Abstract
Hepatitis B virus (HBV) with T-1856 of the precore region is always associated with C-1858 (i.e., TCC at nucleotides 1856 to 1858), and it is reported only in genotype C HBV isolates. We aimed to investigate the phylogenetic, virological, and clinical characteristics of HBV isolates bearing TCC at nucleotides 1856 to 1858. We have previously reported on the presence of two major subgroups in genotype C HBV, namely, HBV genotype Cs (Southeast Asia) and HBV genotype Ce (Far East). We have designed a novel 5′ nuclease technology based on the nucleotide polymorphism (C or A) at nucleotide 2733 to differentiate the two genotype C HBV subgroups. The mutations at the basal core promoter and precore regions were analyzed by direct sequencing. Among 214 genotype C HBV-infected patients, 31% had TCC, 37% had CCC, 3% had CTC, and 29% had CCT at nucleotides 1856 to 1858. All except one HBV strain with TCC at nucleotides 1856 to 1858 belonged to subgroup Cs, which has been reported only in Hong Kong; Guangzhou, China; and Vietnam. HBV with TCC at nucleotides 1856 to 1858 was associated with the G1898A mutation (64%). Patients infected with HBV harboring TCC had more liver cirrhosis than those infected with HBV harboring CCC (18% versus 5%; P = 0.008), and more of the patients infected with HBV harboring TCC were positive for HBeAg (58% versus 36%; P = 0.01) and had higher median alanine aminotransferase levels (65 IU/liter versus 49 IU/liter; P = 0.006); but similar proportions of patients infected with HBV harboring TCC and those infected with HBV harboring CCT had liver cirrhosis (18% versus 13%; P = 0.43). In summary, we report that HBV with TCC at nucleotides 1856 to 1858 of the precore region might represent a specific HBV strain associated with more aggressive liver disease than other genotype C HBV strains.
Hepatitis B virus (HBV) can be differentiated into eight different genotypes based on at least 8% heterogeneity of the entire viral genome (3, 25, 30). HBV genotypes B and C are the most common genotypes in most Asian countries (6, 14, 17, 26). Ample evidence suggests that HBeAg seroconversion is delayed in those infected with genotype C HBV compared to the time to serconversion in those infected with genotype B HBV (12) and that genotype C HBV infection results in more aggressive hepatitis (8, 12, 16), worse liver histology (11), and a higher risk of hepatocellular carcinoma (6, 15, 17, 32) than genotype B HBV infection. On phylogenetic analysis, genotype C HBV can be further classified into two subgroups, as defined by a more than 4% difference in the entire HBV genomic sequence (10). One subgroup, HBV genotype Cs, is more prevalent in Southeast Asia (Vietnam, Thailand, Myanmar, and southern China), while the other genotype, HBV genotype Ce, is more prevalent in the Far East (Japan, Korea, and northern China). Eighty percent of the genotype C HBV strains in Hong Kong belong to HBV genotype subgroup Cs and 20% belong to HBV genotype subgroup Ce.
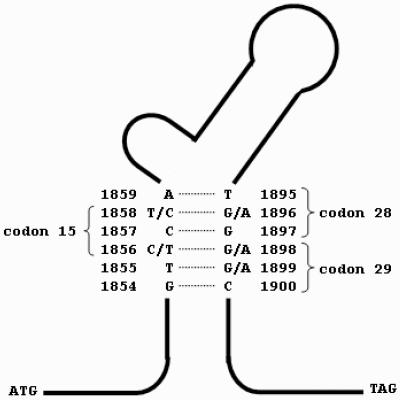
The configuration of nucleotides 1856 to 1858 of the HBV precore region directly determines the development of a precore stop codon mutation due to the stability of the stem-loop structure of the encapsidation sequence (23) (Fig. 1). The precore stop mutation (G1896A) develops only with T and not with C at nucleotide position 1858 (7, 9). This phenomenon indirectly affects the development of a basal core promoter mutation, which is an alternative mechanism for HBeAg seroconversion (9). In vitro studies have demonstrated that HBV mutants with the precore stop codon and basal core promoter mutations have higher levels of viral replication compared to the levels encountered with wild-type virus (4, 29). The basal core promoter mutation lies in the X region of the HBV genome, and its presence may increase the risk of hepatocellular carcinoma (15, 18). Therefore, the configuration of nucleotides 1856 to 1858 of the HBV precore region may have direct and indirect influences on the activity of hepatitis and HBeAg seroconversion in patients with chronic HBV infection.
FIG. 1.
Stem-loop structure of the encapsidation sequence of the HBV precore genome.
Nucleotide 1858 can be either C or T in genotype C HBV strains, but it is always T in genotype B HBV strains (6). HBV strains with T at nucleotide 1856 have been reported only with C at nucleotide 1858 to give a TCC configuration, which can be found only in genotype C HBV strains (20, 23). The impact of different configurations at nucleotides 1856 to 1858 has not been adequately studied. In this study, we set out to investigate the phylogenetic, virological, and clinical characteristics of HBV strains bearing TCC at nucleotides 1856 to 1858 of the precore region compared to those of HBV strains with other nucleotide configurations by using strains from a large cohort of genotype C HBV-infected patients. The phylogenetic characteristics were analyzed in terms of the genotype C subgroup (Cs and Ce) and the geographical distributions of the different HBV isolates. The virological characteristics at the basal core promoter and precore regions as well as the clinical features of patients harboring different HBV isolates were also studied.
MATERIALS AND METHODS
Patients.
Serum samples that had been stored at −80°C and that had been collected from a cohort of patients with chronic hepatitis B who had been recruited since December 1997 by the Hepatitis Clinic, Prince of Wales Hospital, Hong Kong,were studied (9). All patients were infected by genotype C HBV, as identified by restriction fragment length polymorphism (6). None of the patients had coinfection with hepatitis C virus or other agents that are causes of liver disease. Six patients had previously received interferon treatment; one patient had previously received famciclovir treatment; and one patient had previously received sequential treatment with interferon, lamivudine, and famciclovir. For all patients, antiviral treatment had been stopped for at least 6 months at the time of recruitment into the study. Liver cirrhosis was defined as ultrasonic features of liver cirrhosis plus evidence of hypersplenism (splenomegaly with a platelet count of less than 100 × 109/liter or a white blood cell count of less than 4 × 109/liter), clinical ascites, varices, and/or hepatic encephalopathy.
Rapid detection of genotype C HBV subgroup.
Based on the entire genomic sequences of the genotype C HBV strains in 49 samples from Hong Kong as well as the 64 published sequences in GenBank, two subgroups, namely, Cs (Southeast Asia) and Ce (Far East) could be identified (10). Consistent nucleotide differences that could best differentiate the two genotype C HBV subgroups were identified by aligning the entire nucleotide sequences of the HBV strains. At nucleotide 2733, the nucleotide configuration of HBV genotype Cs was C and that of HBV genotype Ce was A in the sequences of the strains in all 49 samples from Hong Kong and 62 of the 64 published genotype C HBV sequences from GenBank. This nucleotide polymorphism was selected for the differentiation of these two genotype C HBV subgroups in a novel subtyping assay.
Primer Express software version 2.0 (PE Applied Biosystems, Foster City, CA) was used to find suitable primers and probes. TaqMan real-time PCR technology was used to differentiate the nucleotide variant. Briefly, HBV DNA was extracted from serum by using a QIAamp DNA Blood Mini kit (QIAGEN GmbH, Hilden, Germany), according to the manufacturer's instructions. The oligonucleotide primers and TaqMan probes used for the detection of the variant subgroups were designed according to the sequence available from the GenBank database and, according to our recent study, (10) the sequence adjacent to nucleotide 2733. The PCR primers were 5′-AAAGGCAT(C/T)AAACC(A/T/G)TATTATCCTGAA-3′ (sense; nucleotides 2685 to 2711) and 5′-CCAGCCTTCCACAGAGTATGTAAA-3′ (antisense; nucleotides 2747 to 2770). The TaqMan probe was 5′-CATTACTTC(A/C)AAACTAG-3′ (sense; nucleotides 2724 to 2740) and had a fluorescent reporter dye (6-carboxyfluorescein and VIC) covalently linked to its 5′ end and a downstream nonfluorescent quencher linked to its 3′ end.
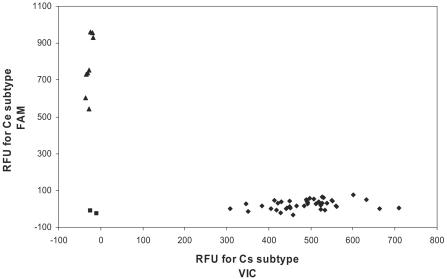
In the conventional real-time PCR system, the reaction mixture, which had a total volume of 25 ml, contained 50 ng DNA template, 50 pmol of each primer, 200 nM of TaqMan probe, and 1× TaqMan universal master mix (PE Applied Biosystems). Target and standard samples were assayed simultaneously. Amplification and detection of specific products were carried out in an iCycler iQ real-time PCR detection system (Bio-Rad Laboratories Inc., Hercules, CA) by using an amplification protocol that consisted of 1 cycle at 50°C for 2 min, 1 cycle at 95°C for 10 min, and 40 cycles at 95°C for 15 s and 60°C for 1 min. Nucleotide discrimination analysis was performed by using the built-in software (version 3.0) (Fig. 2).
FIG. 2.
Scatter plot illustrating the 6-carboxyfluorescein and VIC fluorescence signals from the TaqMan assay with HBV genotype C subgroups Cs and Ce. ▪, signals from the no-template controls; ⧫, signals from Cs subgroup; ▴, signals from the Ce subgroups.
The subgroups of the strains in 10% of the samples with positive results by the real-time PCR were validated by direct sequencing of the amplicon region. For serum samples with negative results by the real-time PCR, the amplicon region of HBV DNA was amplified by PCR and was then directly sequenced to determine the configuration of nucleotide 2733.
Mutations at basal core promoter and precore regions.
HBV DNA was extracted from 100 μl of serum by using the QIAamp DNA Blood Mini kit (QIAGEN GmbH), according to the manufacturer's instructions. PCR and direct sequencing of the amplified products were performed as described previously by using primers that flanked the core promoter and precore regions (7). The configurations at nucleotides 1856 to 1858 of the precore region as well as mutations at the basal core promoter and precore regions were determined.
Laboratory assays.
The samples were tested for hepatitis B surface antigen with commercially available enzyme-linked immunosorbent assay kits (Abbott GmBH Diagnostika, Wiesbaden-Delkenheim, Germany). HBeAg and anti-HBe were measured by enzyme-linked immunosorbent assay (Sanofi Diagnostics, Pasteur, France). HBV DNA was measured by a TaqMan real-time PCR assay, based on the Eurohep HBV standard, with a range of detection of HBV DNA from 102 to 109 copies/ml (5, 22).
Statistical analysis.
All statistical analyses were performed by using the Statistical Package for Social Sciences (version 11.0; SPSS, Inc., Chicago, IL). Continuous variables were expressed as the means ± standard deviations or medians (ranges), as appropriate. Categorical variables were compared by the chi-square test, and continuous variables were compared by Student's t test or the Mann-Whitney U test, as appropriate. All statistical tests were two sided. Statistical significance was taken as a P value of <0.05.
RESULTS
Detection of genotype C HBV subgroup by real-time PCR assay.
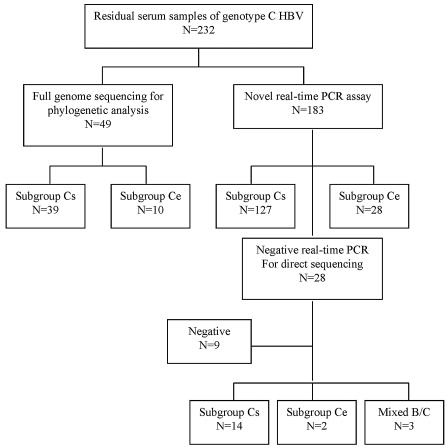
Stored serum samples from 232 patients infected with genotype C HBV were available for testing. A previous phylogenetic study determined the HBV genotype C subgroup in 49 patients, and so the samples from these patients were not retested in this study (14). One hundred eighty-three samples were tested by the novel real-time PCR assay for determination of the HBV genotype C subgroup (Fig. 3). Genotypes Cs and Ce could be differentiated in 155 (85%) samples. Among the 28 remaining samples negative by the real-time PCR, 16 samples had either genotype subgroup Cs or Ce, as differentiated by direct sequencing; 3 samples had genotype B and C coinfection; and 9 samples had negative PCR results, as confirmed by direct sequencing. Among the 16 samples with negative signals for genotype C HBV by real-time PCR, 7 samples had one nucleotide difference between the HBV DNA sequence and the TaqMan probe sequence. Stored serum samples from 15 patients infected with genotype B HBV, as confirmed by direct sequencing, were tested as controls; and all had negative results by the real-time PCR. Overall, this assay has a sensitivity of 85% and a specificity of 100% for detection of the genotype C HBV subgroups.
FIG. 3.
HBV genotype C subgroups as determined by the rapid real-time PCR method and direct sequencing.
Phylogenetic features.
Among the 232 serum samples that were available for testing, the genotype C subgroup could be classified in 220 samples by the rapid detection method and direct sequencing. The samples from six patients had mixed patterns at codon 15 and were excluded from analysis (Table 1). Among the remaining 214 patients, the mean age was 39 ± 13 years, and 136 (64%) of them were male. One hundred four (49%) patients were positive for HBeAg. The median alanine aminotransferase (ALT) level was 59.5 IU/liter (range, 13 to 1,011 IU/liter). Twenty-four (11%) patients had liver cirrhosis. Sixty-seven (31%) patients had TCC, 79 (37%) patients had CCC, 62 (29%) patients had CCT, and 6 (3%) patients had CTC at nucleotides 1856 to 1858.
TABLE 1.
Genotype C subgroup distribution of 64 HBV sequences from GenBank and 220 HBV sequences from Hong Kong with different configurations at nucleotides 1856 to 1858
| Sequence at nucleotides 1856 to 1858 | No. of sequences
|
|||
|---|---|---|---|---|
| GenBank
|
Hong Kong
|
|||
| HBV Cs | HBV Ce | HBV Cs | HBV Ce | |
| TCC | 5 | 0 | 66 | 1 |
| CCC | 12 | 0 | 79 | 0 |
| CCT | 11 | 35 | 23 | 39 |
| CTC | 0 | 0 | 6 | 0 |
| Mixed | 1 | 0 | 6 | 0 |
Among the HBV subgroup Cs sequences from GenBank and the Hong Kong cohort, all except one HBV strain had TCC at nucleotides 1856 to 1858 (Table 1). All HBV sequences with TCC at nucleotides 1856 to 1858 were derived from patients who originated in Hong Kong; Guangzhou, China; and Vietnam. The HBV strains with CCC at nucleotides 1856 to 1858 also belonged to subgroup Cs, but the origins of these patients were spread throughout Asia. HBV strains with CCT at nucleotides 1856 to 1858 were found in HBV strains of both subgenotype Ce and subgenotype Cs.
Virological features.
The prevalence of basal core promoter mutations was similar among HBV strains with TCC and those with CCC or CTC at nucleotides 1856 to 1858 (Table 2). The G1764A mutation tended to occur more frequently among HBV strains with TCC at nucleotides 1856 to 1858 (72%) than among those with CCT (57%), but the difference fell just short of statistical significance. The conventional precore stop codon mutation (G1896A) was not found in any of the HBV strains containing C-1858. The G1897A mutation was found in all six HBV isolates with CTC, and this mutation gave rise to a stop codon TGA at codon 28. The prevalence of the G1898A mutation was significantly higher among HBV strains with TCC (64%) than among the other HBV isolates. The G1899A mutation was found in a significantly higher proportion of HBV strains with CCT (16%) than those with TCC (5%).
TABLE 2.
Virological characteristic of genotype C HBV strains with different configurations at nucleotides 1856 to 1858a
| Mutation | No. (%) of isolates with:
|
P value for TCC vs CCC | No. (%) of isolates with CTC (n = 6) | P value for TCC vs CTC | No. (%) of isolates with CCT (n = 62) | P value for TCC vs CCT | |
|---|---|---|---|---|---|---|---|
| TCC (n = 67) | CCC (n = 79) | ||||||
| A1762T | 45 (67) | 58 (73) | 0.41 | 4 (67) | 0.98 | 34 (55) | 0.15 |
| G1764A | 48 (71) | 60 (76) | 0.55 | 5 (83) | 0.54 | 35 (57) | 0.072 |
| G1896A | 0 (0) | 0 (0) | 0 (0) | 28 (45) | <0.001 | ||
| G1897A | 0 (0) | 0 (0) | 6 (100) | <0.001 | 0 (0) | ||
| G1898A | 43 (64) | 1 (1) | <0.001 | 0 (0) | 0.002 | 0 (0) | <0.001 |
| G1899A | 3 (5) | 3 (4) | 0.84 | 0 (0) | 0.60 | 10 (16) | 0.028 |
The prevalence of mutations among different HBV isolates was compared to that among HBV isolates with TCC at nucleotides 1856 to 1858.
Clinical features.
Patients infected with HBV strains with TCC at nucleotides 1856 to 1858 had a higher prevalence of liver cirrhosis (18%) compared to the prevalence among those infected with HBV strains bearing CCC (5%), and the prevalence among those infected with HBV strains with TCC was comparable to that among those infected with HBV bearing CCT (13%) (Table 3). In a comparison of patients infected with HBV strains bearing TCC and those infected with HBV strains bearing CCT at nucleotides 1856 to 1858, there was a higher prevalence of positivity for HBeAg (58% versus 36%) and a higher level of ALT (65 IU/liter versus 49 IU/liter) among the individuals in the former group (Table 3). The lower prevalence of HBeAg positivity among patients infected with HBV strains with CCT was likely related to the development of the precore stop codon mutation (G1896A). The proportion of patients with ALT levels more than two times the upper limit of laboratory normal was also higher among patients infected with HBV strains with TCC (33% versus 13%).
TABLE 3.
Clinical characteristics of patients infected with genotype C HBV isolates with different configurations at nucleotides 1856 to 1858a
| Characteristic | TCC (n = 67) | CCC (n = 79) | P value for TCC vs CCC | CCT (n = 62) | P value for TCC vs CCT |
|---|---|---|---|---|---|
| Age (yr)c | 38 ± 15 | 38 ± 12 | 0.60 | 41 ± 12 | 0.41 |
| No. (%) male | 44 (66) | 56 (71) | 0.50 | 31 (50) | 0.071 |
| No. (%) with previous antiviral treatment | 4 (6) | 2 (3) | 0.44 | 2 (3) | 0.64 |
| No. (%) HBeAg positive | 39 (58) | 43 (54) | 0.65 | 22 (36) | 0.01 |
| No. (%) with cirrhosis | 12 (18) | 4 (5) | 0.013 | 8 (13) | 0.43 |
| Log HBV DNA level (no. of copies/ml)c | 6.56 ± 1.92 | 6.47 ± 1.91 | 0.77 | 6.12 ± 1.92 | 0.20 |
| No. (%) with HBV DNA level >10,000 copies/ml | 60 (90) | 72 (91) | 0.75 | 51 (82) | 0.23 |
| ALT level (range)d | 65 (13-1,011) | 62 (14-710) | 0.37 | 49 (17-753) | 0.006 |
| No. (%) with ALT level: | |||||
| Normal | 29 (43) | 37 (47) | 36 (58) | ||
| One to two times ULNb | 16 (24) | 27 (34) | 0.22 | 18 (29) | 0.044 |
| Two to five times ULN | 15 (22) | 9 (11) | 7 (11) | ||
| More than five times ULN | 7 (11) | 6 (8) | 1 (2) |
As the number of patients with CTC at nucleotides 1856 to 1858 was small, the clinical features of patients harboring HBV with TCC were compared to those harboring HBV with CCC and CCT.
ULN, upper limit of normal.
Values are means ± SD.
Values are in international units per liter.
G1898A mutation.
As the G1898A mutation was almost unique to HBV strains with TCC at nucleotides 1856 to 1858, we further analyzed the impact of this mutation on the clinical manifestations of chronic hepatitis B. This mutation was translated to an amino acid change of glycine to serine at codon 29 of the precore region. Patients who developed the G1898A mutation were generally older (44 ± 15 years) than those who did not develop this mutation (38 ± 13 years) (P = 0.013). Eleven of 44 (25%) patients with the G1898A mutation had liver cirrhosis, whereas 13 of 170 (8%) patients without the G1898A mutation had liver cirrhosis (P = 0.001). Otherwise, there was no difference between patients infected with HBV strains with and without the G1898A mutation by gender (male patients, 59% versus 65%; P = 0.49), HBeAg positivity (46% versus 49%; P = 0.64), log HBV DNA level (6.09 ± 1.81 versus 6.43 ± 1.93; P = 0.28), or median ALT level (65 IU/liter versus 58 IU/liter; P = 0.17).
DISCUSSION
Most previous studies on the HBV encapsidation sequence focused only on the configuration of nucleotide 1858 (21, 27). Although the polymorphism of C or T at nucleotide 1858 affects the development of the precore stop codon mutation, it does not seem to influence disease activity (6, 15). In this study, we have demonstrated that the configuration of nucleotides 1856 to 1858 affects both the virological and the clinical manifestations experienced by genotype C HBV-infected patients. Genotype C HBV with TCC at nucleotides 1856 to 1858 probably represents a virological strain that is associated with liver disease more aggressive than that caused by strains with CCC and CCT.
Based on phylogenetic analysis, different HBV genotypes and subgroups have different configurations at nucleotides 1856 to 1858, but there is also substantial overlap. CCC at nucleotides 1856 to 1858 is found predominantly in HBV genotypes A, C, and F; and CCT at nucleotides 1856 to 1858 is found predominantly in HBV genotypes B, C, D, and E (2, 19, 20). In contrast, TCC at nucleotides 1856 to 1858 has been reported only in HBV genotype C isolates from Hong Kong, Guangzhou, and Vietnam (13, 20, 24, 28). In our study, it was found almost exclusively within the Cs subgroup of genotype C HBV. In previous studies of families, the TCC configuration at nucleotides 1856 to 1858 is conserved in the HBV isolates among all members of the same family, and therefore, it should represent a genuine HBV strain rather than a mutation (1). The characterization of this specific HBV strain will be important in future epidemiological studies of HBV infection in this region.
The higher prevalence of liver cirrhosis among patients infected with HBV with TCC at nucleotides 1856 to 1858 than among those infected with HBV with CCC cannot be explained by age, gender, HBeAg status, or HBV DNA or ALT level. In fact, patients infected with HBV with TCC and CCT at nucleotides 1858 to 1858 have similar prevalences of liver cirrhosis, despite higher ALT levels in the former group. As patients in Asia usually acquire the infection during infancy, the difference in the prevalence of liver cirrhosis cannot be explained by a difference in the duration of infection. However, as HBeAg status, HBV DNA levels, and ALT levels change with time, a longer-term follow-up that evaluates these parameters may reveal the reason(s) for the difference. The impact of other direct viral factors on liver cirrhosis, such as the G1898A mutation in association with TCC at nucleotides 1856 to 1858, will need further investigation (23).
Patients infected with HBV with TCC at nucleotides 1856 to 1858 tend to have a higher prevalence of HBeAg positivity and higher ALT levels than those infected with HBV with CCT. Due to the nucleotide pairing at the stem-loop structure of the precore region, the G1986A mutation develops only in the presence of T and not C at nucleotide 1858 (23). Genotype C HBV with CCT at nucleotides 1856 to 1858 can therefore develop a precore stop codon mutation (G1896A), as in genotype B HBV. In other words, patients infected with genotype C HBV isolates with CCT at codon 15 may also develop HBeAg seroconversion and ALT normalization earlier than patients infected with genotype C HBV with C-1858 (17). Despite an earlier HBeAg seroconversion, patients infected with genotype C HBV with CCT at nucleotides 1856 to 1858 do not seem to have less liver cirrhosis than patients infected with genotype C HBV with TCC. Other investigators (31) have observed the G1899A mutation, which is associated with HBV bearing T-1858, among patients with more active liver disease. Whether this HBV mutant causes more accelerated liver damage is still unknown.
Owing to the stem-loop structure of the precore genome, nucleotide 1856 forms a covalent bond with nucleotide 1898 (Fig. 1). A T at nucleotide 1856 favors a G-to-A mutation at nucleotide 1898 (23). The G1898A mutation probably develops with time, as it appears to be more common among older subjects. The higher prevalence of this mutation among cirrhotic patients may therefore be confounded by the aging effect. The association of G1898A and higher ALT levels, as found in previous studies (23), cannot be confirmed in this study; and we could not demonstrate higher HBV DNA levels associated with G1898A mutation. Whether the G1898A mutation increases the replicative competency of HBV or is merely a compensatory mutation that stabilizes the stem-loop structure of the encapsidation sequence will await further experimental evidence.
A sensitive and robust laboratory method is essential for the study of HBV genotypes and subgroups in epidemiological studies. In this study, we used the 5′-nuclease technology of the TaqMan real-time PCR, which can detect HBV DNA at levels down to the order of 100 to 1,000 viral copies/ml, to differentiate the two genotype C subgroups (5, 22). The polymorphism at nucleotide 2733 is very specific for the differentiation of HBV genotypes Cs and Ce, and the adjacent genomic sequence covered by the TaqMan probe is well conserved in genotype C HBV. This feature allows a very high yield (85%) of detection, despite the high sensitivity of the TaqMan probe down to a single nucleotide change of the DNA sequence. On the other hand, the genomic sequence covered by the TaqMan probe has 1 to 3 nucleotide differences between the sequence of HBV genotype C and those of HBV genotypes A, B, E, F, and G. We anticipate that a negative signal will be obtained for these HBV genotypes, as a nucleotide mismatch, particularly near the center position of the probe, will disrupt the probe-template hybrid stability. In the cases of our genotype B HBV controls, none of the 15 samples in this study had a positive signal by our novel assay. The three samples with mixed genotype B and C HBV isolates also had negative signals in this study. As we do not have non-B, non-C HBV genotypes in our locality, the specificity of our novel assay for these genotypes requires further validation. This test may potentially facilitate future epidemiological studies of genotype C HBV.
Our study has several limitations. First, we did not assess the liver histology of the patients, and therefore, we cannot comment on the degree of fibrosis and may have missed cases of early liver cirrhosis. On the other hand, we avoided mislabeling of the lack of clinical liver cirrhosis, which often occurs because of the sampling error of the liver biopsy procedure. Second, none of the patients had hepatocellular carcinoma at the time of recruitment. Further case-control studies or large-scale studies with long-term follow-up are needed to address the carcinogenic potential of HBV bearing TCC at nucleotides 1856 to 1858. Last, our finding of an association of HBV bearing TCC with more active liver disease is only preliminary and is based on the findings of a cross-sectional study. Further longitudinal studies are required to confirm the causal relationship of this HBV strain and the development of liver cirrhosis.
In conclusion, we have characterized an HBV strain with TCC at nucleotides 1856 to 1858 of the precore region. This HBV strain belongs to a specific subgroup, subgroup Cs, of genotype C HBV, which is reported only in Hong Kong, Guangzhou, and Vietnam. Its clinical behavior appears to be more aggressive than those of genotype C HBV isolates with other nucleotide configurations at nucleotides 1856 to 1858. Further studies on the basic mechanism of liver damage by this HBV strain and its association with hepatocellular carcinoma are warranted.
Acknowledgments
This study was supported by Innovation and Technology Fund (ITS/188/01) to J.J.-Y.S. and the Michael Kadoorie Cancer Genetics Research Programme (project ID 4720054) to H.L.-Y.C., Hong Kong Special Administrative Region, China.
REFERENCES
- 1.Akarca, U. S., S. Greene, and A. S. F. Lok. 1994. Detection of precore hepatitis B virus mutants in asymptomatic HBsAg-positive family members. Hepatology 19:1366-1370. [PubMed] [Google Scholar]
- 2.Alestig, E., C. Hannoun. P. Horal, and M. Lindh. 2001. Phylogenetic origin of hepatitis B virus strains with precore C-1858 variant. J. Clin. Microbiol. 39:3200-3203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arauz-Ruiz, P., H. Norder, B. H. Robertson, and L. O. Magnius. 2002. Genotype H: a new Amerindian genotype of hepatitis B virus revealed in Central America. J. Gen. Virol. 83:2059-2073. [DOI] [PubMed] [Google Scholar]
- 4.Buckwold, V. E., Z. Xu, M. Chen, T. S. B. Yen, and J. H. Ou. 1996. Effects of a naturally occurring mutation in the hepatitis virus basal core promoter on precore gene expression and viral replication. J. Virol. 70:5845-5851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chan, H. L. Y., A. K. K. Chui, W. Y. Lau, F. K. L. Chan, M. L. Wong, C. H. Tse, A. R. N. Rao, J. Wong, and J. J. Y. Sung. 2002. Factors associated with viral breakthrough in lamivudine monoprophylaxis of hepatitis B virus recurrence after liver transplantation. J. Med. Virol. 68:182-187. [DOI] [PubMed] [Google Scholar]
- 6.Chan, H. L. Y., A. Y. Hui, M. L. Wong, A. M. L. Tse, L. C. T. Hung, V. W. S. Wong, and J. J. Y. Sung. 2004. Genotype C hepatitis B virus infection is associated with increased risk of hepatocellular carcinoma. Gut 53:1494-1498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chan, H. L. Y., M. Hussain, and A. S. F. Lok. 1999. Different hepatitis B virus genotypes are associated with different mutations in the core promoter and precore regions during hepatitis B e antigen seroconversion. Hepatology 29:976-984. [DOI] [PubMed] [Google Scholar]
- 8.Chan, H. L. Y., M. L. Wong, A. Y. Hui, L. C. T. Hung, F. K. L. Chan, and J. J. Y. Sung. 2003. Genotype C hepatitis B virus takes a more aggressive disease course than genotype B hepatitis B virus in hepatitis B e antigen-positive patients. J. Clin. Microbiol. 41:1277-1279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chan, H. L. Y., N. W. Y. Leung, M. Hussain, M. L. Wong, and A. S. F. Lok. 2000. Hepatitis B e antigen-negative chronic hepatitis B in Hong Kong. Hepatology 31:763-768. [DOI] [PubMed] [Google Scholar]
- 10.Chan, H. L. Y., S. K. W. Tsui, C. H. Tse, E. Y. T. Ng, T. C. C. Au, L. Yuen, A. Bartholomeusz, K. S. Leung, K. H. Kee, S. Locarnini, and J. J. Y. Sung. 2005. Epidemiological and virological characteristics of two subgroups of genotype C hepatitis C virus. J. Infect. Dis. 191:2022-2032. [DOI] [PubMed] [Google Scholar]
- 11.Chan, H. L. Y., S. W. C. Tsang, C. T. Liew, C. H. Tse, M. L. Wong, J. Y. L. Ching, N. W. Y. Leung, J. S. L. Tam, and J. J. Y. Sung. 2002. Viral genotype and hepatitis B virus DNA levels are correlated with histological liver damage in HBeAg-negative chronic hepatitis B virus infection. Am. J. Gastroenterol. 97:406-412. [DOI] [PubMed] [Google Scholar]
- 12.Chu, C. J., M. Hussain, and A. S. F. Lok. 2002. Hepatitis B virus genotype B is associated with earlier HBeAg seroconversion compared with hepatitis B virus genotype C. Gastroenterology 122:1756-1762. [DOI] [PubMed] [Google Scholar]
- 13.Huy, T. T. T., H. Ushijima, V. X. Quant, K. M. Win, P. Luengrojanakul, K. Kikuchi, T. Sata, and K. Abe. 2004. Genotype C of hepatitis B virus can be classified into at least two subgroups. J. Gen. Virol. 85:283-292. [DOI] [PubMed] [Google Scholar]
- 14.Huy, T. T. T., and K. Abe. 2004. Molecular epidemiology of hepatitis B and C virus infections in Asia. Pediatr. Int. 46:223-230. [DOI] [PubMed] [Google Scholar]
- 15.Kao, J. H., P. J. Chen, M. Y. Lai, and D. S. Chen. 2003. Basal core promoter mutations of hepatitis B virus increase the risk of hepatocellular carcinoma in hepatitis B carriers. Gastroenterology 124:327-334. [DOI] [PubMed] [Google Scholar]
- 16.Kao, J. H., P. J. Chen, M. Y. Lai, and D. S. Chen. 2002. Clinical and virological aspects of blood donors infected with hepatitis B genotype B and C. J. Clin. Microbiol. 40:22-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kao, J. H., P. J. Chen, M. Y. Lai, and D. S. Chen. 2000. Hepatitis B genotypes correlate with clinical outcome in patients with chronic hepatitis B. Gastroenterology 118:554-559. [DOI] [PubMed] [Google Scholar]
- 18.Kuang, S. Y., P. E. Jackson, J. B. Wang, P. X. Lu, A. Munoz, G. S. Qian, T. W. Kenslet, and J. D. Groopman. 2004. Specific mutations of hepatitis B virus in plasma predict liver cancer development. Proc. Natl. Acad. Sci. USA 101:3575-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li, J. S., S. P. Tong, Y. M. Wen, L. Vitvitski, Q. Shang, and C. Trepo. 1993. Hepatitis B virus genotype A rarely circulates as an HBe-minus mutant: possible contribution of a single nucleotide in the precore region. J. Virol. 67:5402-5410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lindh, M., A. S. Andersson, and A. Gusdal. 1997. Genotypes, nt 1858 variants, and geographical origin of hepatitis B virus—large-scale analysis using a new genotyping method. J. Infect. Dis. 175:1285-1293. [DOI] [PubMed] [Google Scholar]
- 21.Lindh, M., Y. Furuta, A. Vahlne, G. Norkrans, and P. Horal. 1995. Emergence of precore TAG mutation during hepatitis e seroconversion and its dependence on pregenomic base pairing between nucleotide 1858 and 1896. J. Infect. Dis. 172:1343-1347. [DOI] [PubMed] [Google Scholar]
- 22.Loeb, K. R., K. R. Jerome, J. Goddard, M. L. Huang, A. Cent, and L. Corey. 2000. High-throughput quantitative analysis of hepatitis B virus DNA in serum using the TaqMan fluorogenic detection system. Hepatology 32:626-629. [DOI] [PubMed] [Google Scholar]
- 23.Lok, A. S. F., U. Akarca, and S. Greene. 1994. Mutations in the pre-core region of hepatitis B virus serve to enhance the stability of the secondary structure of the pre-genome encapsidation signal. Proc. Natl. Acad. Sci. USA 91:4077-4081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Luo, K., Z. Liu, H. He, J. Peng, W. Liang, W. Kai, and J. Hou. 2004. The putative recombination of hepatitis B virus genotype B with pre-C-C region of genotype C. Virus Genes 29:31-41. [DOI] [PubMed] [Google Scholar]
- 25.Norder, H., A. M. Courouce, and L. O. Magnius. 1994. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology 198:489-503. [DOI] [PubMed] [Google Scholar]
- 26.Orito, E., T. Ichida, H. Sakugawa, M. Sata, N. Horike, K. Hino, K. Okita, TR. Okanoue, S. Iino, E. Tanaka, K. Suzuki, H. Watanabe, S. Hige, and M. Mizokami. 2001. Geographical distribution of hepatitis B virus (HBV) genotype in patients with chronic HBV infection in Japan. Hepatology 34:590-594. [DOI] [PubMed] [Google Scholar]
- 27.Rodriguez-Frias, F., M. Biti, R. Jardi, M. Cotrina, L. Viladomiu, R. Esteban, and J. Guardia. 1995. Hepatitis B virus infection: precore mutants and its relation to viral genotypes and core mutations. Hepatology 22:1641-1647. [DOI] [PubMed] [Google Scholar]
- 28.Sakurai, M., F. Sugauchi, N. Tsai, S. Suzuki, I. Hasegawa, K. Fujiwara, E. Orito, R. Ueda, and M. Mizokami. 2004. Genotype and phylogenetic charactrerization of hepatitis B virus among multi-ethnic cohort in Hawaii. World J. Gastroenterol. 10:2218-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scaglioni, P. P., M. Melegari, and J. R. Wands. 1997. Biological properties of hepatitis B viral genomes with mutations in the precore promoter and precore open reading frame. Virology 233:374-381. [DOI] [PubMed] [Google Scholar]
- 30.Stuyver, L., S. De Gendt, C. Van Geyt, F. Zoulim, M. Fried, R. F. Schinazi, and R. Rossau. 2000. A new genotype of hepatitis B virus: complete genome and phylogenetic relatedness. J. Gen. Virol. 81:67-74. [DOI] [PubMed] [Google Scholar]
- 31.Tillmann, H., C. Trautwein, D. Walker, K. Michitaka, S. Kubicka, K. Boker, and M. Manns. 1995. Clinical relevance of mutations in the precore genome of the hepatitis B virus. Gut 37:568-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu, M. W., S. H. Yeh, P. J. Chen, Y. F. Liaw, C. L. Lin, C. J. Liu, W. L. Shih, J. H. Kao, D. S. Chen, and C. J. Chen. 2005. Hepatitis B virus genotype and DNA level and hepatocellular carcinoma: a prospective study in men. J. Natl. Cancer Inst. 97:265-272. [DOI] [PubMed] [Google Scholar]