSM 16000T | ||
| Saturated | ||||||||||||
| 12:0 | trd (1) | 2.7 ± 0.2 | 1.8 ± 0.1 | 1.5 ± 0.1 | 2.0 ± 0.1 | 1.7 ± 0.2 | 0.1 (1) | ne | 0.5 | 2.5 | NDf | |
| 14:0 | 3.9 ± 0.2 | 0.3 ± 0.0 | 0.5 ± 0.1 | 0.7 ± 0.0 | 0.4 ± 0.0 | 0.5 ± 0.1 | 4.4 ± 0.2 | 2.8 | 2.1 | 0.7 | ND | |
| 16:0 | 24.9 ± 0.8 | 17.8 ± 0.8 | 24.7 ± 1.3 | 21.0 ± 0.8 | 22.7 ± 0.7 | 21.3 ± 1.3 | 26.7 ± 2.1 | 24.5 | 34.7 | 33.2 | 1.4 | |
| 17:0 | 0.2 ± 0.1 | 0.5 ± 0.1 | 0.2 (1) | 0.3 ± 0.0 | 0.3 ± 0.0 | 0.3 ± 0.0 | 0.5 ± 0.1 | 1.6 | n | n | 4.2 | |
| 18:0 | 1.1 ± 0.2 | 0.9 ± 0.0 | 0.5 ± 0.1 | 0.9 ± 0.1 | 1.0 ± 0.1 | 0.6 ± 0.1 | 0.4 ± 0.0 | 0.4 | 2.6 | n | 1.3 | |
| Unsaturated | ||||||||||||
| 13:1 AT 12-13 | 0.5 ± 0.6 | 0.7 ± 0.6 | 0.6 ± 0.6 | 1.2 ± 1.2 (2) | 0.7 ± 0.7 | 0.3 ± 0.3 | ND | n | n | n | ND | |
| 16:1ω5c | ND | 0.3 ± 0.1 | 0.3 ± 0.1 | 0.2 (1) | 0.3 (1) | 0.4 ± 0.1 | 0.2 ± 0.0 (2) | n | n | n | 0.3 | |
| 16:1ω7cg | 16.0 ± 4.0 | 22.4 ± 3.9 | 23.8 ± 2.1 | 20.2 ± 1.8 | 20.8 ± 2.0 | 13.5 ± 1.4 | 33.7 ± 1.1 | 35.5 | 31.4 | 40.1 | 3.4 | |
| 18:1ω7c | 28.0 ± 2.73 | 33.6 ± 1.6 | 27.6 ± 1.6 | 34.1 ± 1.1 | 34.0 ± 0.8 | 30.8 ± 4.7 | 20.2 ± 1.6 | 22.3 | 9.7 | 17.2 | 46.7 | |
| Hydroxy | ||||||||||||
| 10:0 3OH | ND | ND | ND | ND | ND | ND | ND | n | n | 2.5 | ND | |
| 12:0 2OH | ND | 0.3 ± 0.1 | 0.9 ± 0.0 | 0.8 ± 0.1 | 0.7 ± 0.0 | 1.2 ± 0.1 | ND | n | 2.5 | n | ND | |
| 14:0 2OH | ND | ND | ND | 0.2 (1) | 0.1 (1) | 0.1 ± 0.0 | 0.8 ± 0.3 | 2.5 | 2.2 | n | ND | |
| 14:0 3OHh | 4.5 ± 0.3 | 5.7 ± 0.4 | 5.2 ± 0.0 | 5.0 ± 0.5 | 5.4 ± 0.2 | 5.4 ± 0.0 | 6.4 ± 0.1 | 9.2 | 7.6 | n | 1.2 | |
| 16:0 2OH | 0.9 ± 0.2 | 0.2 ± 0.1 (2) | 0.2 (1) | 0.3 ± 0.1 | 0.2 (1) | 0.5 ± 0.1 | 0.2 (1) | n | n | n | 0.8 | |
| 16:0 3OH | 3.9 ± 0.6 | 4.1 ± 0.5 | 4.4 ± 0.6 | 3.8 ± 1.0 | 4.5 ± 0.5 | 4.5 ± 0.2 | 0.1 (1) | n | n | n | 1.3 | |
| 16:1 2OH | 1.0 ± 0.1 | 0.4 ± 0.1 | 0.5 ± 0.2 | 0.5 ± 0.1 | 0.3 (1) | 0.4 ± 0.2 | 1.2 ± 0.5 | n | n | n | ND | |
| 18:1 2OH | 1.3 ± 0.2 | 0.8 ± 0.2 | 0.8 ± 0.2 | 1.8 ± 0.5 | 0.6 ± 0.1 | 1.9 ± 0.4 | 3.0 ± 0.6 | n | n | n | 15.9 | |
| Cyclo | ||||||||||||
| 17:0 cyclo | 8.5 ± 3.1 | 6.1 ± 3.3 | 6.2 ± 2.0 | 5.6 ± 0.8 | 4.2 ± 1.5 | 10.8 ± 3.0 | 0.8 ± 0.2 | n | 5.6 | 3.3 | ND | |
| 19:0 cyclo ω8c | 5.1 ± 3.1 | 2.7 ± 1.6 | 2.0 ± 1.0 | 2.5 ± 0.3 | 2.1 ± 1.0 | 5.4 ± 2.0 | ND | n | n | n | 9.6 | |
| Library comparison | ||||||||||||
| TSBA50j | B. gladioli/ B. cepacia | B. pyrrocinia/ P. pnomenusa | P. apista | P. pnomenusa | P. pnomenusa/ P. pulmonicola | B. cepacia | R. pickettii/ Erwinia chrysanthemi biotype II | Library match | Library match | Library match | No match | |
Fatty acid compositions of BCC type strains are given in Table 1. Fatty acids present in all species in an amount lower than 0.5% were omitted. Cultivation conditions for the TSBA50 standard library were used.
In a designation, the first value is number of C atoms in the fatty acid. The number following the colon is the number of double-bound C atoms. The value following the letter ω signifies the position in the carbon chain (counted from opposite the carboxyl group). The number preceding OH indicates the number of OH groups. cyclo, cyclopropane circle; c, cis conformation.
Values are means ± standard deviations. Values in parentheses show how many times the fatty acid appeared. Mean values were calculated either for three independent analyses (type strains) or for several isolates (one isolate each for B. multivorans, B. cenocepacia, and B. stabilis) by using FAME data from the standard MIS Sherlock composition report. With the aid of MS Excel software, the standard deviations were calculated according to the formula [sgrt (Σ(x − x′)2/(n − 1))]. Values in boldface type clearly differentiate individual species or genera.
tr, trace amount (fatty acid was present in an amount lower than 0.1%).
n, not mentioned.
ND, not detected.
Detected as Summed feature 3, which can comprise C15:0 iso 2OH, C16:1ω7c, or any combination of these fatty acids. Summed feature 3 probably corresponds to C16:1ω7c, as this fatty acid has been reported for Burkholderia species (2).
Detected as Summed feature 2, which can comprise C14:0 3OH, C16:1 iso I, an unidentified fatty acid with an equivalent chain length value of 10.928, C12:0 ALDE, or any combination of these fatty acids. Summed feature 2 probably corresponds to C14:0 3OH, as this fatty acid has been reported for Burkholderia species (2).
Data were taken from the TSBA50 library of MIS Sherlock. Data for Cupriavidus necator are included in the library as Ralstonia eutropha.
All similarity indexes were calculated as a good library match (0.7 or higher). The similarity index is a software-generated calculation of the distance, in multidimensional space, between the profile of the unknown and the mean profile of the closest library entry. Organisms given could come out as a good library match solely or together as the two closest matches. Apart from individual BCC species, Pandoraea norimbergensis, Pandoraea sputorum, and Inquilinus limosum are not included in the TSBA50 library.
Because the fatty acid composition in the bacterial cell is influenced to a great extent by different culture conditions, the manufacturer's culture conditions given for each MIS library must always be followed to get usable identification results. To prepare whole-cell fatty acid methyl esters (FAME), one loop of fresh growing cultures of studied strains was harvested. Fatty acids were then saponified by means of heated alkaline hydrolysis, converted to the methyl esters, and extracted following the instructions of the MIS Sherlock operating manual (MIS operating manual, version 4.0; MIDI, Inc. Newark, Del.; partly available online). Samples were analyzed on a 25-m by 0.2-mm phenylmethyl siloxane capillary column (HP Ultra 2) using an HP 6890 gas chromatograph equipped with a flame-ionization detector. Gained FAME profiles could generally be compared in two standard libraries, CLIN50 and TSBA50. According to the quadrant plate-streaking technique, where four quadrants with different densities of cells are created, the third quadrant, which usually contains cells in the desired late log phase of growth, was harvested. For the CLIN50 library, strains were grown on trypticase soy broth (BBL, Becton Dickinson) solidified with agar (Difco, Becton Dickinson) and 5% sheep blood for 24 h at 35°C; for the TSBA50 library, strains were grown on trypticase soy broth solidified with agar for 24 h at 28°C. From our experience, some BCC strains (B. ambifaria type strain and some B. cenocepacia clinical isolates) are not able to grow sufficiently after 24 h (in all four quadrants). These strains may require a longer period to obtain sufficient growth in comparison with other members of the BCC. To see how much it can influence the fatty acid results, we prolonged cultivation conditions for the TSBA50 library up to 48 h. All incubations were performed in ambient air.
As with other commercially available BCC differentiation systems, the usefulness of standard libraries included in MIS Sherlock software is limited. From nine species belonging to the BCC, it offers entries of only B. cepacia (CLIN50 and TSBA50 libraries) and B. pyrrocinia (TSBA50 library). Generally, application of both libraries is appropriate for differentiation of phenotypically related R. pickettii and Pandoraea spp. from BCC. The TSBA50 comprises a wider database supplemented with more recently described new species than the CLIN50 library, so the TSBA50 library is more favorable for the standard library comparison than the CLIN50. Also, the addition of blood to the CLIN50 standard medium enhances the variability of the fatty acid patterns. After the comparison of fatty acid profiles with the TSBA50 library, all BCC strains were identified, mostly as B. cepacia, B. gladioli, or B. pyrrocinia (Tables 1 and 2). Interestingly, only the B. multivorans type strain was misidentified as Serratia odorifera or Raoultella terrigena; all other B. multivorans clinical isolates were “correctly” recognized as B. cepacia. That was probably because the B. multivorans type strain showed a larger amount of C16:1ω7c than of C18:1ω7c fatty acid, which was just the opposite of results obtained with B. multivorans clinical isolates.
The possibility of identifying BCC species was further evaluated by the detailed inspection of FAME profiles. There, such fatty acids which would be characteristic for BCC as a group or even for one particular BCC species and whose amount would be minimally influenced by cultivation conditions were searched. Analysis showed that the chosen incubation period and medium affected mostly the amounts of unsaturated and cyclopropane fatty acids (data not shown). This could be the reason why we did not confirm previously published results regarding B. ambifaria (2). In our study, the amounts of the unsaturated C16:1ω7c and C18:1ω7c fatty acids and of the cyclopropane fatty acids C17:0 and C19:0ω8c detected in B. ambifaria were comparable with amounts detected in other BCC members. Differences can be easily explained by the biosynthesis pathway of the cyclopropane fatty acids. In the stationary phase of growth, a part of the unsaturated fatty acids is transferred into the corresponding cyclopropane fatty acid, i.e., C18:1ω7c to C19:0 cyclo ω7c, by insertion of a methyl group through S-adenosylmethionine into the double bond of the fatty acid, leading to the cyclic cis-11,12-methylenoctadecanoic acid (lactobacillic acid). The sums of unsaturated fatty acids and the corresponding cyclopropane fatty acids were nearly the same amounts in both studies, and these summed values were independent of the length of cultivation. Identification of BCC to the species level due to the amounts of unsaturated or cyclopropane fatty acids proved to be inappropriate, notably in view of potentially diverse laboratory practices. If insufficient growth of a strain appears, it is recommended to prolong the cultivation period or to change the harvested quadrant. Which possibility will be chosen depends on the laboratory's way of carrying out the standard cultivation.
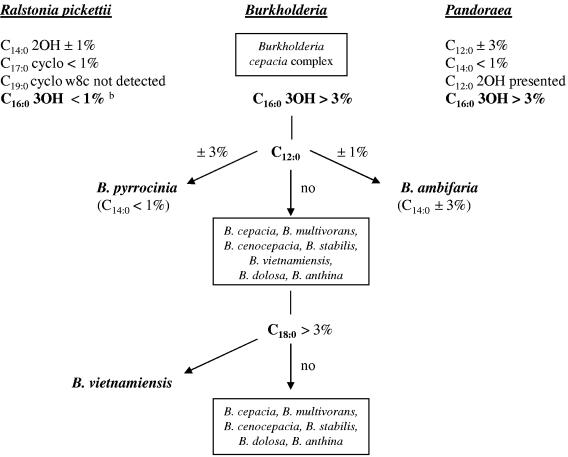
Saturated C12:0, C14:0, and C18:0 fatty acids were considered better markers because they were stable and did not alter significantly with different cultivation conditions. Based on the quantitative differences of these diagnostic fatty acids, we tried to construct an identification key (Fig. 1). The key was constructed to be able to evaluate FAME profiles gained after all cultivation methods were used.
FIG. 1.
Key for identification of presumptive BCC isolates according to their fatty acid compositions. Fatty acid compositions obtained by the cultivation of strains along all conditions described (TSBA50 and CLIN50 standard libraries and TSBA50 prolonged up to 48 h) can be evaluated using this identification key. “b” indicates that this composition is also valid for Cupriavidus spp. (C. necator, C. gilardii, C. pauculus, C. taiwanensis, and C. respiraculi) previously found in the respiratory tracts of CF patients (formerly described as Ralstonia or Wautersia) (see references 3, 11, 16, and 19).
On the basis of the obtained results, we must rectify the previous assumption about the genera Burkholderia and Pandoraea. Significant percentages of C12:0 are no longer essential for their differentiation (1), because B. pyrrocinia contained an amount of C12:0 fatty acid comparable with all Pandoraea type strains (Tables 1 and 2). C12:0 was also detected in B. ambifaria (13) and noticed in B. hospita (5) and B. sordidicola (7). Furthermore, C14:0 fatty acid was detected in B. pyrrocinia in an amount comparable with amounts detected in all Pandoraea species. Thus, the previously described absence of C14:0 cannot easily distinguish members of the genus Pandoraea from the genera Ralstonia and Burkholderia (1). In our study, differentiation of Pandoraea spp. and BCC from R. pickettii was possible due to the presence of C16:0 3OH. For Pandoraea spp., the presence of C12:0 2OH, which was not detected in BCC, B. gladioli, or R. pickettii at all, was typical. Also, the fatty acids C16:0 2OH and C16:1 2OH in Pandoraea spp. were commonly below 0.5%, whereas amounts of named fatty acids found in BCC were higher (about 1%) and corresponded with B. gladioli or other described Burkholderia species (for an example, see reference 7).
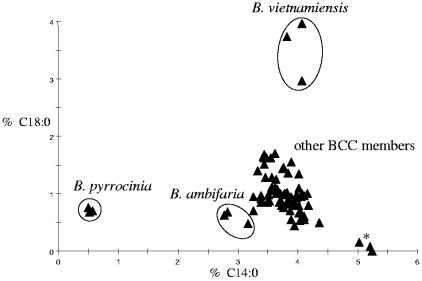
According to the proposed identification key, the possibility of differentiating B. pyrrocinia, B. ambifaria, and B. vietnamiensis species was tested by the two-dimensional plot cluster analysis technique (Fig. 2), which uses principal component analysis to separate groups of samples in n-dimensional spaces to find relationships among fatty acid profiles (Library Generation Software LGS user's manual, version 4.5; MIDI, Inc., Newark, Del.). According to the created two-dimensional plot, it is obvious that observed differences in the amounts of both C14:0 and C18:0 fatty acids distinguish B. pyrrocinia, B. ambifaria, and B. vietnamiensis from the remaining BCC species. In the two-dimensional plot, the fatty acid profile comparison was not based on C12:0, because this fatty acid was not present in all strains tested. Just the presence of C12:0 fatty acid additionally improved the distinction of B. ambifaria and B. pyrrocinia from other BCC members.
FIG. 2.
Relationships among fatty acid profiles depicted in the two-dimensional plot according to the diagnostic fatty acids C14:0 and C18:0, which were present in all BCC type strains and 47 clinical isolates. The asterisk indicates the B. multivorans type strain. For fatty acid profiles used in the two-dimensional plot, strains were cultivated along conditions for the TSBA50 standard library. Reproducibility was ensured by three independent analyses of each strain tested.
Only three of the most frequently isolated BCC species, B. multivorans, B. cenocepacia, and B. stabilis, were obtained from clinical samples. Some minor fatty acids which will be helpful to differentiate B. multivorans, B. cenocepacia, and B. stabilis from the rest of the BCC were noticed (Tables 1 and 2). B. multivorans contained the smallest amount of C18:1 2OH; in B. cenocepacia, C14:0 2OH was strikingly often detected; and in B. stabilis, a repeated occurrence of C17:0 was observed. Because clinical isolates of only three species were obtained, construction of their own library was not practicable. Type strain data are not satisfactory for the judgment if the suggested differentiation of BCC species is valid. To confirm the BCC identification key requires the inclusion of a much wider set of well-characterized strains isolated from various sources and not only from human clinical material (8). To prove our results, testing of a full set of clinical isolates of all BCC species will be needed. Even if B. pyrrocinia, B. ambifaria, or B. vietnamiensis were not present in the tested set of clinical strains studied, their differentiation from other BCC members according to the amount of diagnostic fatty acids was feasible also on the basis of the previously reported results (2, 13, 14).
If there is reason to hesitate about BCC identification, an identification key which offers verification through simple checking of single diagnostic fatty acids could be applied as an additional tool to the standard MIS Sherlock library comparison. FAME analysis could be helpful to confirm B. ambifaria or B. pyrrocinia, which can generate aberrant 16S rRNA gene-based restriction profiles (17). The appearance of C12:0 fatty acid in recA restriction fragment length polymorphism types Se6 and AR of B. pyrrocinia (10) enables the use of fatty acid analysis as a method of choice for differentiation from B. cepacia. The two named species can positively react with the same recA gene primers (18). These primers were originally designed for B. cepacia; a recA gene-based PCR assay specific only for B. pyrrocinia has not been published yet.
Nevertheless, the differentiation of BCC members to the species level using the identification key is limited; only B. vietnamiensis, B. ambifaria, and B. pyrrocinia could be recognized solely on the basis of the fatty acid analysis. Identification of other BCC members, e.g., the most transmissible species among CF patients, B. cenocepacia, B. multivorans, and B. stabilis, frequently isolated from the respiratory tracts of CF patients must be achieved by more accurate genotypic methods.
REFERENCES
- 1.Coenye, T., E. Falsen, B. Hoste, M. Ohlén, J. Goris, J. R. W. Govan, K. Kersters, and P. Vandamme. 2000. Description of Pandoraea gen. nov. with Pandoraea apista sp. nov., Pandoraea pulmonicola sp. nov., Pandoraea pnomenusa sp. nov., Pandoraea sputorum sp. nov. and Pandoraea norimbergensis comb. nov. Int. J. Syst. Evol. Microbiol. 50:887-899. [DOI] [PubMed] [Google Scholar]
- 2.Coenye, T., E. Mahenthiralingam, D. Henry, J. J. LiPuma, S. Laevens, M. Gillis, D. P. Speert, and P. Vandamme. 2001. Burkholderia ambifaria sp. nov., a novel member of the Burkholderia cepacia complex including biocontrol and cystic fibrosis-related isolates. Int. J. Syst. Evol. Microbiol. 51:1481-1490. [DOI] [PubMed] [Google Scholar]
- 3.Coenye, T., P. Vandamme, and J. LiPuma. 2003. Ralstonia respiraculi sp. nov., isolated from respiratory tract of cystic fibrosis patients. Int. J. Syst. Evol. Microbiol. 53:1339-1342. [DOI] [PubMed] [Google Scholar]
- 4.Dřevínek, P., H. Hrbáčková, O. Cinek, J. Bartošová, O. Nyč, A. Nemec, and P. Pohunek. 2002. Direct PCR detection of Burkholderia cepacia complex and identification of its genomovars by using sputum as source of DNA. J. Clin. Microbiol. 40:3485-3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Goris, J., W. Dejonghe, E. Falsen, E. De Clerck, B. Geeraerts, A. Willems, E. M. Top, P. Vandamme, and P. De Vos. 2002. Diversity of transconjugants that acquired plasmid pJP4 or pEMT1 after inoculation of a donor strain in the A- and B-horizon of an agricultural soil and description of Burkholderia hospita sp. nov. and Burkholderia terricola sp. nov. Syst. Appl. Microbiol. 25:340-352. [DOI] [PubMed] [Google Scholar]
- 6.Henry, D. A., E. Mahenthiralingam, P. Vandamme, T. Coenye, and D. P. Speert. 2001. Phenotypic methods for determining genomovar status of the Burkholderia cepacia complex. J. Clin. Microbiol. 39:1073-1078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim, Y. W., K. S. Baik, K. S. Han, S. B. Kim, and K. S. Bae. 2003. Burkholderia sordidicola sp. nov., isolated from the white-rot fungus Phanerochaete sordida. Int. J. Syst. Evol. Microbiol. 53:1631-1636. [DOI] [PubMed] [Google Scholar]
- 8.Mahenthiralingam, E., T. Coenye, J. W. Chung, D. P. Speert, J. R. W. Govan, P. Taylor, and P. Vandamme. 2000. Diagnostically and experimentally useful panel of strains from the Burkholderia cepacia complex. J. Clin. Microbiol. 38:910-913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Speert, D. P., D. Henry, P. Vandamme, M. Corey, and E. Mahenthiralingam. 2002. Epidemiology of Burkholderia cepacia complex in patients with cystic fibrosis, Canada. Emerg. Infect. Dis. 8:181-187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Storms, V., N. Van Den Vreken, T. Coenye, E. Mahenthiralingam, J. J. LiPuma, M. Gillis, and P. Vandamme. 2004. Polyphasic characterisation of Burkholderia cepacia-like isolates leading to the emended description of Burkholderia pyrrocinia. Syst. Appl. Microbiol. 27:517-526. [DOI] [PubMed] [Google Scholar]
- 11.Vandamme, P., and T. Coenye. 2004. Taxonomy of the genus Cupriavidus: a tale of lost and found. Int. J. Syst. Evol. Microbiol. 54:2285-2289. [DOI] [PubMed] [Google Scholar]
- 12.Vandamme, P., B. Holmes, T. Coenye, J. Goris, E. Mahethiralingam, J. J. LiPuma, and J. R. W. Govan. 2003. Burkholderia cenocepacia sp. nov.—a new twist to an old story. Res. Microbiol. 154:91-96. [DOI] [PubMed] [Google Scholar]
- 13.Vandamme, P., D. Henry, T. Coenye, S. Nzula, M. Vancanneyt, J. J. LiPuma, D. P. Speert, J. R. W. Govan, and E. Mahenthiralingam. 2002. Burkholderia anthina sp. nov. and Burkholderia pyrrocinia, two additional Burkholderia cepacia complex bacteria, may confound results of new molecular diagnostic tools. FEMS Immunol. Med. Microbiol. 33:143-149. [DOI] [PubMed] [Google Scholar]
- 14.Vandamme, P., E. Mahenthiralingam, B. Holmes, T. Coenye, B. Hoste, P. De Vos, D. Henry, and D. P. Speert. 2000. Identification and population structure of Burkholderia stabilis sp. nov. (formerly Burkholderia cepacia genomovar IV). J. Clin. Microbiol. 38:1042-1047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vandamme, P., B. Pot, M. Gillis, P. De Vos, K. Kersters, and J. Swings. 1996. Polyphasic taxonomy, a consensus approach to bacterial systematics. Microbiol. Rev. 60:407-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaneechoutte, M., P. Kämpfer, T. De Baere, E. Falsen, and G. Verschraegen. 2004. Wautersia gen. nov., a novel genus accommodating the phylogenetic lineage including Ralstonia eutropha and related species, and proposal of Ralstonia [Pseudomonas] syzygii (Roberts et al. 1990) comb. nov. Int. J. Syst. Evol. Microbiol. 54:317-327. [DOI] [PubMed] [Google Scholar]
- 17.Vermis, K., C. Vandekerckhove, H. J. Nelis, and P. A. R. Vandamme. 2002. Evaluation of restriction fragment length polymorphism analysis of 16S rDNA as a tool for genomovar characterization within the Burkholderia cepacia complex. FEMS Microbiol. Lett. 214:1-5. [DOI] [PubMed] [Google Scholar]
- 18.Vermis, K., T. Coenye, E. Mahenthiralingam, H. J. Nelis, and P. Vandamme. 2002. Evaluation of species-specific recA-based PCR tests for genomovar level identification within the Burkholderia cepacia complex. J. Med. Microbiol. 51:937-940. [DOI] [PubMed] [Google Scholar]
- 19.Yabuuchi, E., Y. Kosako, I. Yano, H. Hotta, and Y. Nishiuchi. 1995. Transfer of two Burkholderia and an Alcaligenes species to Ralstonia gen. nov.: proposal of Ralstonia pickettii (Ralston, Palleroni and Doudoroff 1973) comb. nov., Ralstonia solanacearum (Smith 1896) comb. nov. and Ralstonia eutropha (Davis 1969) comb. nov. Microbiol. Immunol. 39:897-904. [DOI] [PubMed] [Google Scholar]