Abstract
The number of infections attributable to community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) in Singapore is progressively increasing. Most cases in the past 2 years were caused by Panton-Valentine leukocidin-positive isolates belonging to sequence type 30, according to multilocus sequence typing. This has clearly become the predominant sequence type among CA-MRSA isolates in Singapore.
Community-associated methicillin-resistant Staphylococcus aureus (CA-MRSA) has become a global phenomenon (2, 10, 16, 18). CA-MRSA isolates are characterized by the presence of the smaller and theoretically more mobile staphylococcal chromosome cassette mec (SCCmec) types IV and V (7, 18), as well as the presence of Panton-Valentine leukocidin (PVL) genes, in the majority of reported cases (18).
The discovery of sporadic CA-MRSA isolates in Singapore (4) led to the development of a basic surveillance program aimed towards monitoring CA-MRSA trends locally. In essence, microbiologists and infectious disease physicians belonging to local public hospitals were asked to identify MRSA isolates that had antibiotic susceptibility profiles which differed from those of the dominant local healthcare-associated isolates (5) and/or had been cultured under circumstances matching local epidemiologic criteria for CA-MRSA, namely, isolation of the organism within 48 h of hospitalization from patients who had neither been hospitalized nor placed in a chronic nursing facility for >1 year (4). Referring doctors obtained clinical and epidemiological data for patients infected by these isolates via chart review, and the referring microbiology laboratories provided the antibiotic resistance profile of each isolate.
The isolates were processed at a central facility. They were confirmed to be S. aureus via coagulation of citrated rabbit plasma with EDTA (BBL Becton Dickinson and Co., Cockeysville, MD) and by the production of clumping factor and protein A (BactiStaph; Remel, Lenexa, KS). The molecular epidemiology of isolates was determined via pulsed-field gel electrophoresis (PFGE) using SmaI endonuclease (12), multilocus sequence typing (1), SCCmec typing (7, 10, 14), and multiple-locus variable-number tandem-repeat analysis (MLVA) (11). PFGE and MLVA gel images were collated using Molecular Analyst v1.6 (Bio-Rad) and were analyzed using the Dice coefficient and the unweighted-pair group method using average linkages. The presence of PVL genes was determined using previously established protocols (9).
A total of 37 isolates were submitted between May 2004 and June 2005. The sequence types and antibiotic resistance profiles of these isolates, determined using the disk diffusion method following CLSI (formerly NCCLS) guidelines (13), are shown in Table 1, with the majority being susceptible to all non-beta-lactam antibiotics tested. Most (30 of 37) CA-MRSA isolates were cultured from patients with cutaneous abscesses. There were three patients with bacteremia, one of whom had endocarditis, and a case of tibial osteomyelitis. The last two cases, along with four cases of cutaneous abscesses, had been described elsewhere (6). One isolate was obtained from the skin swab of a patient with exfoliative dermatitis. Two isolates were obtained from patients with wound infections shortly following elective surgery—these were the only two cases that did not meet epidemiologic criteria for community acquisition. However, molecular typing revealed that these were CA-MRSA isolates, as both isolates tested positive for PVL genes; one was sequence type 30 (ST30)-MRSA-IVc, and the other was ST8-MRSA-V. None of the 37 cases could be linked epidemiologically.
TABLE 1.
Antibiotic resistance profiles of 37 community-associated methicillin-resistant Staphylococcus aureus isolates stratified by year of isolation and multilocus ST
| Profile | Resistance to non-β-lactam antibiotics testeda | ST (no. of isolates)
|
|
|---|---|---|---|
| 2004 (May-December) | 2005 (January-June) | ||
| I | 30 (13) | 8 (2) | |
| 571 (1) | 30 (13) | ||
| 88 (1) | |||
| II | S, T | 188 (1) | |
| III | E, Cc | 7 (1) | |
| 78 (1) | |||
| IV | E, T | 59 (1) | |
| V | Ci, E, G | 45 (1) | |
| VI | E, Ci, T | 573 (2) | |
Cc, constitutive clindamycin resistance; Ci, inducible clindamycin resistance; E, erythromycin resistance; G, gentamicin resistance; S, sulfamethoxazole resistance; T, tetracycline resistance. All isolates were susceptible to ciprofloxacin, fusidic acid, rifampin, and vancomycin upon testing.
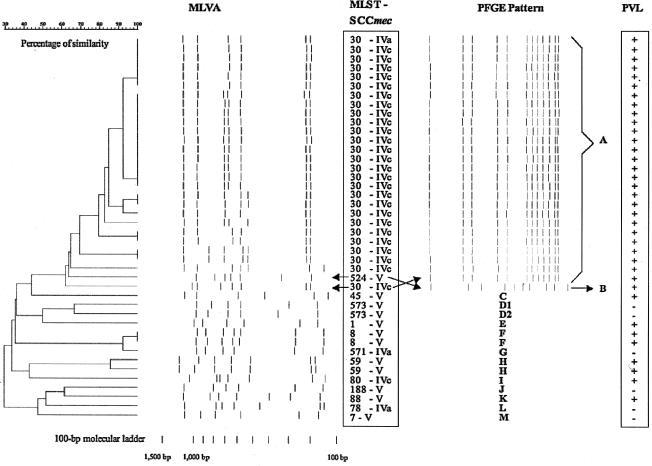
The distributions of PFGE patterns, MLVA patterns, sequence types (STs), and SCCmec subtypes among all 42 local CA-MRSA strains isolated since 2001, as well as the presence of PVL genes, are shown in Fig. 1. The majority of isolates since 2004 belonged to the ST30-MRSA-IVc group. In general, MLVA clusters correlated well with PFGE clusters and multilocus STs. However, all ST30 isolates had identical PFGE patterns, whereas MLVA patterns demonstrated significant variance, although the ST30 cluster did not overlap with other STs, except ST524, on the MLVA dendrogram.
FIG. 1.
Comparison of 42 local community-associated methicillin-resistant Staphylococcus aureus isolates (2001 to June 2005), using MLVA, multilocus sequence typing (MLST), SCCmec typing, PFGE, and determination of the presence of PVL genes. Isolates were considered to be related according to PFGE if gel patterns demonstrated 80% or greater similarity upon analysis using the Dice coefficient and the unweighted-pair group method using average linkages.
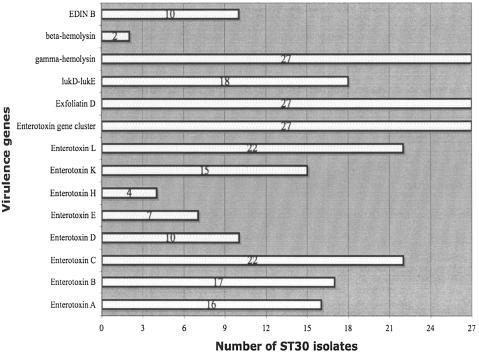
This discordance between MLVA and PFGE patterns prompted ancillary typing of all ST30 isolates via determination of the presence of virulence genes (enterotoxins A, B, C, D, E, H, K, and L; the enterotoxin gene cluster; gamma-, gamma-variant, and beta-hemolysin; exfoliatin toxins A, B, and D; toxic shock syndrome toxin 1; lukE-lukD; and EDIN A and B), as previously described (8). There was significant variance in the distributions of virulence genes among ST30 isolates (Fig. 2), which did not correspond to MLVA profiles and which differed from those in previous studies, where ST30 isolates showed remarkable consistency in possessing mainly the enterotoxin gene cluster, gamma-hemolysin, and the lukD-lukE genes only (3, 17, 18). The heterogeneity of enterotoxin gene distributions among our isolates is unique and suggests an exchange of virulence genes between these ST30 isolates and other S. aureus strains locally.
FIG. 2.
Distribution of virulence genes among 27 ST30 community-associated methicillin-resistant Staphylococcus aureus isolates. All isolates tested negative for the following genes: toxic shock syndrome toxin 1 (tsst-1), exfoliatin A and B, epidermal cell differentiation inhibitor (EDIN) A, and variant gamma-hemolysin. Positive PCR controls were not available for tsst-1, exfoliatin A and B, and EDIN-A.
Our findings suggest that the incidence of CA-MRSA is increasing in Singapore, notwithstanding the obvious bias caused by increasing recognition of this phenomenon over the past 2 years. The numbers reported here are likely to be an underestimation of the true incidence of CA-MRSA locally, as cases presenting to primary healthcare facilities are not captured by this surveillance program, and our criteria for identifying CA-MRSA are unlikely to be sensitive enough to pick up all such isolates.
All local CA-MRSA strains possessed either SCCmec type IV or V, and the great preponderance of cases of cutaneous abscesses is explained by the fact that most (36 of 42) isolates also had PVL genes.
The majority (27 of 42) of all local CA-MRSA isolates belonged to ST30 and had identical PFGE patterns, qualifying under the usual circumstances as clonal dissemination of a highly transmissible isolate. However, MLVA, which purports to have a discriminatory power similar to that of PFGE (11, 15), consistently demonstrated significant differences among several ST30 isolates (Fig. 1). This substantiates the findings in a recent study comparing MLVA and PFGE, where three clonal complex 30 isolates deemed related by PFGE were distinct according to MLVA (11). One explanation for this is that the hypervariable tandem-repeat loci selected for MLVA mutate more frequently and rapidly in ST30 isolates than in other previously tested S. aureus clonal types, and hence this is one clone which has spread rapidly and mutated as it was transmitted. However, the other circumstantial findings—a varied distribution of virulence genes and the presence of SCCmec IVa in one ST30 isolate (as opposed to SCCmec IVc in the rest)—appear to contradict this somewhat.
An alternative explanation is that PFGE may not be sufficiently discriminatory with regards to differentiating certain subtypes of ST30 and that our ST30 CA-MRSA isolates had arisen as a result of the transference of SCCmec to multiple ST30 methicillin-susceptible S. aureus isolates, some of which had subsequently disseminated. Why SCCmec V—which was found in more than a quarter of local CA-MRSA isolates—was not found in any ST30 isolate is a matter of conjecture. Although the implications differ, the truth can only be determined with further longitudinal studies.
In summary, the local epidemiology of CA-MRSA has evolved such that a predominant sequence type has emerged. This is in keeping with worldwide trends of CA-MRSA endemicity (5). In view of the progressive local increase in cases, a more comprehensive surveillance program is required to better gauge and monitor the problem of CA-MRSA such that timely interventions may be planned and prepared in advance. MLVA was found to be a discriminatory and rapid method for typing CA-MRSA isolates.
Acknowledgments
This study was supported by a grant from the National Medical Research Council (grant number NMRC/0903/2004).
We thank the staff of the Microbiology Laboratory at the Singapore General Hospital, especially Lan-Huay Ong and Grace Wang, for their assistance in this study.
REFERENCES
- 1.Enright, M. C., N. P. Day, C. E. Davies, S. J. Peacock, and B. G. Spratt. 2000. Multilocus sequence typing for characterization of methicillin-resistant and methicillin-sensitive clones of Staphylococcus aureus. J. Clin. Microbiol. 38:1008-1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ho, P. L., C. W. Tse, G. C. Mak, K. H. Chow, and T. K. Ng. 2004. Community-acquired methicillin-resistant Staphylococcus aureus arrives in Hong Kong. J. Antimicrob. Chemother. 54:845-846. [DOI] [PubMed] [Google Scholar]
- 3.Holmes, A., M. Ganner, S. McGuane, T. L. Pitt, B. D. Cookson, and A. M. Kearnes. 2005. Staphylococcus aureus isolates carrying Panton-Valentine leucocidin genes in England and Wales: frequency, characterization, and association with clinical disease. J. Clin. Microbiol. 43:2384-2390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hsu, L. Y., A. Tristan, T. H. Koh, M. Bes, J. Etienne, A. Kurup, T. T. Tan, and B. H. Tan. 2005. Community-associated methicillin-resistant Staphylococcus aureus, Singapore. Emerg. Infect. Dis. 11:341-342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hsu, L. Y., T. H. Koh, K. Singh, M. L. Kang, A. Kurup, and B. H. Tan. 2005. Dissemination of multisusceptible methicillin-resistant Staphylococcus aureus in Singapore. J. Clin. Microbiol. 43:2923-2925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hsu L. Y., T. H. Koh, T. Y. Tan, T. Ito, X. X. Ma, R. T. Lin, and B. H. Tan. 2006. Emergence of community-associated methicillin-resistant Staphylococcus aureus in Singapore: a further six cases. Singapore Med. J. 47:20-26. [PubMed] [Google Scholar]
- 7.Ito, T., X. X. Ma, F. Takeuchi, K. Okuma, H. Yuzawa, and K. Hiramatsu. 2004. Novel type V staphylococcal cassette chromosome mec driven by a novel cassette chromosome recombinase, ccrC. Antimicrob. Agents Chemother. 48:2637-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarraud, S., C. Mougel, J. Thioulouse, G. Lina, H. Meugnier, F. Forey, X. Nesme, J. Etienne, and F. Vandenesch. 2002. Relationships between Staphy-lococcus aureus genetic background, virulence factors, agr groups (alleles), and human disease. Infect. Immun. 70:631-641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lina, G., Y. Piemont, F. Godail-Gamot, M. Bes, M. O. Peter, V. Gauduchon, F. Vandenesch, and J. Etienne. 1999. Involvement of Panton-Valentine leukocidin-producing Staphylococcus aureus in primary skin infections and pneumonia. Clin. Infect. Dis. 29:1128-1132. [DOI] [PubMed] [Google Scholar]
- 10.Ma, X. X., A. Galiana, W. Pedreira, M. Mowszowicz, I. Christophersen, S. Machiavello, L. Lope, S. Benaderet, F. Buela, W. Vincentino, M. Albini, O. Bertaux, I. Constenla, H. Bagnulo, L. Llosa, T. Ito, and K. Hiramatsu. 2005. Community-acquired methicillin-resistant Staphylococcus aureus, Uruguay. Emerg. Infect. Dis. 11:973-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malachowa, N., A. Sabat, M. Gniadkowski, J. Krzyszton-Russjan, J. Empel, J. Miedzobrodzki, K. Kosowska-Shick, P. C. Appelbaum, and W. Hryniewicz. 2005. Comparison of multilocus variable-number tandem-repeat analysis with pulsed-field gel electrophoresis, spa typing, and multilocus sequence typing for clonal characterization of Staphylococcus aureus isolates. J. Clin. Microbiol. 43:3095-3100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maslow, J., A. Slutsky, and R. Arbeit. 1993. The application of pulsed-field gel electrophoresis to molecular epidemiology, p. 563-572. In H. Persing, T. Smith, F. Tenover, and T. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 13.NCCLS. 2004. Performance standards for antimicrobial susceptibility testing: fourteenth informational supplement. Document M100-S14, vol. 24. NCCLS, Wayne, Pa.
- 14.Okuma, K., K. Iwakawa, J. D. Turnidge, W. B. Grubb, J. M. Bell, F. G. O'Brien, G. W. Coombs, J. W. Pearman, F. C. Tenover, M. Kapi, C. Tiensasitorn, T. Ito, and K. Hiramatsu. 2002. Dissemination of new methicillin-resistant Staphylococcus aureus clones in the community. J. Clin. Microbiol. 40:4289-4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sabat, A., J. Krzyszton-Russjan, W. Strzalka, R. Filipek, K. Kosowska, W. Hryniewicz, J. Travis, and J. Potempa. 2003. New method for typing Staphylococcus aureus strains: multiple-locus variable-number tandem repeat analysis of polymorphism and genetic relationships of clinical isolates. J. Clin. Microbiol. 41:1801-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Taiwo, S. S., M. Bamidele, E. A. Omonigbehin, K. A. Akinside, S. I. Smith, B. A. Onile, and A. O. Olowe. 2005. Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Ilorin, Nigeria. West Afr. J. Med. 24:100-106. [PubMed] [Google Scholar]
- 17.Takizawa, Y., I. Taneike, S. Nakagawa, T. Oishi, Y. Nitahara, N. Iwakura, K. Ozaki, M. Takano, T. Nakayama, and T. Yamamoto. 2005. A Panton-Valentine leucocidin (PVL)-positive community-acquired methicillin-resistant Staphylococcus aureus (MRSA) strain, another such strain carrying a multiple-drug resistance plasmid, and other more-typical PVL-negative MRSA strains found in Japan. J. Clin. Microbiol. 43:3356-3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vandenesch, F., T. Naimi, M. C. Enright, G. Lina, G. R. Nimmo, H. Heffernan, N. Liassine, M. Bes, T. Greenland, M. E. Reverdy, and J. Etienne. 2003. Community-acquired methicillin-resistant Staphylococcus aureus carrying Panton-Valentine leukocidin genes: worldwide emergence. Emerg. Infect. Dis. 9:978-984. [DOI] [PMC free article] [PubMed] [Google Scholar]