Abstract
The key parameter for diagnosis and management of hepatitis C virus (HCV) infection is HCV RNA. Standardization of HCV RNA assays to IU is mainly based on genotype 1 panels. Little is known about the variability of commercially available HCV RNA assays for quantification of different genotypes. Two real-time reverse transcription (RT)-PCR assays (COBAS TaqMan HCV Test for use with the High-Pure System [HPS/CTM] and COBAS Ampliprep/COBAS TaqMan HCV Test [CAP/CTM]), one standard RT-PCR assay (COBAS Amplicor HCV Monitor 2.0 [CAM]), and one signal amplification assay (Versant Quantitative 3.0 [branched DNA {bDNA}]) were compared for quantification of genotypes 1 to 5 (n = 108). Using CAM as a reference assay for genotype 1-infected patients, the mean interassay differences compared with CAP/CTM, HPS/CTM, and bDNA were 0.16, −0.13, and −0.48 log10 IU/ml HCV RNA, respectively. Comparison of CAM with CAP/CTM, HPS/CTM, and bDNA for the remaining genotypes showed the following results, respectively: 2a/c, −0.24, −0.78, and −0.49; 2b, −0.21, −0.18, and −0.64; 3a, 0.13, −1.04, and −0.55; 4, −0.52, −1.51, and −0.05; and 5, −0.28, −1.00, and −0.24 log IU/ml HCV RNA. A correct decision for treatment discontinuation in genotype 1 patients at week 12 was possible only when the same assay was used at baseline and week 12. Comparison of CAM with the CAP/CTM assay showed equal quantifications of genotype 1, 2, 3, and 5 samples, while genotype 4 samples were slightly underestimated. For the HPS/CTM assay, a significant underestimation of the HCV RNA concentrations of genotypes 2a/c, 3, 4, and 5 was observed. For the bDNA assay, a constant lower quantification of genotypes 1 to 3 was detected.
Present recommendations for the management of alpha interferon-based treatment in patients with chronic hepatitis C virus (HCV) infection are based on HCV RNA measurements before, during, and after antiviral therapy (6, 22). Changes in HCV RNA serum concentrations during the early phase of interferon-based therapy have been analyzed based on complex models of viral kinetics and applied to the prediction of treatment outcomes (13, 16, 31). In different studies, a high predictive value for virologic nonresponse (98 to 100%) was observed for HCV genotype 1- and genotype 4- to 6-infected patients, with a decline in the HCV RNA serum concentration of less than 2 log steps between baseline and week 12 of (pegylated) alpha interferon-ribavirin combination therapy (3, 5, 8). Alternatively, an absolute HCV RNA concentration above 30,000 IU/ml may be used for decisions about early treatment discontinuation at week 12 (3). In addition, for genotype 1- and genotype 4- to 6-infected patients at week 24 of treatment, it is recommended that therapy be discontinued on the basis of detectable HCV RNA in serum by qualitative PCR-based assays (3, 5, 19). Recently, for patients infected with genotype 2 or 3, the HCV RNA concentration at baseline and viral decline at week 4 have been described as highly predictive for virologic response to pegylated alpha interferon-ribavirin combination therapy (4, 18, 30, 32). Furthermore, for assessment of virologic response to currently developed direct antiviral drugs (i.e., protease and polymerase inhibitors), proper HCV RNA quantification for the different HCV genotypes is critical (1, 14, 25, 26).
For measurements of HCV RNA, different qualitative and quantitative tests based on target (reverse transcription [RT]-PCR and transcription-mediated amplification) and signal amplification (branched DNA [bDNA]) techniques with different lower detection limits and linear ranges of amplification are commercially available. All quantitative HCV RNA assays are standardized to IU on the basis of the first WHO HCV international standard, 96/790 (2, 7, 11, 17, 23, 24, 27). However, continuing limitations are the lack of complete automation (22), the necessity for dilutions for quantification by standard PCR-based assays (6), the relatively low sensitivity of quantitative HCV RNA assays (31), and the need for different test systems for qualitative and quantitative HCV RNA measurements (31). Furthermore, standardization of results to IU are mainly based on HCV genotype 1 panels, and little is known about the variability of commercially available HCV RNA assays for quantification of different HCV genotypes.
Real-time PCR methods for the quantification of HCV RNA have the advantage of linear amplification over a broad dynamic range, together with an integrated, automated detection system. With efficient HCV RNA extraction, they have the potential to achieve lower detection limits of less than 10 IU/ml.
In the present study, we compared two real-time RT-PCR-based assays (HCV RNA extraction with the manual High Pure System, together with the COBAS TaqMan 48 Analyzer [HPS/CTM], and HCV RNA extraction with the automated COBAS Ampliprep instrument, together with the COBAS TaqMan 48 Analyzer [CAP/CTM]), one standard RT-PCR-based assay (COBAS Amplicor HCV Monitor 2.0 [CAM]), and one signal amplification assay (Versant HCV Quantitative 3.0 [bDNA]) for HCV RNA quantification of clinical samples of HCV genotype 1a-, 1b-, 2a/c-, 2b-, 3a-, 4-, and 5-infected patients in Europe. For genotype 1- and genotype 2- and 3-infected patients, in addition to baseline samples, week 12 and week 4 samples, respectively, were analyzed. For estimation of assay precision in the lower range and investigation of apparent underquantification of certain HCV genotypes by the HPS/CTM assay, HCV RNA measurements of samples after 1:20 dilution were performed.
MATERIALS AND METHODS
Comparative analyses of serum or plasma samples from patients with chronic hepatitis C were performed with the following quantitative HCV RNA assays: (i) HPS/CTM for real-time HCV RNA amplification and detection (Roche Diagnostics), (ii) CAP/CTM for real-time HCV RNA amplification and detection (Roche Diagnostics), (iii) CAM as a standard nucleic acid extraction/RT-PCR-based assay (Roche Diagnostics), and (iv) bDNA as a signal amplification assay based on branched DNA technology (Bayer Diagnostics).
The COBAS TaqMan HCV Test for use with the High Pure System, the COBAS AmpliPrep/COBAS TaqMan HCV Test, and the COBAS Amplicor HCV Monitor Test v2.0 are currently not available for in vitro diagnostic use in the United States.
Undiluted clinical serum or plasma samples from European patients with chronic hepatitis C infected with HCV genotype 1a/b (n = 40), 2a/c (n = 14), 2b (n = 11), 3a (n = 24), 4 (n = 9), or 5 (n = 10) were used for parallel testing with the four different assays in a single determination. To guarantee identical conditions, serum or plasma samples stored at −80°C were thawed to generate appropriate aliquots (50 to 850 μl) for the different assays. Prior to being tested with the four different assays, all aliquots were stored again at −80°C. The same procedure was carried out for testing of 1:20 dilutions in a subset of samples with sufficient volumes available. For testing at week 12 and week 4, 24 samples from genotype 1a/b- and 24 samples from genotype 2- and 3-infected patients were available. All HCV RNA measurements with the four different assays were performed in the virologic laboratories of the Saarland University Hospital.
All patients had been enrolled in prospective studies and had been treated in the hepatology outpatient clinics of the University Hospitals of Homburg and Frankfurt, Germany. Treatment was performed with polyethylene glycol-alpha interferon 2a at 180 μg per week subcutaneously, plus ribavirin orally (800 mg for genotype 2- and 3- and 1,000/1,200 mg for genotype 1- and genotype 4- and 5-infected patients). Genotype 2- and 3-infected patients were treated for 24 weeks, and genotype 1- and genotype 4- and 5-infected patients were treated for a maximum of 48 weeks.
Virologic response in the trials was assessed by a qualitative HCV RNA assay with a lower sensitivity of 50 IU/ml (COBAS Amplicor HCV 2.0; Roche Diagnostics). According to the qualitative HCV RNA results, patients were defined as (i) virologic nonresponders (NR) (HCV RNA positive at the end of treatment), (ii) end-of-treatment responders with relapse (REL) (HCV RNA negative at the end of treatment but positive thereafter), and (iii) virologic sustained responders (SR) (HCV RNA negative at the end of treatment and at the end of follow-up). Genotype 1-infected patients with a positive qualitative HCV RNA test at week 24 were defined as nonresponders, and treatment was discontinued according to the study protocol.
Written informed consent was obtained from each patient, and the studies were approved by the Ethics Committees of Medical Research in Homburg and Frankfurt in accordance with the 1975 Declaration of Helsinki. All specimens in this evaluation represented leftover samples from the studies described above; no bleeds were obtained from patients specifically for this HCV RNA quantification study of different HCV gentoypes.
Genotyping of HCV according to the classification of Simmonds et al. (29) was performed by reverse hybridization assay (INNO LiPA HCV-II; Innogenetics, Gent, Belgium).
Real-time RT-PCR-based assays (HPS/CTM and CAP/CTM).
For testing with the HPS/CTM assay, HCV RNA was isolated from a 0.5-ml aliquot of controls and clinical specimens using the manual High Pure System Viral Nucleic Acid kit procedure. A known amount of HCV quantification standard RNA was introduced into each specimen, along with the lysis reagent. For adsorption of HCV RNA and quantification standard to a glass fiber surface, isopropanol was added to the lysis mixture before centrifugation through a column with a glass fiber filter insert. After removal of unbound substances, HCV RNA and HCV quantification standard RNA were eluted from the glass fiber particles.
For testing with the CAP/CTM assay, HCV RNA was isolated from 0.85-ml aliquots of controls and clinical specimens using the automated COBAS Ampliprep instrument. HCV quantification standard was added to the sample in order to achieve full process control. After a protease incubation step, the lysis reagent, together with the magnetic glass particles, was introduced into each specimen. HCV RNA and HCV quantification standard RNA were bound to the surfaces of magnetic glass particles. After completion of several washing steps, the adsorbed nucleic acids were eluted at elevated temperature with an aqueous solution.
After HPS- and CAP-based extraction of nucleic acids, samples and controls were processed for amplification and detection using the COBAS TaqMan 48 Analyzer according to the instructions of the manufacturer. While the compositions of the reagents for amplification and detection are not identical, both assays (HPS/CTM and CAP/CTM) rely on the same test principles. Reverse transcription and amplification were carried out using primers that bind within the highly conserved 5′ nontranslated region of HCV; deoxynucleoside triphosphates, including deoxyuridine and Thermus species strain Z05 polymerase (a single-tube, single-enzyme, single-primer set process). In the presence of manganese (Mn2+), Z05 has both reverse transcription and DNA polymerase activities. The quantification standard represents a noninfectious RNA construct containing fragments of HCV sequences with primer binding regions identical to those of the HCV 5′ nontranslated region, leading to an amplification product of the same length and base composition as the HCV target RNA. For destruction of potential contaminating DNA from previous amplifications, the AmpErase system (AmpErase) was used. For detection of amplification products, the assays utilize real-time PCR technology with two different dual-labeled fluorescent oligonucleotide probes, which are able to bind HCV target amplicon and quantification standard amplicon, respectively, within the regions spanned by the primers. The two different probes for the HCV target and the quantification standard are labeled with two different fluorescent reporter dyes. The reporter fluorescence is suppressed in the intact probe by the proximity of the quencher dye due to inductive-resonance-based energy transfer (Förster-type energy transfer). During elongation, the hybridized dual-labeled oligonucleotide probe is cleaved by the 5′-3′ exonuclease activity of Z05 polymerase, leading to the separation of reporter and quencher dyes. Within each cycle during the annealing and elongation phase of PCR, the increasing emission of fluorescence light from such cleaved dual-labeled oligonucleotides is collected independently for the HCV target and quantification standard at different wavelengths. The larger the original HCV RNA amount of a specimen, the earlier the fluorescence of the reporter dye rises above certain assigned fluorescence levels (the critical-threshold value), whereas for the constant titer of quantification standard RNA, the fluorescence of the reporter dye should appear at the same cycle for all specimens. By comparison of critical-threshold values obtained for the target HCV RNA and the quantification standard RNA, the original HCV RNA concentration of the specimen is calculated.
Due to the large dynamic range of the HPS/CTM and CAP/CTM assays, none of the specimens investigated in the first part of this study, addressing the analysis of undiluted pretreatment samples, had to be diluted.
Standard RT-PCR-based assay.
Extraction of nucleic acids for testing with the CAM assay was carried out from a 0.1-ml aliquot of controls and clinical specimens according to the manufacturer's instructions. Briefly, nucleic acids (HCV RNA, together with the quantification standard RNA) were isolated by lysis of virus particles with a chaotropic reagent, followed by a standard precipitation of RNA with alcohol. Subsequently, reverse transcription and PCR amplification by Thermus thermophilus polymerase and HCV RNA quantification with the CAM assay were performed as previously described in detail (9). Predilutions were performed for HCV RNA concentrations above 500,000 IU/ml. The lower detection limit of the CAM version 2.0 assay, according to the manufacturer, is 600 IU/ml.
The CAM, HPS/CTM, and CAP/CTM assays are standardized against the first WHO HCV international standard (96/790), and titer results are automatically reported in international units (IU/ml).
Signal amplification-, bDNA-based assay.
HCV RNA quantification with a signal amplification-based assay was performed with the third generation of the branched DNA nucleic acid probe test (bDNA). Several modifications to enhance signal amplification and to reduce nonspecific binding in comparison to version 2.0 were introduced; the bDNA 3.0 test procedure has been described in detail elsewhere (27).
The bDNA assay version 3.0 is standardized for IU, and the assay has been reported to be linear over its entire dynamic range from the lower detection limit of 615 IU/ml up to 8 million IU/ml (27).
Dilutions.
In samples of different HCV genotypes (1a/b, n = 5; 2a/c, n = 3; 2b, n = 4; 3a, n = 3; 4, n = 3; and 5, n = 3) with sufficient volume remaining after initial testing, 1:20 dilutions were performed, and six replicates of each sample were tested by each of the four quantitative HCV RNA assays. The results for diluted samples were compared with the HCV RNA concentrations derived from a single determination of the original undiluted specimens divided by 20.
Data analysis.
Results are expressed as mean, median, and standard deviation (SD) as appropriate. Correlation coefficients (R) were calculated for the linearity of the assays.
RESULTS
Analyses of undiluted pretreatment samples.
For analyses of the different assays throughout their complete dynamic ranges of quantification, undiluted HCV RNA concentrations of the different HCV genotypes ranging from approximately 1 × 104 to 1 × 105 or 1 × 107 IU/ml were available (Table 1). For comparison of the different HCV RNA quantification assays, the results of the CAM test were used as a reference. The mean HCV RNA concentration and range results for the CAM assay are shown in Table 1.
TABLE 1.
HCV RNA concentrations of genotype 1- to 5-infected patients by four different assays
| Genotype | n | CAM
|
CAP/CTM
|
HPS/CTM
|
bDNA
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Mean concn (IU/ml log10) | Range (IU/ml log10) | Mean concn (IU/ml log10) | Difference from CAM | Mean concn (IU/ml log10) | Difference from CAM | Mean concn (IU/ml log10) | Difference from CAM | ||
| 1a/b | 40 | 6.18 | 4.08-7.19 | 6.34 | 0.16 | 6.05 | −0.13 | 5.70 | −0.48 |
| 2a/c | 14 | 6.02 | 4.43-7.36 | 5.78 | −0.24 | 5.24 | −0.78 | 5.54 | −0.49 |
| 2b | 11 | 6.57 | 5.08-7.16 | 6.36 | −0.21 | 6.39 | −0.18 | 5.93 | −0.64 |
| 3a | 24 | 6.03 | 4.04-7.27 | 6.16 | 0.13 | 4.99 | −1.04 | 5.48 | −0.55 |
| 4 | 9 | 5.46 | 4.42-5.96 | 4.94 | −0.52 | 3.95 | −1.51 | 5.41 | −0.05 |
| 5 | 10 | 6.00 | 5.61-6.34 | 5.72 | −0.28 | 5.00 | −1.00 | 5.76 | −0.24 |
Comparison of CAM with CAP/CTM.
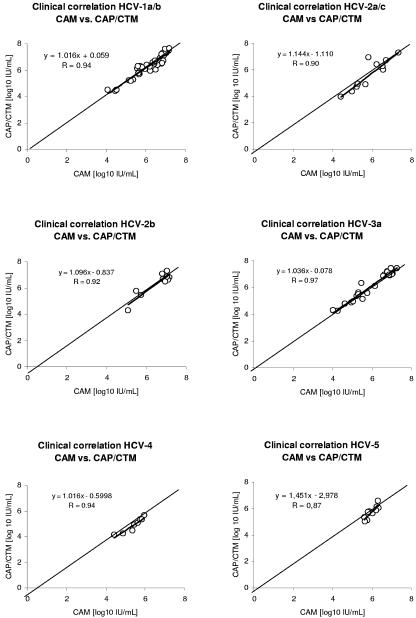
Comparison of CAM with CAP/CTM showed a good correlation for genotypes 1, 2a/c, 2b, 3a, and 5 with differences ranging from −0.28 to +0.16 log10 IU/ml (Table 1). For HCV genotype 4 samples, a slightly higher mean deviation of −0.52 log10 IU/ml was observed. The correlation coefficients (R) were calculated individually for all of the different genotypes (Fig. 1). The results for R for genotypes 1, 2b, 3a, and 4 ranged from 0.92 to 0.97 (Fig. 1). A slightly lower correlation coefficient for genotype 2a/c (R = 0.90) is explained by a higher variability of individual results of HCV RNA concentrations by the CAP/CTM assay than by CAM (Fig. 1). For genotype 5 samples, only patients with relative high viral loads were available (5.61 to 6.34 log10 IU/ml), which also led to a lower correlation coefficient (R = 0.87) (Fig. 1).
FIG. 1.
Correlation of HCV RNA concentrations of clinical samples between the CAM and the CAP/CTM assays. Results are shown separately for HCV genotype 1a/b, 2a/c, 2b, 3a, 4, and 5 samples. In addition to the single HCV RNA concentrations, the identity line and the regression line (boldface) are shown.
Comparison of CAM with HPS/CTM.
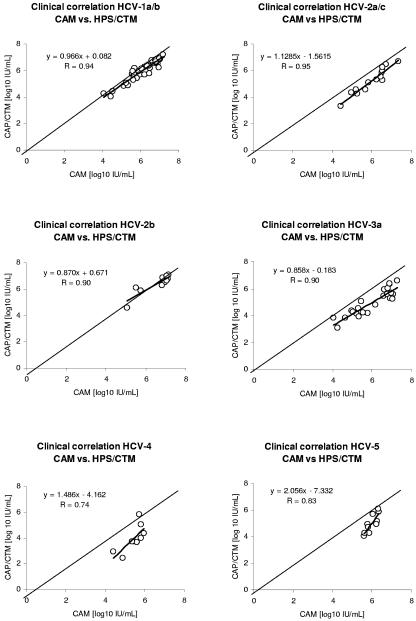
Comparison of CAM with HPS/CTM showed a good correlation for genotype 1 and 2b samples only, with differences of −0.13 and −0.18 log10 IU/ml, respectively (Table 1). For HCV genotypes 2a/c, 3a, 4, and 5, a significant underquantification was observed, ranging from −0.78 to −1.51 log10 IU/ml (Table 1 and Fig. 2). The correlation coefficients were calculated individually for all the different genotypes and ranged from 0.74 to 0.95 (Fig. 2).
FIG. 2.
Correlation of HCV RNA concentrations of clinical samples between the CAM and the HPS/CTM assays. Results are shown separately for HCV genotype 1a/b, 2a/c, 2b, 3a, 4, and 5 samples. In addition to the single HCV RNA concentrations, the identity line and the regression line (boldface) are shown.
Comparison of CAM with bDNA.
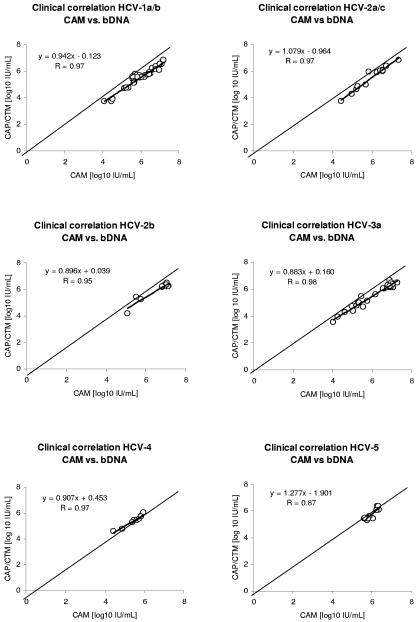
Comparison of CAM with bDNA showed constant lower HCV RNA concentrations for genotype 1, 2a, 2b, and 3a samples, with mean differences of −0.48 to −0.65 log10 IU/ml (Table 1). For HCV genotypes 4 and 5, a good correlation between CAM and bDNA assays was observed, with differences of −0.05 and −0.24 log10 IU/ml, respectively (Table 1). The correlation coefficients for genotypes 1 to 4 were high (R = 0.95 to 0.97) (Fig. 3). Due to the restriction to relatively highly concentrated specimens (5.61 to 6.34 log10 HCV RNA IU/ml), the correlation coefficient for comparison with HCV genotype 5 samples was relatively low (R = 0.87) (Fig. 3).
FIG. 3.
Correlation of HCV RNA concentrations of clinical samples between the CAM and the bDNA assays. Results are shown separately for HCV genotype 1a/b, 2a/c, 2b, 3a, 4, and 5 samples. In addition to the single HCV RNA concentrations, the identity line and the regression line (boldface) are shown.
Analyses of prediluted samples.
For estimation of the precision of the different assays in the lower range and for further analyses of apparent underquantification of HCV genotype 2a/c, 3a, 4, and 5 samples by the HPS/CTM assay, 1:20 dilutions were performed for three to five samples of each genotype/subtype (Table 2). The 1:20 dilutions were tested in six replicates with the CAM, the CAP/CTM, the HPS/CTM, and the VERSANT bDNA tests. The mean titers of the 1:20 dilutions were compared to the expected titers; expected titers were obtained by dividing the titer of the undiluted specimen which had been tested in a single determination by the dilution factor of 20.
TABLE 2.
HCV RNA concentrations of genotype 1- to 5-infected patients by four different assays after 1:20 dilution
| Genotype | n | CAM
|
CAP/CTM
|
HPS/CTM
|
bDNA
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean titer (IU/ml log10)a | Mean 1:20 (IU/ml log10) | Mean SD | Difference (IU/ml log10) | Mean titer (IU/ml log10) | Mean 1:20 (IU/ml log10) | Mean SD | Difference (IU/ml log10) | Mean titer (IU/ml log10) | Mean 1:20 (IU/ml log10) | Mean SD | Difference (IU/ml log10) | Mean titer (IU/ml log10) | Mean 1:20 (IU/ml log10) | Mean SD | Difference (IU/ml log10) | ||
| 1a/b | 5 | 4.22 | 4.07 | 0.10 | −0.15 | 4.22 | 4.07 | 0.05 | −0.15 | 4.15 | 3.91 | 0.12 | −0.14 | 3.78 | 3.84 | 0.03 | 0.06 |
| 2a/c | 3 | 4.54 | 4.33 | 0.08 | −0.21 | 4.03 | 3.92 | 0.07 | −0.11 | 3.37 | 3.14 | 0.08 | −0.23 | 3.99 | 3.98 | 0.04 | −0.01 |
| 2b | 4 | 5.34 | 5.06 | 0.05 | −0.28 | 5.27 | 4.82 | 0.09 | −0.45 | 5.00 | 4.70 | 0.17 | −0.30 | 4.72 | 4.70 | 0.02 | −0.02 |
| 3a | 3 | 5.33 | 4.96 | 0.06 | −0.37 | 5.23 | 4.76 | 0.07 | −0.37 | 3.69 | 4.07 | 0.31 | 0.38 | 4.55 | 4.58 | 0.03 | 0.03 |
| 4 | 3 | 4.14 | 3.92 | 0.10 | −0.22 | 3.58 | 3.42 | 0.09 | −0.16 | 2.06 | 2.66 | 0.27 | 0.60 | 4.03 | 3.95 | 0.03 | −0.08 |
| 5 | 3 | 4.61 | 5.01 | 0.04 | 0.40 | 4.44 | 4.47 | 0.08 | 0.03 | 3.20 | 3.82 | 0.08 | 0.62 | 4.50 | 4.56 | 0.02 | 0.06 |
The mean calculated titer is derived from a single measurement of the respective undiluted specimens divided by 20.
For the CAM assay, a mean SD of a sixfold measurement of each sample from 0.04 to 0.10 log10 IU/ml was observed for HCV genotype 1 to 5 samples (Table 2). The differences between observed and calculated results fell between −0.37 and 0.40 log10 IU/ml (Table 2). For the CAP/CTM assay, a mean SD of 0.05 to 0.09 log10 IU/ml and differences between observed and calculated results of −0.45 to 0.03 were detected (Table 2). For the HPS/CTM assay, relatively high mean SDs of 0.08 to 0.37 were observed (Table 2). Interestingly, especially for genotype 3a-, 4-, and 5-infected patients, underquantification in comparison with CAM was partially compensated for by 1:20 dilution. Mean HCV RNA concentrations for genotype 3a, 4, and 5 samples after 1:20 dilution were 0.38 to 0.62 log10 IU/ml higher, as was to be expected (Table 2), thereby partially compensating for the underquantification of 1.00 to 1.51 log10 IU/ml (Table 1). Finally, for the bDNA assay, very high accuracy of results after 1:20 dilution was observed, with mean SDs of 0.02 to 0.04 log10 IU/ml and differences between observed and expected results ranging from −0.08 to 0.06 log10 IU/ml.
Analyses of week 4 and 12 samples.
For genotype 1-infected patients, HCV RNA concentrations at baseline and week 12 were analyzed with the four different assays. A mean HCV RNA decline of ≥2 log10 IU/ml was observed for all SR and REL patients by the CAP/CTM and HPS/CTM assays due to the lower detection limit of approximately 10 IU/ml for these assays (28). However, also for the CAM and bDNA assays, results at week 12 were either ≥2 log units below baseline or below the detection limit of 600 to 615 IU/ml for all SR and REL patients (Table 3). Prediction of a virologic relapse after the end of treatment was not possible on the basis of the HCV RNA decline between baseline and week 12. The mean differences of HCV RNA concentrations between baseline and week 12 were even higher in REL patients than in SR patients (Table 3).
TABLE 3.
HCV RNA concentrations of genotype 1-infected patients: baseline compared with week 12 during therapy
| Virologic response | n | CAM
|
CAP/CTM
|
HPS/CTM
|
bDNA
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Bl vs. week 12 (IU/ml log10)a | Range | Bl vs. week 12 (IU/ml log10) | Range | Bl vs. week 12 (IU/ml log10) | Range | Bl vs. week 12 (IU/ml log10) | Range | ||
| SR | 8 | 3.29 | 1.76-4.15 | 5.00 | 3.42-6.29 | 4.54 | 3.01-5.81 | 2.72 | 0.93-3.74 |
| REL | 8 | 3.87 | 2.97-4.49 | 5.17 | 2.91-6.63 | 5.17 | 3.69-6.18 | 3.23 | 2.35-4.04 |
| NR | 8 | 1.34 | 0.04-2.64 | 1.50 | 0.44-2.79 | 1.36 | 0.12-2.60 | 1.40 | 0.42-2.77 |
Mean differences between HCV RNA concentrations at baseline (Bl) and week 12 of antiviral therapy.
For NR patients, the mean difference between baseline and week 12 HCV RNA concentrations was <2 log IU/ml by all assays (Table 3). However, three of eight NR patients had a decline of ≥2 log IU/ml at week 12 but were HCV RNA positive at week 24.
Furthermore, measurement of HCV RNA concentrations at baseline and week 12 with different assays (e.g., CAP/CTM at baseline and bDNA at week 12) would have led to an aberrant continuation of therapy in up to three NR patients.
For all HCV genotype 2- and 3-infected patients, a sharp HCV RNA decline was observed from baseline to week 4, and only one patient exhibited virologic nonresponse due to breakthrough during further antiviral therapy (Table 4). As for genotype 1-infected patients, prediction of virologic relapse was not possible by analyses of HCV RNA decline between baseline and week 4 in patients with genotype 2 and 3 infections.
TABLE 4.
HCV RNA concentrations of genotype 2- and 3-infected patients: baseline compared with week 4 during therapy
| Virologic response | n | CAM
|
CAP/CTM
|
HPS/CTM
|
bDNA
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Bl vs. week 4 (IU/ml log10)a | Range | Bl vs. week 4 (IU/ml log10) | Range | Bl vs. week 4 (IU/ml log10) | Range | Bl vs. week 4 (IU/ml log10) | Range | ||
| SR | 20 | 2.99 | 1.20-4.58 | 4.69 | 1.92-6.44 | 3.69 | 1.36-5.59 | 2.46 | 0.74-3.84 |
| REL | 3 | 3.93 | 3.28-4.29 | 4.99 | 2.63-6.44 | 3.71 | 2.18-4.68 | 3.30 | 2.83-3.70 |
| NR (BT)b | 1 | 4.10 | 6.15 | 5.00 | 3.51 | ||||
Mean differences between HCV RNA concentrations at baseline (Bl) and week 4 of antiviral therapy.
BT, breakthrough.
For all patients, a highly precise estimation of HCV RNA decline early during therapy was possible by the CAP/CTM and HPS/CTM assays due to their lower detection limits of around 10 IU/ml (28). The range of decline was approximately 1 to 2 log steps higher for the CAP/CTM and HPS/CTM assays than with the CAM and bDNA tests with lower detection limits of 600 to 615 IU/ml (Tables 3 and 4).
For genotype 1 and genotypes 2 and 3, mean HCV RNA concentrations at baseline were higher in REL than in SR patients (7.0 × 106 versus 3.8 × 106 IU/ml and 6.4 × 106 versus 4.1 × 106 IU/ml, respectively). However, the differences did not reach statistical significance (Mann-Whitney U test).
DISCUSSION
HCV RNA is the key parameter for management of acute and chronic hepatitis C. Different methods have been developed for commercially available assays for the measurement of HCV RNA in blood samples. Polymerase and ligase chain reactions, as well as transcription-mediated amplification, amplify the HCV RNA before detection by colorimetric measurements using specifically labeled primers or DNA probes. As an alternative to amplification of the target, the original concentration within a given sample can be quantified by enhancing the fluorescent signal of specifically hybridized probes above a detectable limit (bDNA). However, the different techniques have their restrictions, which have led to diversification of assays suitable either for sensitive qualitative or less sensitive quantitative detection of HCV RNA. Real-time PCR technology has the potential to overcome these restrictions by linear online detection of HCV RNA from very low to extremely high concentrations and therefore is considered the technique of choice for highly sensitive quantification of DNA or RNA targets (10, 15, 20).
A dual-labeled probe-based real-time RT-PCR assay (TaqMan) has been automated and standardized for detection of different RNA and DNA targets, including HCV RNA (CTM). For measurement of HCV RNA concentrations, the COBAS TaqMan test was combined with a manual (HPS) sample preparation assay or an automated (CAP) sample preparation step. In the present study, these new real-time RT-PCR-based assays (HPS/CTM and CAP/CTM) were compared with the widely distributed quantitative HCV RNA assays CAM and bDNA.
For comparison of the CAP/CTM assay with CAM as a reference test, a high correlation for HCV RNA concentrations of genotype 1, 2a/c, 2b, 3a, and 5 samples was observed. The mean differences were always below ±0.3 log10 IU/ml (i.e., twofold), and thus, a linear correlation between the two assays is present. Only in genotype 4 samples was a slight underquantification (mean HCV RNA concentrations, −0.52 log IU/ml) detected, which requires further investigation on the basis of a larger number of samples.
Comparison of the HPS/CTM assays with the CAM test showed a good correlation for genotype 1 and 2b samples, with a mean difference of −0.13 and −0.18 log10 IU/ml. For genotype 2a/c, 3a, 4, and 5 samples, a significant underquantification of −0.78 to −1.51 log10 IU/ml was observed. Due to this underquantification, version 1 of the HPS/CTM assay is restricted by the manufacturer to use in patients with genotype 1 and 6 infections only. Unfortunately for the present study, no specimens of genotype 6-infected patients, which are very rare in Europe, were available. The reasons for underquantification of certain HCV genotypes by the HPS/CTM assay are unknown. Because real-time RT-PCR-based assays represent a complex optimization of the different steps of nucleic acid preparation, amplification, and detection for the different HCV isolates, subtypes, and genotypes, multiple reasons for the underquantification of specific HCV genotypes have to be considered. General mismatch of primers or the TaqMan probe located within a highly conserved part of the 5′ nontranslated region of the HCV genome, which was already used for amplification by the CAM assay, was excluded by the manufacturer. However, suboptimal binding of oligonucleotides due to the secondary structure of the internal ribosome entry site located within the 5′ nontranslated region may be possible. In the present study, we demonstrate that the underquantification in the HPS/CTM assay can be partially compensated for by 1:20 predilution of the samples. Thus, transfer of an inhibitor for PCR amplification or detection from nucleic acid extraction appears to be a conceivable explanation for the underquantification by the HPS/CTM assay. Restriction to certain HCV genotypes (2a/c, 3a, 4, and 5) may be explained by differences in the amplification efficiencies between HCV genotypes.
For the comparison of the bDNA assay with CAM, relatively stable lower HCV RNA concentrations were observed in genotype 1, 2a/c, 2b, and 3a samples (−0.48 to −0.64 log10 IU/ml), despite the standardization of both assays to the first international WHO HCV standard (96/790). For genotype 4 and 5 samples, a good correlation of HCV RNA concentrations between bDNA and CAM was detected. It is known that linearity of quantification is lost for CAM for HCV RNA concentrations above 1 × 105 to 5 × 105 IU/ml (2, 11). However, in previous studies, predilution for the CAM assay was performed for samples of >850,000 IU/ml only (2, 11). In the present study, HCV RNA titers above 500,000 IU/ml were considered to be outside the linear range of the CAM assay and were retested after dilution. This may explain differences from previous CAM/bDNA comparative studies. Interestingly, after predilution (1:20) for samples of all genotypes, the correlation between CAM and bDNA improved to a mean difference of approximately 0.3 log10 IU/ml. This is in accordance with recent findings, with a recommendation of predilution for all samples before measurement with CAM due to saturation effects (21). However, also in the prediluted samples, a principal difference of a significantly lower estimation of HCV RNA concentrations in genotype 1 to 3 samples by the bDNA assay compared to the CAM and CAP/CTM assays was detectable throughout the present study, which may reflect differences in the process for calibration to the WHO standard of the assays.
In patients with chronic hepatitis C genotype 1 infection treated with (pegylated) interferon plus ribavirin, decisions about early treatment discontinuation are currently based on quantitative HCV RNA measurements at baseline and week 12 of therapy and a qualitative HCV RNA measurement at week 24 (3, 5, 8, 22). In the present study, reliable treatment decisions at week 12 could be made on the basis of all HCV RNA assays tested (HPS/CTM, CAP/CTM, CAM, and bDNA). However, it is important to note that a change between different quantitative HCV RNA assays from baseline to week 12 may lead to incorrect treatment decisions and therefore should be avoided. Furthermore, the importance of the week 24 discontinuation rule was demonstrated by the fact that a significant number of NR patients achieved a >2-log-unit decline at week 12 but were HCV RNA positive at week 24.
Taken together, comparison of the standard PCR-based CAM assay with the real-time PCR-based, automated CAP/CTM assay showed a good correlation of HCV RNA quantification for HCV genotypes 1 to 5. By comparison of the CAM assay with the HPS/CTM test, a significant underquantification of HCV genotypes 2a/c, 3a, 4, and 5 was observed, while for genotype 1 and 2b samples, a high concordance of HCV RNA concentrations was detected. For the bDNA assay, constant ≈0.5-log-unit-lower HCV RNA concentrations were observed in comparison with the CAM assay for genotype 1 to 3 samples.
Because of the high significance of HCV RNA concentrations in the management of the current (pegylated) interferon-ribavirin combination therapies and for future antiviral treatment options based on direct antiviral drugs (3, 12, 14, 26, 32), HCV RNA assays must be carefully analyzed for reliable quantification of all of the different HCV genotypes.
Acknowledgments
We are grateful to Eva Herrmann for statistical calculations and to Stella Traver and Claudia Boeck for their excellent technical assistance.
REFERENCES
- 1.Afdhal, N., E. Godofsky, J. Dienstag, V. Rustgi, L. Schick, D. McEniry, X. J. Zhou, G. Chao, C. Fang, B. Fielman, M. Myers, and N. Brown. 2004. Final phase I/II trial results for NM283, a new polymerase inhibitor for hepatitis C: antiviral efficacy and tolerance in patients with HCV-1 infection, including previous interferon failures. Hepatology 40(Suppl. 1):726A. [Google Scholar]
- 2.Beld, M., R. Sentjens, S. Rebers, C. Weegink, J. Weel, C. Sol, and R. Boom. 2002. Performance of the new Bayer VERSANT HCV RNA 3.0 assay for quantitation of hepatitis C virus RNA in plasma and serum: conversion to international units and comparison with the Roche COBAS Amplicor HCV Monitor, version 2.0, assay. J. Clin. Microbiol. 40:788-793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Berg, T., C. Sarrazin, E. Herrmann, H. Hinrichsen, T. Gerlach, R. Zachoval, B. Wiedenmann, U. Hopf, and S. Zeuzem. 2003. Prediction of treatment outcome in patients with chronic hepatitis C: significance of baseline parameters and viral dynamics during therapy. Hepatology 37:600-609. [DOI] [PubMed] [Google Scholar]
- 4.Dalgard, O., K. Bjoro, K. B. Hellum, B. Myrvang, S. Ritland, K. Skaug, N. Raknerud, and H. Bell. 2004. Treatment with pegylated interferon and ribavarin in HCV infection with genotype 2 or 3 for 14 weeks: a pilot study. Hepatology 40:1260-1265. [DOI] [PubMed] [Google Scholar]
- 5.Davis, G. L., J. B. Wong, J. G. McHutchison, M. P. Manns, J. Harvey, and J. Albrecht. 2003. Early virologic response to treatment with peginterferon alfa-2b plus ribavirin in patients with chronic hepatitis C. Hepatology 38:645-652. [DOI] [PubMed] [Google Scholar]
- 6.Davis, G. L. 2002. Monitoring of viral levels during therapy of hepatitis C. Hepatology 36:S145-S151. [DOI] [PubMed] [Google Scholar]
- 7.Elbeik, T., J. Surtihadi, M. Destree, J. Gorlin, M. Holodniy, S. A. Jortani, K. Kuramoto, V. Ng, R. Valdes, Jr., A. Valsamakis, and N. A. Terrault. 2004. Multicenter evaluation of the performance characteristics of the bayer VERSANT HCV RNA 3.0 assay (bDNA). J. Clin. Microbiol. 42:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fried, M. W., M. L. Shiffman, K. R. Reddy, C. Smith, G. Marinos, F. L. Gonzales, D. Häussinger, M. Diago, G. Carosi, D. Dhumeaux, A. Craxi, A. Lin, J. Hoffmann, and J. Yu. 2002. Peginterferon alfa-2a plus ribavirin for chronic hepatitis C virus infection. N. Engl. J. Med. 347:975-982. [DOI] [PubMed] [Google Scholar]
- 9.Gerken, G., T. Rothaar, M. G. Rumi, R. Soffredini, M. Trippler, M. J. Blunk, A. Butcher, S. Soviero, and G. Colucci. 2000. Performance of the COBAS AMPLICOR HCV MONITOR test, version 2.0, an automated reverse transcription-PCR quantitative system for hepatitis C virus load determination. J. Clin. Microbiol. 38:2210-2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Germer, J. J., W. S. Harmsen, J. N. Mandrekar, P. S. Mitchell, and J. D. Yao. 2005. Evaluation of the COBAS TaqMan HCV test with automated sample processing using the MagNA pure LC instrument. J. Clin. Microbiol. 43:293-298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Germer, J. J., P. J. Heimgartner, D. M. Ilstrup, W. S. Harmsen, G. D. Jenkins, and R. Patel. 2002. Comparative evaluation of the VERSANT HCV RNA 3.0, QUANTIPLEX HCV RNA 2.0, and COBAS AMPLICOR HCV MONITOR version 2.0 assays for quantification of hepatitis C virus RNA in serum. J. Clin. Microbiol. 40:495-500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hadziyannis, S. J., H. Sette, Jr., T. R. Morgan, V. Balan, M. Diago, P. Marcellin, G. Ramadori, H. C. Bodenheimer, Jr., D. Bernstein, M. Rizzetto, S. Zeuzem, P. J. Pockros, A. Lin, and A. M. Ackrill. 2004. Peginterferon-alfa2a and ribavirin combination therapy in chronic hepatitis C. A randomized study of treatment duration and ribavirin dose. Ann. Intern. Med. 140:346-355. [DOI] [PubMed] [Google Scholar]
- 13.Herrmann, E., A. U. Neumann, J. M. Schmidt, and S. Zeuzem. 2000. Hepatitis C virus kinetics. Antivir. Ther. 5:85-90. [PubMed] [Google Scholar]
- 14.Hinrichsen, H., Y. Benhamou, H. Wedemeyer, M. Reiser, R. E. Sentjens, J. L. Calleja, X. Forns, A. Erhardt, J. Cronlein, R. L. Chaves, C. L. Yong, G. Nehmiz, and G. G. Steinmann. 2004. Short-term antiviral efficacy of BILN 2061, a hepatitis C virus serine protease inhibitor, in hepatitis C genotype 1 patients. Gastroenterology 127:1347-1355. [DOI] [PubMed] [Google Scholar]
- 15.Konnick, E. Q., S. M. Williams, E. R. Ashwood, and D. R. Hillyard. 2005. Evaluation of the COBAS Hepatitis C Virus (HCV) TaqMan analyte-specific reagent assay and comparison to the COBAS Amplicor HCV Monitor V2.0 and Versant HCV bDNA 3.0 assays. J. Clin. Microbiol. 43:2133-2140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Layden, J. E., T. J. Layden, K. R. Reddy, R. S. Levy-Drummer, J. Poulakos, and A. U. Neumann. 2002. First phase viral kinetic parameters as predictors of treatment response and their influence on the second phase viral decline. J. Viral Hepat. 9:340-345. [DOI] [PubMed] [Google Scholar]
- 17.Lee, S. C., A. Antony, N. Lee, J. Leibow, J. Q. Yang, S. Soviero, K. Gutekunst, and M. Rosenstraus. 2000. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J. Clin. Microbiol. 38:4171-4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mangia, A., R. Santoro, N. Minerva, G. L. Ricci, V. Carretta, M. Persico, F. Vinelli, G. Scotto, D. Bacca, M. Annese, M. Romano, F. Zechini, F. Sogari, F. Spirito, and A. Andriulli. 2005. Peginterferon alfa-2b and ribavirin for 12 vs. 24 weeks in HCV genotype 2 or 3. N. Engl. J. Med. 352:2609-2617. [DOI] [PubMed] [Google Scholar]
- 19.Manns, M. P., J. G. McHutchison, S. C. Gordon, V. K. Rustgi, M. Shiffman, R. Reindollar, Z. D. Goodman, K. Koury, M. Ling, and J. K. Albrecht. 2001. Peginterferon alfa-2b plus ribavirin compared with interferon alfa-2b plus ribavirin for initial treatment of chronic hepatitis C: a randomised trial. Lancet 358:958-965. [DOI] [PubMed] [Google Scholar]
- 20.Martell, M., J. Gomez, J. I. Esteban, S. Sauleda, J. Quer, B. Cabot, R. Esteban, and J. Guardia. 1999. High-throughput real-time reverse transcription-PCR quantitation of hepatitis C virus RNA. J. Clin. Microbiol. 37:327-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morishima, C., M. Chung, K. Ng, D. J. Brambilla, and D. R. Gretch. 2004. Strengths and limitations of commercial tests for hepatitis C virus RNA quantification. J. Clin. Microbiol. 42:421-425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.National Institutes of Health. 2002. National Institutes of Health Consensus Development Conference statement: management of hepatitis C: 2002-June 10-12, 2002. Hepatology 36:S3-S20. [DOI] [PubMed] [Google Scholar]
- 23.Nolte, F. S., M. W. Fried, M. L. Shiffman, A. Ferreira-Gonzalez, C. T. Garrett, E. R. Schiff, S. J. Polyak, and D. R. Gretch. 2001. Prospective multicenter clinical evaluation of AMPLICOR and COBAS AMPLICOR hepatitis C virus tests. J. Clin. Microbiol. 39:4005-4012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pawlotsky, J. M., M. Bouvier-Alias, C. Hezode, F. Darthuy, J. Remire, and D. Dhumeaux. 2000. Standardization of hepatitis C virus RNA quantification. Hepatology 32:654-659. [DOI] [PubMed] [Google Scholar]
- 25.Reesink, H. W., S. Zeuzem, C. J. Weegink, N. Forestier, A. van Vliet, J. van de Wetering de Rooij, L. McNair, S. Purdy, H.-M. Chu, and P. L. M. Jansen. 2005. Initial results of a phase 1b multiple dose study of VX950, a hepatitis C virus protease inhibitor. Gastroenterology 128(Suppl. 2):527A. [Google Scholar]
- 26.Reiser, M., H. Hinrichsen, J. P. Benhamou, H. W. Reesink, H. Wedemeyer, C. Avendano, N. Riba, C. L. Yong, G. Nehmiz, and G. Steinmann. 2005. Antiviral efficacy of NS3-serine protease inhibitor BILN-2061 in patients with chronic genotype 2 and 3 hepatitis C. Hepatology 41:832-835. [DOI] [PubMed] [Google Scholar]
- 27.Ross, R. S., S. Viazov, S. Sarr, S. Hoffmann, A. Kramer, and M. Roggendorf. 2002. Quantitation of hepatitis C virus RNA by third generation branched DNA-based signal amplification assay. J. Virol. Methods 101:159-168. [DOI] [PubMed] [Google Scholar]
- 28.Sarrazin, C., B. Gärtner, M. Welker, S. Traver, and S. Zeuzem. 2004. Evaluation of a new, highly sensitive, real time PCR based assay for quantification of HCV RNA. J. Hepatol. 40(Suppl. 1):150A. [DOI] [PubMed] [Google Scholar]
- 29.Simmonds, P., E. C. Holmes, T. A. Cha, F. McOmish, B. Irvine, E. Beall, P. L. Yap, J. Kolberg, and M. S. Urdea. 1993. Classification of hepatitis C virus into six major genotypes and a series of subtypes by phylogenetic analysis of the NS-5 region. J. Gen. Virol. 74:2391-2399. [DOI] [PubMed] [Google Scholar]
- 30.von Wagner, M., M. Huber, T. Berg, H. Hinrichsen, J. Rasenack, T. Heintges, A. Bergk, C. Bernsmeier, D. Haussinger, E. Herrmann, and S. Zeuzem. 2005. Peginterferon-alpha-2a (40KD) and ribavirin for 16 or 24 weeks in patients with genotype 2 or 3 chronic hepatitis C. Gastroenterology 129:522-527. [DOI] [PubMed] [Google Scholar]
- 31.Zeuzem, S., E. Herrmann, J. H. Lee, J. Fricke, A. U. Neumann, M. Modi, G. Colucci, and W. K. Roth. 2001. Viral kinetics in patients with chronic hepatitis C treated with standard or peginterferon alpha2a. Gastroenterology 120:1438-1447. [DOI] [PubMed] [Google Scholar]
- 32.Zeuzem, S., R. Hultcrantz, M. Bourliere, T. Goeser, P. Marcellin, J. Sanchez-Tapias, C. Sarrazin, J. Harvey, C. Brass, and J. Albrecht. 2004. Peginterferon alfa-2b plus ribavirin for treatment of chronic hepatitis C in previously untreated patients infected with HCV genotypes 2 or 3. J. Hepatol. 40:993-999. [DOI] [PubMed] [Google Scholar]