Abstract
Many clinical laboratories are familiar with a sizeable group of “unserotypeable Yersinia enterocolitica” strains. Due to identification problems, this group may hide Y. bercovieri, Y. mollaretii, and Y. rohdei strains. We present a simple scheme to distinguish between pathogenic Y. enterocolitica and potentially nonpathogenic Y. enterocolitica-like strains.
Yersinia enterocolitica, Y. pseudotuberculosis, and Y. pestis have clearly been shown to cause human disease, while characterization of the remaining eight Yersinia species often referred as “Yersinia enterocolitica-like” strains has been more limited. Recently, however, these species thought to be nonpathogenic to humans have been found to possess novel virulence mechanisms, and some of them have been associated with human disease (3, 4, 14, 16, 17). Among these species, identification of Y. bercovieri, Y. mollaretii, and Y. rohdei is a problem for clinical microbiology laboratories because the widely used commercial identification systems (for example, API 20 E, API Rapid 32 IDE, Micronaut E, and the Vitek GNI Card) do not list these species in their databases and usually misidentify them as Y. enterocolitica (10, 11). The identification of Yersinia in clinical microbiology laboratories is generally based on the combination of results from these biochemical identification systems and commercially available antisera against pathogenic serotypes of Y. enterocolitica. In addition, a 16S rRNA gene-based PCR assay for identifying and separating Y. enterocolitica isolates of European and American origin has recently been developed (12). However, this method involves sequencing, which is not easily applicable in most routine clinical microbiology laboratories. In the present study, we provide readers with simple means of recognizing and identifying Y. enterocolitica-like strains, some of which may have pathogenic potential.
In this study, unserotypeable Y. enterocolitica strains (n = 67) isolated and identified by Finnish hospital laboratories were retested at the Enteric Bacteria Laboratory (EBL), National Public Health Institute, with API 20 E (bioMérieux, France) at 30°C and by slide agglutination with antisera against Y. enterocolitica O:3, O:5, O:8, and O:9 (Denka Seiken, Japan). The strains were also biotyped according to Wauters et al. (18), and 55 strains belonged to biotype (BT) 1A (data not shown). Of the remaining 12 strains, 11 were not biotypeable (at least two reactions diverged from the established biotypes), and one strain (IH 111767) was BT 3 but was unserotypeable (Table 1). Thus, their identification as Y. enterocolitica strains was considered doubtful. These strains were further tested for fermentation of sorbose and fucose (at 25°C for 24 and 48 h) and on Congo red-magnesium oxalate agar (CR-MOX test) (13). For comparison, five strains of BTs 1A, 3, and 4 were included (Table 1). The colony morphology of all 17 strains through a stereomicroscope (Olympus [Japan] SZH10 zoom stereomicroscope with an SZH-ILLK illumination base) was examined on cefsulodin-irgasan-novobiocin (CIN) agar (Oxoid) incubated at 30°C for 22 to 24 h and compared to those of Y. enterocolitica (ATCC 9610, NCTC 11176, and RH 4823 [BT 1A control strain of EBL, National Public Health Institute]), Y. mollaretii (ATCC 43969), Y. bercovieri (ATCC 43970), and Y. rohdei (ATCC 43380 and 43872) cultured and incubated in parallel.
TABLE 1.
Further characterization of Y. enterocolitica strains identified by clinical laboratories
| Strain | Yr | Original IDa (37°C) | API 20E profile (30°C) | STb | Morphologyc | CR-MOXd | Reactions (25°C) in BTe
|
srbf | fuc | YeO:3RS profileg | ID after sequencingh | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| esc | sal | pyz | lip | xyl | tre | ind | VP | BT | |||||||||||
| IH 40555 | 1988 | Y. enterocolitica | 1114523 (99.7%) | NA | Y. mollaretii | − | − | − | + | − | + | + | − | + | NBT | + | − | W | Y. mollaretii |
| IH 41571 | 1995 | Y. enterocolitica? | 1014763 (53%) | NA | Mostly like Y. kristensenii | − | − | − | + | − | + | + | − | (+) | NBT | − | − | ND | Y. rohdei |
| IH 110292 | 1997 | Y. enterocolitica | 1114523 (99.7%) | NT | Y. mollaretii | − | − | − | + | − | + | + | − | − | NBT | + | − | W | Y. mollaretii |
| IH 111322 | 1999 | Y. enterocolitica | 0114523 (99.9%) | NA | Mostly like Y. mollaretii | Ca+ | − | − | + | − | + | + | − | + | NBT | − | + | W | Y. bercovieri |
| IH 111501 | 1999 | Y. enterocolitica | 1114523 (99.7%) | NA | Y. bercovieri | − | − | − | + | − | + | + | + | + | NBT | − | + | W | Y. bercovieri |
| IH 111541 | 1999 | Y. enterocolitica | 1014522 (93.9%) | NA | Mostly like Y. mollaretii | (+) | − | − | (+) | − | + | + | − | + | NBT | + | − | W | Y. mollaretii |
| IH 116003 | 2000 | Y. enterocolitica? | 1114703 (26.2%) | NA | Unique | − | − | − | + | − | + | + | − | (+) | NBT | + | − | W | Y. bercovieri |
| IH 116025 | 2000 | Y. enterocolitica | 0114523 (99.9%) | NA | Mostly like Y. bercovieri | − | − | − | + | − | + | + | − | − | NBT | − | + | W | Y. bercovieri |
| IH 116028 | 2000 | Y. enterocolitica | 0014523 (97.7%) | NA | Mostly like Y. bercovieri | − | − | − | + | − | + | + | − | − | NBT | − | + | W | Y. bercovieri |
| IH 111767 | 2000 | Y. enterocolitica | 1014522 (93.9%) | NT | Mostly like Y. mollaretii | − | − | − | − | − | + | + | − | − | BT3 | + | − | W-I | Y. mollaretii |
| IH 111778 | 2000 | Y. enterocolitica | 0154723 (99.2%) | NT | Atypical Y. enterocolitica BT1A | − | + | + | + | + | + | + | − | − | NBT | + | + | W | Y. enterocolitica |
| IH 111799 | 2000 | Y. enterocolitica? | 1014122 (13.8%) | NT | Y. mollaretii | Ca+ | − | − | (+) | − | + | + | − | + | NBT | + | − | W | Y. mollaretii |
| Comparison strains | |||||||||||||||||||
| IH 111430 | 1999 | Y. enterocolitica | 1154763 (97.8%) | NT | Mostly like Y. enterocolitica | (+) | + | + | + | + | + | + | + | + | BT1A | ND | ND | W | Y. enterocolitica |
| IH 111517 | 1999 | Y. enterocolitica | 1014523 (96.4%) | O:3 | Y. enterocolitica O:3 | + | − | − | − | − | − | + | − | − | BT 4 | ND | ND | 3.2a | Y. enterocolitica |
| IH 116007 | 2000 | Y. enterocolitica | 0114723 (99.9%) | O:3 | Y. enterocolitica O:3 | + | − | − | − | − | − | + | − | − | BT 4 | ND | ND | 3.11k | Y. enterocolitica |
| IH 41122 | 1991 | Y. enterocolitica | 1154523 (92.3%) | O:9 | Y. enterocolitica O:9 | + | − | − | − | − | + | + | − | + | BT 3 | ND | ND | 9.4b | Y. enterocolitica |
| IH 41334 | 1993 | Y. enterocolitica | 1015723 (92.8%) | O:5 | Y. enterocolitica O:5,27 | + | − | − | − | − | + | + | − | + | BT 3 | ND | ND | 5.6b | Y. enterocolitica |
Identified by different hospital laboratories.
Serotype of the strain determined by agglutination with sera O:3, O:5, O:8, and O:9 by Denka Seiken (Tokyo, Japan). NA, not agglutinating with the sera used; NT, not typeable, cross-reacting with the sera used.
Colony morphology on CIN agar (incubation at 30°C for 22 to 24 h) observed through a stereomicroscope and compared to ATCC reference strains.
Congo red magnesium oxalate agar test (13) for the presence of the virulence plasmid at 37°C for 24 and 48 h. −, negative; (+), weak positive; +, positive; Ca+, strain has pinpoint colonies showing calcium dependence (5) but does not bind Congo red.
Y. enterocolitica strains divided into BT 1 to BT 5 according to Wauters et al. (18). BT consists of tests for esculin (esc), salicin (sal), pyrazinamidase (pyz), lipase (Tween-esterase) (lip), xylose (xyl), trehalose (tre), indole (ind), and Voges-Proskauer at 25°C for 24 and 48 h. NBT, not biotypeable.
Srb, sorbose; fuc, fucose tests at 25°C for 24 and 48 h. ND, not determined.
Genotyping using Y. enterocolitica O:3 repeated sequence probe (6). W, weak hybridization; I, incomplete typing pattern; ND, not determined.
Genotyping was performed using hybridization with a YeO:3RS probe as described previously (6). For sequencing, DNA was isolated from bacterial cells by being boiled. Primers (forward, FD1 MOD; and reverse, 533r) were used to amplify the beginning (450 bp) of the 16S rRNA gene sequence (8, 9). Sequences with the 533r primer were determined with an ABIPrism 310 Genetic Analyzer using BigDye fluorescent terminator chemistry (Applied Biosystems, Warrington, United Kingdom).
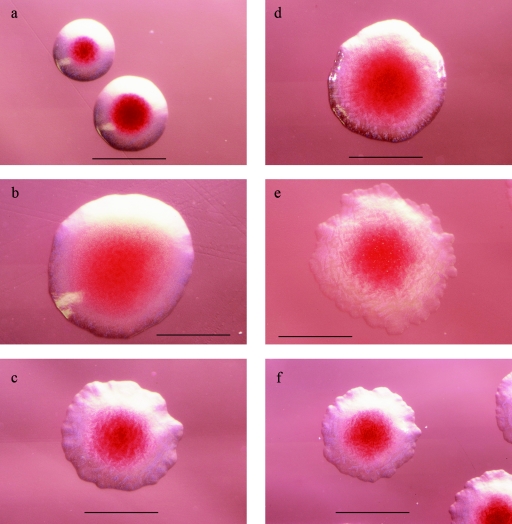
API 20 E identified 9 of the 12 doubtful strains as Y. enterocolitica with >90% certainty (Table 1). In contrast, sequencing revealed only one Y. enterocolitica strain (identical to ATCC 9610 and NCTC 11176) but five Y. mollaretii strains (0- to 1-nucleotide difference from ATCC 43969), five Y. bercovieri strains (0- to 3-nucleotide difference from ATCC 43970), and one Y. rohdei strain (identical to ATCC 43380). Based on the colony morphology on CIN agar, all of the Y. mollaretii strains and three of the five Y. bercovieri strains (IH 111501, IH 116025, and IH 116028) were tentatively identified as Y. mollaretii or Y. bercovieri, respectively, compared to the reference strains prior to any other testing (Table 1; Fig. 1). The microscopic examination of Y. mollaretii and Y. bercovieri colonies (approximately 1.5 mm in diameter) revealed characteristic erose edges and ground-glass appearance of the translucent zone surrounding the red center of the colonies (best visible in slightly oblique illumination). These features distinguished them from the colonies of Y. enterocolitica BT 1A (approximately 2 mm in diameter, larger center of the colony, and the surrounding zone devoid of ground-glass appearance), serotype O:3 (approximately <1 mm in diameter, smaller, deeper red center of the colony with a sharper border (Fig. 1), and serotype O:9 (data not shown). The rest of the non-Y. enterocolitica strains (IH 41571, IH 116003, and IH 111799) had variable colony morphology (Table 1) that clearly differed from the morphology of Y. enterocolitica. Of the 11 non-Y. enterocolitica strains, it was possible to avoid misidentification as Y. enterocolitica for all 11 strains by colony morphology, but only for 3 strains (IH 41571, IH 116003, and IH 111799) with API 20 E.
FIG. 1.
Differentiating between Y. enterocolitica, Y. bercovieri, and Y. mollaretii on CIN agar (incubation at 30°C, 22 to 24 h) through a stereomicroscope (black bar, 1 mm). (a) Y. enterocolitica O:3 (pathogenic serotype, BT 4) appears as characteristically small (approximately <1 mm in diameter), circular colonies with entire edge. The colonies have a small, deep red center (bull's eye) with a sharp border surrounded by a translucent or transparent zone. (b) Y. enterocolitica BT 1A (nonpathogenic biotype). Large (approximately 2 mm in diameter), circular colonies with slightly lighter red center surrounded by a translucent to milk-white zone are shown. The center of the colony is large compared to the surrounding zone and has a blurred border. (c) Y. bercovieri. Erose-edged, slightly irregular circular colonies (approximately 1.5 mm in diameter) are shown, with a medium-red (sometimes pitting) center with an erose border. The surrounding translucent zone has a characteristic ground-glass appearance (best visible by slightly oblique illumination). (d and e) Y. mollaretii. Slightly irregular circular colonies (approximately 1.5 mm in diameter) are shown; a medium-red, diffuse center with no sharp borderline is visible. The surrounding translucent to milk-white zone has a characteristic ground-glass appearance (best visible in slightly oblique illumination). Two types exist: mucoid, with a smoother and more convex appearance (d) and a flat, dry, more irregular- and erose-edged colony (e). It has been noticed in EBL that approximately half of the incoming Y. mollaretii strains have a smooth, mucoid colony type and the other half have a dry, flat colony type (unpublished data). (f) Y. enterocolitica O:5,27 (pathogenic serotype). Circular, usually erose-edged (almost starlike) colonies (approximately 1 to 1.5 mm in diameter) are shown. A small, deep-red center with a slightly blurred border is visible. The surrounding translucent or transparent zone is large, compared to the center (as with Y. enterocolitica O:3), and has a ground-glass appearance.
In EBL, it has been found useful to start the identification of API 20 E-verified Y. enterocolitica strains received from clinical microbiology laboratories by examining the microscopic colony morphology on CIN agar and by CR-MOX testing. The strains are then forwarded for serotyping (i.e., the appearance of pathogenic serotypes O:3 and O:9), biotyping (the appearance of other strains), and additional biochemical testing (Y. enterocolitica-like appearance and presence of nonbiotypeable strains) if necessary. For example, Y. bercovieri and Y. mollaretii strains, misidentified as Y. enterocolitica, are easily revealed by colony morphology and by negative reactions to esculine, salicine, and lipase and a positive reaction to pyrazinamidase. Y. enterocolitica serotype O:5,27, which is generally less frequently isolated, has a colony morphology sometimes similar to that of Y. bercovieri and Y. mollaretii (Fig. 1), but it can be distinguished by a negative pyrazinamidase reaction with biotyping. In this study, the only exception was the strain IH 111767 with a negative pyrazinamidase reaction, placing it to Y. enterocolitica BT 3, but confirmed as Y. mollaretii after sequencing. Distinguishing between Y. bercovieri and Y. mollaretii can then be made by tests for fucose and sorbose (Table 1) (2, 15). Sequencing is necessary only for a few strains remaining unidentified after these steps, including rare cases of Y. rohdei, pyrazinamidase-negative Y. bercovieri or Y. mollaretii with colony morphology similar to that of Y. enterocolitica O:5,27, and Y. enterocolitica with coinciding atypical morphology and biotype. After introduction of the microscopic morphology in the identification process in EBL, it has been noticed that approximately half of the incoming Y. mollaretii strains have a smooth, mucoid colony type; the other half have a dry, flat colony type (unpublished data).
The cross-reactions of the commercial antisera in serotyping are typical of Y. enterocolitica-like strains and Y. enterocolitica BT 1A (1, 19). In EBL in 2000, the nonserotypeable strains represented about 40% of the incoming Y. enterocolitica strains (7). Therefore, building the identification of Y. enterocolitica solely on the use of these sera, together with a diagnostic kit like API 20 E, is inadequate. For laboratories that have limited capacity for biotyping, the simplest way to avoid misidentifications is to compare the colony morphology of a API 20 E-identified Y. enterocolitica strain with the Y. enterocolitica control strains representing serotypes and/or biotypes O:3/4, O:5,27/2 or O:5,27/3, O:8/1B, O:9/2, O:9/3, and BT 1A.
The probe YeO:3RS contains a region upstream of the Y. enterocolitica O-antigen cluster, a repeated sequence (RS) that is present in multiple copies in the genome. In our previous study (6), the RS was shown to be present only in the genome of the “European” pathogenic serotypes (namely, O:3, O:5,27, O:9, O:1, and O:2) of Y. enterocolitica. The sequence was absent from the genomes of other Y. enterocolitica serotypes and Yersinia species, resulting in a weak or incomplete typing pattern of those strains in that study. The current results are in accordance with the previous observations; none of the Y. enterocolitica-like strains were typeable with the probe compared to the complete typing pattern of the “European” pathogenic bioserotypes in this study. The latter strains were also clearly positive by a CR-MOX test (Table 1).
To summarize, a simple scheme for identification of Y. enterocolitica-like strains is presented. Without accurate identification, already in the primary diagnostics, it is impossible to gain insight into the true clinical significance of Y. enterocolitica-like species. Although the advanced molecular methods are constantly developed, they still may not be available in many routine clinical microbiology laboratories. Therefore, it was interesting to notice how easily the straightforward comparison of the colony morphology of Yersinia isolates can effectively prevent the misidentification of a strain as Y. enterocolitica.
REFERENCES
- 1.Aleksic, S. 1995. Occurrence of Y. enterocolitica antigens O:3, O:9 and O:8 in different Yersinia species, their corresponding H antigens and origin. Contrib. Microbiol. Immunol. 13:89-92. [PubMed] [Google Scholar]
- 2.Aleksic, S., and J. Bockemühl. 1999. Yersinia and other Enterobacteriaceae, p. 483-491. In P. R. Murray, E. J. Baron, M. A. Pfaller, F. C. Tenover, and R. H. Yolken (ed.), Manual of clinical microbiology, 7th ed. ASM Press, Washington, D.C.
- 3.Bottone, E. J. 1997. Yersinia enterocolitica: the charisma continues. Clin. Microbiol. Rev. 10:257-276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Delor, I., A. Kaeckenbeek, G. Wauters, and G. R. Cornelis. 1990. Nucleotide sequence of yst, the Yersinia enterocolitica gene encoding the heat-stable enterotoxin, and the prevalence of the gene among pathogenic and non-pathogenic yersiniae. Infect. Immun. 58:2983-2988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gemski, P., J. R. Lazere, and T. Casey. 1980. Plasmid associated with pathogenicity and calcium dependency of Yersinia enterocolitica. Infect. Immun. 27:682-685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hallanvuo, S., M. Skurnik, K. Asplund, and A. Siitonen. 2002. Detection of a novel repeated sequence useful for epidemiological typing of pathogenic Yersinia enterocolitica. Int. J. Med. Microbiol. 292:215-225. [DOI] [PubMed] [Google Scholar]
- 7.Hallanvuo, S., and A. Siitonen. 2002. The fecal Yersinia enterocolitica isolates—clinically significant or not? Bull. Natl. Publ. Health Inst. Finl. 9:9-10. (In Finnish.) [Google Scholar]
- 8.Jalava, J. 2000. Molecular detection and identification of bacteria based on PCR and rRNA phylogeny—medical applications. Ph.D. thesis. University of Turku, Turku, Finland.
- 9.Kotilainen, P., J. Jalava, O. Meurman, O.-P. Lehtonen, E. Rintala, O.-P. Seppälä, E. Eerola, and S. Nikkari. 1998. Diagnosis of meningococcal meningitis by broad-range bacterial PCR with cerebrospinal fluid. J. Clin. Microbiol. 36:2205-2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Linde, H-J., H. Neubauer, H. Meyer, S. Aleksic, and N. Lehn. 1999. Identification of Yersinia species by the Vitek GNI card. J. Clin. Microbiol. 37:211-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Neubauer, H., T. Sauer, H. Becker, S. Aleksic, and H. Meyer. 1998. Comparison of systems for identification and differentiation of species within the genus Yersinia. J. Clin. Microbiol. 36:3366-3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neubauer, H., A. Hensel, S. Aleksic, and H. Meyer. 2000. Identification of Yersinia enterocolitica within the genus Yersinia. Syst. Appl. Microbiol. 23:58-62. [DOI] [PubMed] [Google Scholar]
- 13.Riley, G., and S. Toma. 1989. Detection of pathogenic Yersinia enterocolitica by using Congo red-magnesium oxalate agar medium. J. Clin. Microbiol. 27:213-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Robins-Browne, R. M., S. Ciancosi, A.-M. Bordun, and G. Wauters. 1991. Pathogenicity of Yersinia kristensenii for mice. Infect. Immun. 59:162-167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stock, I., B. Henrichfreise, and B. Wiedemann. 2002. Natural antibiotic susceptibility and biochemical profiles of Yersinia enterocolitica-like strains: Y. bercovieri, Y. mollaretii, Y. aldovae and ‘Y. ruckeri.’ J. Med. Microbiol. 51:56-69. [DOI] [PubMed] [Google Scholar]
- 16.Sulakvelidze, A. 2000. Yersiniae other than Y. enterocolitica, Y. pseudotuberculosis, and Y. pestis: the ignored species. Microbes Infect. 2:497-513. [DOI] [PubMed] [Google Scholar]
- 17.Sulakvelidze, A., A. Kreger, A. Joseph, et al. 1999. Production of enterotoxin by Yersinia bercovieri, a recently identified Yersinia enterocolitica-like species. Infect. Immun. 67:968-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wauters, G., K. Kandolo, and M. Janssens. 1987. Revised biogrouping scheme of Yersinia enterocolitica. Contrib. Microbiol. Immunol. 9:14-21. [PubMed] [Google Scholar]
- 19.Wauters, G., M. Janssens, A. G. Steigerwalt, and D. J. Brenner. 1988. Yersinia mollaretii sp. nov., Yersinia bercovieri sp. nov., formerly called Yersinia enterocolitica biogroups 3A and 3B. Int. J. Syst. Bacteriol. 38:424-429. [Google Scholar]