Abstract
The envelope glycoprotein of small ruminant lentiviruses (SRLV) is a major target of the humoral immune response and contains several linear B-cell epitopes. We amplified and sequenced the genomic segment encoding the SU5 antigenic site of the envelope glycoprotein of several SRLV field isolates. With synthetic peptides based on the deduced amino acid sequences of SU5 in an enzyme-linked immunosorbent assay (ELISA), we have (i) proved the immunodominance of this region regardless of its high variability, (ii) defined the epitopes encompassed by SU5, (iii) illustrated the rapid and peculiar kinetics of seroconversion to this antigenic site, and (iv) shown the rapid and strong maturation of the avidity of the anti-SU5 antibody. Finally, we demonstrated the modular diagnostic potential of SU5 peptides. Under Swiss field conditions, the SU5 ELISA was shown to detect the majority of infected animals and, when applied in a molecular epidemiological context, to permit rapid phylogenetic classification of the infecting virus.
Caprine arthritis-encephalitis virus (CAEV) of goats and maedi-visna virus (MVV) of sheep belong to the genus Lentivirus of the family Retroviridae (18, 22, 39). Strong evidence indicates that cross-species transmission from sheep to goats and vice versa occurs under field conditions (16, 24, 33). Therefore, these viruses are no longer considered to be species specific and are referred to as small-ruminant lentiviruses (SRLV).
The majority of infected animals mount a strong immune response to these viruses but remain persistently infected. Only one-third of infected goats develop overt clinical disease, and in sheep the percentage of animals with clinical symptoms differs greatly between breeds, strongly suggesting that, in both species, genetic factors play a key role in determining the clinical outcome of infection (11, 28, 29).
In several countries, eradication programs have been initiated to control SRLV-induced diseases with the aim of eliminating these viruses (23). The Swiss CAEV eradication program, initiated in 1984, has reduced the seroprevalence from 60 to 80% to less than 1% and eliminated clinical cases in the goat population (14, 15). However, in the last phase of the eradication program, detection and elimination of the remaining virus carriers appear to be very difficult. The serological tools currently used (37, 38) are of limited use when applied to screen a population with a low seroprevalence. In the absence of reliable tools to directly detect SRLV, consistent serological diagnoses depend on the costly use of a combination of tests run in parallel (4). Major problems in SRLV serology are slow seroconversions and low titers of antibody to Gag in some animals. In this respect, the strong immunogenicity of the envelope glycoprotein (Env) and especially the rapid seroconversion induced by this antigen in infected animals make it an ideal candidate for diagnostic applications (2, 19). Goats and sheep infected with SRLV develop high titers of antibodies to several conformational and linear epitopes of Env (10, 12). However, particularly in goats, these antibodies do not show consistent neutralizing activity (19). The humoral immune response is directed against the surface (SU) and transmembrane (TM) subunits of Env. Antibodies reacting with linear immunodominant epitopes of TM have been associated with disease in infected goats (3, 13, 17). The linear B-cell epitopes of Env were mapped for the CAEV-CO strain (2, 3, 35) and consist of six epitopes in the TM region and at least five in the SU region.
One of the SU epitopes mapped previously (SU5) appeared to be particularly promising regarding its use as a diagnostic tool. When a limited panel of sera was tested for reactivity to SU5, this epitope appeared to be immunodominant and an early target of the antibody response in infected animals. Additionally, the sera tested showed a partly type-specific reaction, suggesting a possible application of these SU5 peptides in SRLV serotyping, as described for human immunodeficiency virus (HIV) with peptides corresponding to the highly variable V3 loop region of Env (1, 25).
The principal objectives of the present study were to amplify by PCR and sequence the genomic region encoding the SU5 antigenic site of several Swiss field isolates, to synthesize peptides corresponding to their deduced amino acid sequences, and to test the hypothesis that these peptides are immunodominant and can be used to develop a novel SRLV serological test.
MATERIALS AND METHODS
Cell isolation. (i) PBMC.
Peripheral blood mononuclear cells (PBMC) were isolated from EDTA-anticoagulated blood (Vacuette K3E EDTA K3; Greiner Labortechnik, Kremsmünster, Austria) by centrifugation (30 min at 700 × g) in Ficoll separation solution (Biochrom AG, Berlin, Germany). The buffy coat was collected in Ca2+- and Mg2+-free phosphate-buffered saline (PBS), pH 7.3, supplemented with 0.3 mM EDTA. The cells were centrifuged at 350 × g for 10 min, resuspended in PBS-EDTA, and pelleted at 350 × g for 10 min (all steps at room temperature). Aliquots of 107 cells per tube were used immediately for DNA isolation and coculture or stored frozen at −80°C.
(ii) Milk cells.
Milk was diluted 1:3 with PBS supplemented with 10% penicillin-streptomycin (Biochrom AG, Berlin, Germany) and centrifuged at 900 × g for 15 min at 4°C. The cells were resuspended in PBS-10% penicillin-streptomycin and centrifuged at 900 × g for 15 min at 4°C. This procedure was repeated once with PBS. Aliquots of 107 cells were prepared for immediate DNA isolation and coculture or stored frozen at −80°C.
Bronchoalveolar lavage.
To harvest alveolar macrophages, 500 ml of 0.9% sodium chloride (NaCl) solution was instilled in the lung upon necropsy and the lavage fluid was aspired and collected in 50-ml conical tubes (Greiner Labortechnik, Kremsmünster, Austria). Fifty to 60% of the applied volume was recovered. The recovered fluid was centrifuged at 300 × g for 10 min, and the pellet was washed once with a 0.9% NaCl solution. Pellets of 107 cells were prepared for DNA isolation and coculture or stored frozen at −80°C.
(iii) Lymph node cells.
The left subscapular lymph node (Ln. subscapularis) of one goat was explanted and mechanically minced, and lymph node cells were resuspended in PBS. Following a washing step, the cells were resuspended in tissue culture medium (modified Eagle's medium-Earle [MEM-Earle]; Biochrom AG, Berlin, Germany) containing 10% fetal bovine serum (Sigma, St. Louis, Mo.).
Cocultures with GSM indicator cells.
Goat synovial membrane (GSM) cells (1.5 × 106) were seeded in a 75-cm2 tissue culture flask (TPP Techno Plastic Products, Trasadingen, Switzerland) and incubated for 4 to 6 h at 37°C in a CO2 incubator. Cells (107 PBMC, milk cells, or lymph node-derived cells) were resuspended in 15 ml of tissue culture medium (MEM-Earle; Biochrom AG, Berlin, Germany) containing 10% fetal bovine serum (Sigma, St. Louis, Mo.), seeded on adhered GSM, and incubated overnight in a CO2 incubator. Nonadherent cells were removed by washing twice with MEM-Earle without additives, and the cultures were further incubated for 10 days. The medium was changed at intervals of 3 to 4 days. Cells were passaged at weekly intervals for a maximum of six passages, and pellets of 107 cells were prepared for DNA or RNA isolation or stored at −80°C.
DNA and/or RNA isolation.
DNA and/or RNA were extracted from different tissues such as PBMC, alveolar macrophages, milk cells, tissue cells, GSM cells, or coculture supernatants by different methods. Total cellular DNA was extracted from 107 cells (milk cells, PBMC, alveolar macrophages, or GSM cells) with DNAzol (Invitrogen, Carlsbad, Calif.) according to the manufacturer's instructions.
The SV total RNA isolation kit (Promega, Madison, Wis.) was used to simultaneously isolate DNA and RNA from 107 PBMC according to the manufacturer's protocol. Total cellular RNA was isolated from the brain tissue of one sheep with the TRIzol reagent (Invitrogen, Carlsbad, Calif.) as recommended by the manufacturer. The same protocol was used to extract RNA from virus pellets obtained by ultracentrifugation of PBMC-GSM cell coculture supernatants of three goats (287,660 × g at 15°C for 30 min, Centrikon 2070; Kontron Medical AG, Basel, Switzerland).
Reverse transcriptase (RT) reaction.
cDNA synthesis was performed with SuperScript II RNase H-free RT (Invitrogen, Carlsbad, Calif.) according to the manufacturer's protocol.
PCR. (i) Primers.
Primers used were designed by aligning several env sequences obtained from GenBank and were synthesized by Microsynth (Balgach, Switzerland). The following primers were used: #423 (sense, position 6885), GGRGCAGARATMATHCCWGAARVYHTGA; #563 (sense, position 7272), GAYATGRYRGARCAYATGAC; #566 (sense, position 7803), GTRAGAGCTTAYACWTATGG; #567 (sense, position 7482), GGIACIAAIACWAATTGGAC; #425 (antisense, position 7960), CCTGCRGCAGCYAYTATHGCCATGAT; #564 (antisense, position 8089), GCYAYATGCTGIACCATGGCATA. Base pair positions refer to the CAEV-CO molecular clone (31). Different primer pair combinations and annealing temperatures were used in direct and nested PCRs (Table 1).
TABLE 1.
Primer pairs used for PCRsa
| Pair | Primers | Position | Product length (bp) | Annealing temp (°C) |
|---|---|---|---|---|
| A | #563/#425 | 7272-7960 | 689 | 45 |
| B | #566/#425 | 7803-7960 | 158 | 54 |
| C | #563/#564 | 7272-8089 | 818 | 47 |
| D | #566/#564 | 7803-8089 | 287 | 47 |
| E | #567/#564 | 7482-8089 | 608 | 49 |
| F | #567/#425 | 7482-7960 | 479 | 47 |
| G | #423/#564 | 6885-8089 | 1,205 | 54 |
| H | #423/#425 | 6885-7960 | 1,076 | 49 |
Primer numbers are assigned according to an internal database, and base pair positions refer to the sequence published by Saltarelli et al. (31).
(ii) Conditions.
env fragments were amplified with different primer combinations (Table 1) and genomic DNA or reverse-transcribed cDNA as the template.
PCR was performed in a T3 Thermocycler (Biometra, Göttingen, Germany). Briefly, 200 ng of template DNA or 2 μl of cDNA was amplified in a total volume of 50 μl of PCR buffer (10 mM Tris-HCl [pH 9.0], 50 mM KCl, 1.5 mM MgCl2, 0.01% [wt/vol] stabilizer, 0.1% Triton X-100) containing 0.25 mM deoxynucleoside triphosphates (Invitrogen, Carlsbad, Calif.), each primer at 0.5 μM, and 1 U of DNA polymerase (Super Taq; HT Biotechnology Ltd., Cambridge, England). For nested or seminested PCR, 1 to 5 μl of the amplified PCR product was used as the template.
After an initial denaturation step of 95°C for 1 min, the following cycling conditions were applied for 39 cycles: 1 min at 95°C, 1 min at the annealing temperature listed in Table 1, followed by 1 min at 72°C. After a final extension step of 10 min at 72°C, the samples were chilled at 4°C. A negative control reaction without template DNA was included for each primer pair to control for potential contamination. In order to minimize the risk of contamination, mixing of reagents, sample preparation, and analysis of the amplified products were performed in three separate rooms. Additionally, all reagents and probes were manipulated in PCR hoods (Cleanspot stations; Coy Laboratory Products, Grass Lake, Mich.) with aerosol-proof pipette tips (Molecular Bio Products, San Diego, Calif.).
Cloning of PCR fragments.
PCR amplicons were separated on 0.8 to 1% low-melting agarose gels (NuSieve GTG agarose; BMA, Rockland, Maine), and bands of the expected sizes were excised. The excised fragments were subsequently cloned with the TOPO TA cloning kit (pCR 2.1-TOPO Vector; Invitrogen Carlsbad, Calif.) according to the manufacturer's protocol. Ten to 20 colonies were picked and grown overnight at 37°C in TB medium (12-g/liter Bacto Tryptone, 24-g/liter yeast extract, 4-ml/liter glycerol, 2.31-g/liter KH2PO4, 12.54-g/liter K2HPO4) with 0.1% ampicillin (Sigma, St. Louis, Mo.). Minipreps were prepared with the Wizard Plus Kit (Promega, Madison, Wis.) according to the manufacturer's instructions.
Sequencing.
Sequencing reactions were performed with commercially available pUC universal primers (Microsynth, Balgach, Switzerland) and the dRhodamine Terminator Cycle Sequencing Ready Reaction DNA-sequencing kit (Applied Biosystems, Foster City, Calif.) and analyzed with the ABIprism 310 (Applied Biosystems, Foster City, Calif.) automatic sequencer.
Phylogenetic analysis.
The sequences obtained were edited and analyzed with the following software: AutoAssembler (Applied Biosystems, Foster City, Calif.), DNASTAR (version 5; DNASTAR Inc., Madison, Wis.), Align Plus 4 (Scientific & Educational Software, Durham, N.C.), ClustalX (version 1.81), and Genedoc (version 2.5). Phylogenetic trees were generated with the MEGA software (Molecular Evolutionary Genetics Analysis, version 1.02) by neighbor joining (30) based on p distances and the TreeExplorer (version 2.01).
Synthetic peptides.
Several synthetic peptides were designed according to novel SU5 sequences or according to sequences obtained from GenBank. Three different types of peptides were synthesized covering the complete SU5 region (SU5-total), the most variable part of SU5 (SU5-variable), or exclusively the constant region (SU5-consensus), respectively. Terminal lysine (K) residues were inserted into peptides missing this amino acid to enhance binding to enzyme-linked immunosorbent assay (ELISA) plates (Table 1, boldface K).
The TM3 peptide (QELDCWHYHQYCITS; CAEV-CO molecular clone [31], env base pairs 8160 to 8204, aa 717 to 731) corresponds to the principal immunodominant determinant of the transmembrane portion of CAEV Env. This peptide was synthesized and chemically cyclized by a covalent bond between the two cysteines (TM3c) in order to mimic a loop structure (3).
Peptides were synthesized and purified by Primm, Milan, Italy, or by Neosystem, Strasbourg, France.
Affinity columns packed with the CAEV-CO- or 1163-SU5-total peptide coupled with cyanogen bromide-activated Sepharose (2 ml) were purchased from Primm, Milan, Italy.
Antibody affinity purification.
Antibody was purified according to the buffer manufacturer's recommendations. Briefly, 10 ml of serum obtained from a goat experimentally infected with the molecular clone CAEV-CO (31) was mixed with 10 ml of binding buffer (ImmunoPure Gentle Binding Buffer; Pierce, Rockford, Ill.), filtered trough a 0.45-μm-pore-size filter (Pierce, Rockford, Ill.), and loaded onto the affinity columns described in the previous paragraph. The flowthrough was collected, and the columns were washed with 30 ml of binding buffer previous to eluting the bound antibody with 10 ml of elution buffer (ImmunoPure Gentle Elution Buffer; Pierce, Rockford, Ill.), collected in 1-ml fractions. All experiments described were performed with a pool of fractions 2 to 4, which showed the highest protein content (absorption at 280 nm).
Chekit ELISA.
The commercially available assay Chekit CAEV/MVVELISA (Bommeli AG/Idexx Laboratories, Liebefeld, Switzerland) is based on a monophasic whole-virus ELISA (38) and was used according to the manufacturer's protocol.
Western blotting (WB).
WB was performed as described earlier (37). Briefly, purified whole-virus antigen was separated on 12% polyacrylamide gels and transferred to nitrocellulose. The membranes were blocked with fat-free milk (5%, wt/vol) in TBT buffer (0.01 M Tris-HCl, 0.15 M NaCl, 0.05% Tween 20 [pH 8.0]). Serum samples were diluted 1:100 in the same buffer and incubated at room temperature under constant shaking. Alkaline phosphatase-conjugated recombinant protein G (Zymed, San Francisco, Calif.), 1.0-mg/ml Fast-Red (Sigma, St. Louis, Mo.), and 0.5-mg/ml Naphthol AS-MX phosphate (Sigma, St. Louis, Mo.) in 0.1 M Tris-HCl, pH 8.0, were used as conjugate and chromogen solutions, respectively. Thorough washing followed each incubation step. A weakly positive serum and a negative control serum were applied to permit direct visual comparison of test sera and control sera.
Peptide ELISA.
Synthetic peptides were dissolved in carbonate-bicarbonate buffer (0.1 M sodium carbonate [pH 9.6]) at 5 μg/ml and allowed to adsorb to ELISA plates (Maxisorp Immunoplate; Nunc, Roskilde, Denmark) overnight at 4°C (100 μl per well). Plates were washed three times with PBS and 0.1% (vol/vol) Tween 20 (Merck-Schuchardt, Hohenbrunn, Germany) (PBS-Tween). Residual adsorption sites were saturated at room temperature for 1 h by incubation with 150 μl of PBS-Tween containing 5% fat-free milk powder (Nestle, Vevey, Switzerland) for 1 h. Plates were washed once with 150 μl of PBS-Tween before adding 100 μl of goat or sheep serum diluted in PBS-Tween supplemented with fat-free milk powder (5%). When not stated differently, sera were diluted 1:25. After 2 h of incubation at room temperature, the plates were washed three times with 150 μl of PBS-Tween. One hundred microliters per well of peroxidase-conjugated protein G diluted 1:5,000 in PBS-Tween was added, and the mixture was incubated for 2 h at room temperature. After two washing steps with PBS-Tween and two washing steps with PBS, bound conjugate was visualized with hydrogen peroxide-2,2′-azinobis(3-ethylbenzthiazolinesulfonic acid) (ABTS) solution. Optical density (OD) was measured at a 405-nm wavelength after 30 min.
Antibody avidity measurements.
The avidity index values of SU5-specific serum immunoglobulin G were measured by testing the stability of antigen-antibody complexes following a wash step with 8 M urea (6, 7). For this experiment, the SU5 ELISA was used in a slightly modified form. Sera diluted to obtain OD values between 1 and 2 were used for this experiment. These sera were incubated on SU5 peptide-coated ELISA plates as described above. Duplicate plates were washed three times for 5 min with either 150 μl of PBS-Tween or 150 μl of 8 M urea in PBS-Tween. All plates were washed an additional four times with PBS-Tween to remove the urea. The avidity index was calculated from the ratio ([A/B] × 100%) of the absorbance value obtained with the urea treatment (A) to that observed with the PBS-Tween 20 control treatment (B). Antibodies with avidity index values of <30% are considered to be of low avidity, those with values of 30 to 50% are considered to be of intermediate avidity, and those with values of >50% are considered to be of high avidity.
RESULTS
SU5 amino acid sequences of Swiss SRLV.
Nineteen SU5 amino acid sequences of SRLV circulating in Switzerland were obtained by PCR or RT-PCR amplification, cloning, and sequencing of the env gene region encoding this antigenic site. DNA and/or RNA was extracted from different cells and tissues such as PBMC, alveolar macrophages, milk cells, lymph nodes, brain or synovial membranes, and cell pellet or cell culture supernatant of GSM cells cocultivated with PBMC of infected animals.
Additionally, 14 SRLV nucleotide sequences, comprising those of three Swiss field isolates described previously (2), were obtained from GenBank and the deduced amino acid sequences of the corresponding SU5 regions were added to our panel. The SU5 region corresponds to base pairs 7720 to 7907 and amino acids 597 to 632 of the CAEV-CO molecular clone (accession number NC_001463) (31).
The novel SU5 sequences were aligned with sequences deposited in GenBank (Fig. 1). Based on this alignment, several synthetic peptides were designed and ordered. The complete panel of peptides used in our experiments is shown in Table 2.
FIG. 1.
Alignment of all of the SU5 sequences analyzed in this study. SU5 sequences deposited in GenBank (underlined) were aligned with newly obtained SU5 sequences (not underlined). Dashed underlines indicate sequences clustering in the MVV group, while continuous underlines mark sequences of the CAEV group. Virus sequences obtained from goats are marked with a bold G, while those obtained from sheep are marked with a bold S.
TABLE 2.
SU5 synthetic peptidesa
| Peptide type and designation | Sequence |
|---|---|
| SU5 variable | |
| NT8716-SU5-variable | ---------KDMPQEYKRRNRKK |
| 1355-SU5-variable | ---------KDLPQSYEKINLKRRK |
| 615-SU5-variable | ---------KEMPKNYEKVSLNRKK |
| 4663-SU5-variable | ---------KEMPKNYERITQNRKK |
| CAEV-CO-SU5-variable | ---------KEMPENYAKTRIINRKK |
| Zuesi-SU5-variable | ---------KEMPKNYNKLKLNRKK |
| SG6393-SU5-variable | ---------KDMPKSYVQNQEKDKR |
| 1163M-SU5-variable | ----------DMPQSYIQNQEKYKK |
| VLVGAGA-SU5-variable | ---------KEMPQSYMEAQGENKR |
| Zn643-SU5-variable | ---------KEMPTSYMESQKRKKK |
| 4745-SU5-variable | ---------KELPKNYMKTISLNRKK |
| SU5 total | |
| NT8716-SU5-total | RVRAYTYGVIDMPQEYKRRNRKK |
| 1355-SU5-total | RVRAYTYGVIDLPQSYEKINLKRRK |
| 615-SU5-total | RVRAYTYGVIEMPKNYEKVSLNRKK |
| 4663-SU5-total | RVRAYTYGVIEMPKNYERITQNRKK |
| CAEV-CO-SU5-total | KVRAYTYGVIEMPENYAKTRIINRKK |
| Zuesi-SU5-total | RVRAYTYGVIEMPKNYNKLKLNRKK |
| 3151-SU5-total | RVRAYTYGVIEMQKNYEQMPRNRRK |
| SG6393-SU5-total | KRVRAYTYGVVDMPKSYVQNQEKDKR |
| 1163M-SU5-total | KVRAYTYGVVDMPQSYIQNQEKYKK |
| VLVGAGA-SU5-total | KRVRAYTYGVVEMPQSYMEAQGENKR |
| Zn643-SU5-total | KVRAYTYGVVEMPTSYMESQKRKKK |
| OLVSAOMVCG-SU5-total | KVRAYTYGVVDMPKAYREKNMRNKR |
| SU5 consensus | |
| SG6393-SU5-consensus | RVRAYTYGVVDMPK |
Three types of synthetic peptides were synthesized: SU5 variable, encompassing the highly variable region of SU5; SU5 total, covering both constant and variable regions; and SU5 consensus, based on the conserved amino-terminal sequence of SU5. Lysine (K) residues (shown in boldface) were added to enhance binding to the ELISA plates.
Kinetics of seroconversion to SU5.
Radioimmunoprecipitation experiments have shown that goats experimentally infected with CAEV seroconvert rapidly to Env, which precedes or parallels seroconversion to Gag (3). The ability to rapidly detect seroconversions is indispensable for an efficient diagnostic test. With sera taken at regular intervals after experimental infection, we determined the kinetics of seroconversion to SU5 and compared it to that of routine diagnostic tests such as the Chekit ELISA and WB.
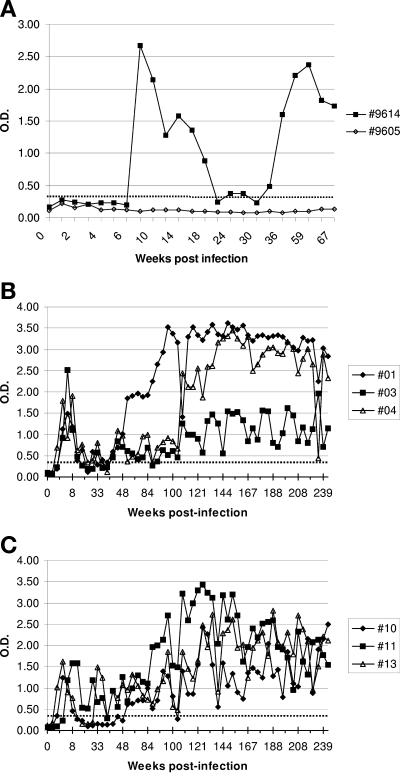
Two young goats, 9605 and 9614, were experimentally infected by intravenous transfer of 20 ml of blood from a naturally infected goat with severe clinical arthritis (goat 1355). Subsequently, serum samples were taken at regular intervals. Goat 9614 seroconverted at 7 weeks postinfection (p.i.) as monitored by WB and at 13 weeks p.i. as monitored by the Chekit ELISA. Goat 9605 remained seronegative during this period. These sera were analyzed in an SU5 ELISA with the peptide 1355-SU5-total. As shown in Fig. 2A, Goat 9614 seroconverted between weeks 6 and 8 p.i., confirming the result obtained by WB. After a first peak, the reactivity decreased and at around 20 weeks p.i. it reached a level close to the arbitrary cutoff set at an OD of 0.3. In the subsequent weeks, the reactivity to the SU5 peptide increased and remained well above the positive cutoff for more than 200 weeks. As shown in Fig. 2A, goat 9605 did not seroconvert in the SU5 ELISA, thus confirming the WB result.
FIG. 2.
Kinetics of seroconversion in an SU5-total ELISA of goats experimentally infected with SRLV. (A) Results of goat 9614 infected with strain 1355. Sera were tested at different time points (black squares) in a 1355-SU5-total ELISA. Goat 9605 (open diamonds) did not seroconvert and served as a negative control. (B and C) Results of goats 01, 03, and 04 (B) and 10, 11, and 13 (C), respectively. Sera were tested at different time points in a CO-SU5-total ELISA. OD at 405 nm was measured at 30 min after addition of the chromophore. The cutoff (dotted line) was set at an OD of 0.3.
Additionally, three female (01, 03, and 04) and three castrated male Saanen goats (10, 11, and 13) were experimentally infected in the left carpal joints and intravenously with 0.5 ml each of cell culture supernatant containing 107 50% tissue culture infective doses of CAEV-CO. The animals seroconverted in the Chekit ELISA between 3 (03, 04, and 13) and 5 (01, 10, and 11) weeks p.i. and were monitored on a regular basis over the next 5 years. All sera were analyzed in the SU5 ELISA with the CO-SU5-total peptide, and the results are shown in Fig. 2B and C. Animals 04, 10, and 13 seroconverted already at week 2, 01 and 03 at week 3, and 11 at week 5 p.i. During the first 5 weeks p.i., the animals showed a first peak of reactivity that decreased in the subsequent weeks, reaching the weak positive range or, in some instances, even falling into the negative range. Subsequently, all sera showed increased reactivity levels that stably settled into the positive range, thus confirming the results obtained with goat 9614 (Fig. 2A). The sera of goats 01 and 04 showed a steady reactivity, with ODs above 2.5 and only small fluctuations, whereas the remaining animals showed more-fluctuating kinetics of reactivity at lower ODs (Fig. 2B and C).
Kinetics of antibody avidity maturation in SU5 and TM3 ELISAs.
As described above and shown in Fig. 2A, goat 9614 seroconverted after experimental infection. Like all of the animals tested so far, goat 9614 showed a first antibody peak at 7 weeks p.i., followed by a decrease in ELISA reactivity and a subsequent increase in OD values that leveled off after week 35 p.i. (Fig. 2A). The avidity of serum samples taken at 7, 36, and 163 weeks p.i., respectively, was analyzed as described in Materials and Methods. Distinct avidity maturation could be observed, with the appearance of low-avidity antibody (24%) at 7 weeks p.i., antibody of intermediate avidity (50%) at 36 weeks p.i., and antibody of high avidity (64%) at 163 weeks p.i.
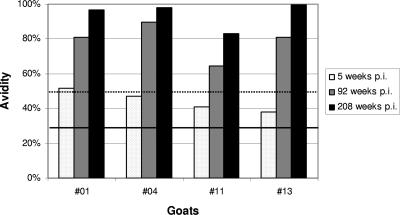
To confirm these results, the avidity maturation of sera from goats experimentally infected with CAEV-CO was analyzed. Based on the antibody kinetics described in the previous section (Fig. 2B and C), sera from goats 01, 04, 11, and 13 obtained during the first antibody peak (5 weeks p.i.), the ensuing phase of rising antibody titers (week 92), and the final phase of the experiment (208 weeks p.i.) were selected and tested. As shown in Fig. 3, all of the goats showed marked maturation of anti-SU5 antibody avidity. At week 5 p.i., three sera contained antibody of intermediate avidity and goat 01 already had high-avidity antibody. All sera taken at 92 or 208 weeks p.i. contained high-avidity antibody (Fig. 3).
FIG. 3.
Kinetics of antibody avidity maturation. Maturation of the avidity of anti-SU5 antibody from goats 01, 04, 11, and 13 taken at 5, 92, and 208 weeks post experimental infection is shown. The avidity index was calculated as described in Materials and Methods. Serum antibodies with avidity index values of <30% are designated low-avidity antibodies (below the solid line), those with values between 30 and 50% are intermediate-avidity antibodies (between the solid and dotted lines), and those with values of >50% are considered high-avidity antibodies (above the dotted line).
The same sera were used to monitor maturation of the avidity of the antibody to the TM3 peptide that encompasses the principal immunodominant epitope of the transmembrane portion of Env (3, 20). Due to the slow seroconversion to this peptide, only sera taken at 92 and 208 weeks p.i. were analyzed. All sera showed avidity indexes of >60%, confirming the presence of high-avidity antibodies to this immunodominant TM epitope (data not shown).
Defining the antibody-binding region of SU5.
Depending on the SRLV strain considered, the SU5-total peptides consist of 25 or 26 amino acids. With peptides derived from different field isolates and goat sera from animals infected with defined SRLV strains, we observed various patterns of reactivity going from strictly type specific to broadly reacting, suggesting that the antigenic site covered by the SU5-total peptide may contain different epitopes. To clarify this point, we tested the abilities of various peptides spanning the highly variable or the constant region of SU5 to compete in an ELISA for antibody binding to SU5-total peptides. Additionally, we tested several sera for reactivity to a peptide spanning the constant region of SU5 (SG6393-SU5-consensus). Only very few sera (<5%) reacted to this peptide, indicating that the antibody binding sequence is located in the variable region or encompasses both variable and constant regions.
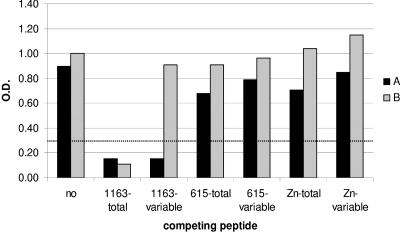
These alternative explanations were tested by using two sera (A and B) showing strictly type-specific reactivity to the 1163 peptide or broad reactivity to several peptides, respectively. The reactivities of these two sera to 1163-SU5-total in the absence (control) or presence of competing peptides encompassing the complete SU5 region of various strains or only the highly variable SU5 region of strain 1163 were compared. The addition of soluble 1163-SU5-total peptide as a competitor to sera A and B very efficiently quenched the reactivity of both sera, confirming the specificity of the reaction (Fig. 4). In contrast, the addition of the short 1163-SU5-variable peptide, which spans the variable portion of SU5, suppressed the reactivity of type-specific serum A without affecting antibody binding to the 1163-SU5-total peptide in serum B.
FIG. 4.
Competition for 1163-SU5 peptide binding. Serum A (black column) and serum B (shaded column) were allowed to react with 1163-SU5-total peptides in an ELISA without (no) or with the following competing peptides: 1163-SU5-total, 1163-SU5-variable, 615-SU5-total, 615-SU5-variable, Zn-SU5-total, and Zn-SU5-variable (Table 1). OD at 405 nm was measured at 30 min after addition of the chromophore. The cutoff (dotted line) was set at an OD of 0.3.
The addition of four peptides derived from different SRLV strains (615-SU5-total, Zn-SU5-total, 615-SU5-variable, and Zn-SU5-variable) or a short peptide spanning the constant region of SU5 did not significantly inhibit the antibody binding of either serum. This experiment demonstrates that serum A contains type-specific antibodies which bind exclusively to the variable part of the 1163-SU5-total peptide, whereas the antibodies contained in serum B bind not to the constant region of SU5 but to a region spanning the constant and variable regions. In contrast to the ELISA results, where serum B showed broad reactivity to several SU5-total peptides, the competition experiments described above suggest that the reactivity of serum B was type specific as well.
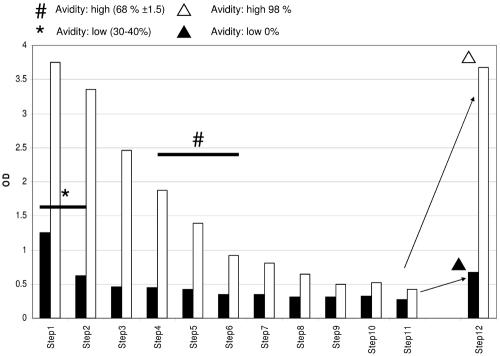
To resolve this apparent contradiction, we purified group-specific anti-SU5 antibodies by affinity chromatography. The serum of goat 13, an animal experimentally infected with the CAEV-CO strain, was applied to an affinity column prepared with the 1163-SU5-total peptide. This peptide is based on the SU5 sequence of an SRLV field isolate phylogenetically associated with the MVV group, and its SU5 variable region is highly different from that of its CAEV-CO counterpart (Fig. 1). As shown in Fig. 5, the eluate of the 1163-SU5-total column contains antibodies reacting with both the 1163- and CO-SU5-total peptides. The reactivity to the 1163-SU5-total peptide was of intermediate avidity (30 to 40% reactivity upon 8 M urea washing), while the reactivity to the CAEV-CO-SU5-total peptide was of high avidity 68% reactivity upon 8 M urea washing). These antibodies can be depleted by sequential adsorption to ELISA plate wells coated with the 1163- or CAEV-CO-SU5-total peptide, respectively. Antibody preparations depleted of 1163-SU5-specific antibodies still reacted very strongly with the CAEV-CO-SU5 peptide in an ELISA. This binding was extremely resistant to washing with a solution containing 8 M urea (98% binding compared to the control plate), demonstrating the high avidity of these antibodies. In contrast, the antibodies remaining after sequential depletion of CAEV-CO-SU5 specificity showed weak binding to the 1163-SU5 peptide in an ELISA, which was completely abolished by the 8 M urea washing (0% reactivity compared to the control plate), indicating that these antibodies were of very low avidity (Fig. 5).
FIG. 5.
Analysis of SU5 cross-reacting antibody. Antibodies cross-reacting between phylogenetically distinct SU5 peptides were purified by affinity chromatography from the serum of goat 13, which was experimentally infected with the molecular clone CAEV-CO, on a column prepared with an SU5-total peptide derived from the sequence of distantly related strain 1163. The histogram shows the absorption (OD) measured in an ELISA with the 1163-SU5-total peptide (black columns) or the CO-SU5-total peptide (open columns). All sera were incubated for 30 min in duplicate and subsequently transferred to the next wells. This operation was repeated 11 times (steps) on the matching peptide (1163- or CO-SU5-total, respectively) and resulted in the depletion of peptide-specific reactivity. In the last step (step 12), the sera depleted of CO-SU5-total specific antibody were tested in an 1163-SU5-total ELISA (black column). Conversely, sera depleted of 1163-SU5-total specificity were tested in a CO-SU5-total ELISA (white column). The avidities of the sera marked with thick black lines above the histogram columns were determined. The symbols *, #, ▴, and ▵ indicate the measured avidities to specific samples or sample groups.
These results resolve the contradiction described above and confirm that broadly reacting sera do not bind exclusively to the constant region of SU5 but make contact with amino acids comprised in both the constant and variable regions. The amino acid composition of the variable region determines the avidity of the reaction.
Additionally, the specificity of affinity-purified antibody was tested in competition experiments analogous to those described above. The binding of antibody purified on a CAEV-CO-SU5-total affinity column was tested in the homologous ELISA (OD, 2.61; 100%). This reaction was efficiently quenched by addition of the CAEV-CO-SU5-total peptide (OD, 0.26; 10.2%) and reduced by addition of the CAEV-CO-SU5-variable (OD, 1.27; 49%) and 1163-SU5-total (OD, 2.0; 77%) peptides, while addition of the 1163-SU5-variable peptide (OD, 2.52; 96.6%) did not affect binding.
Taken together, these results reinforce the concept presented above that, in addition to antibody specific for the variable portion of the SU5 peptides, an important fraction of antibody binds to both the constant and variable regions of the SU5 peptides.
Furthermore, the binding of antibody purified on the 1163-SU5-total affinity column was tested in an 1163-SU5-total ELISA (OD, 0.7; 100%). The binding was completely abrogated by the addition of an excess of free 1163-SU5-total (OD, 0.2; 32%) or -variable (OD, 0.2; 33.6%) peptide but was unaffected by the addition of the CAEV-CO-SU5-total peptide (OD, 0.69; 99.%). This demonstrates the peptide specificity of antibody binding to the variable portion of the 1163-SU5 peptide, whose amino acid sequence is unrelated to the SU5 sequence of the CAEV-CO strain used to infect the serum donor. This also proves the unspecific nature, relating to the infecting virus, of this low-avidity antibody.
Analysis of a panel of sera obtained from animals with defined SRLV PCR status.
To explore the suitability of the SU5 peptides for diagnostic purposes, we tested a well-defined panel of goat and sheep sera comprising 72 goat and 9 sheep sera. For all animals, we had access to PCR results and purified genomic DNA extracted from PBMCs, generously provided by the Swiss National Center of Retroviruses. A phylogenetic analysis of the SRLV sequences obtained from 62 of these animals has been published elsewhere (32).
All sera were positive by WB, while the commercially available Chekit ELISA detected only 85% of the sera.
For all of the peptides, the ELISA cutoff was set at an OD of 0.3. This cutoff value was determined by testing a panel of 208 WB-negative goat sera with the 615-SU5-total ELISA, setting a target specificity of ≥99%.
Initially, the entire panel was tested on 11 SU5-total peptides. The serum of an offspring of a goat experimentally infected with SRLV strain 4663 did not react with the corresponding SU5-total peptides but gave a positive ELISA result with the 4663-SU5-variable peptide.
The serum of goat 3151 was negative with all of the SU5-total and -variable peptides tested. A PCR fragment encompassing the SU5 region of the virus infecting this animal was amplified, cloned, and sequenced starting from DNA isolated from its PBMC. The deduced amino acid sequence of the highly variable region of SU5 clustered in the CAEV-like phylogenetic group on a distinct branch (data not shown), possibly due to a mutation of a highly conserved proline (P) into a glutamine (Q) at position 13. Based on this sequence, a corresponding peptide (3151-SU5-total) was synthesized (Table 2). The serum of goat 3151 reacted to this specific peptide, which was therefore added to the panel of SU5-total peptides. Finally, the 81 sera were tested in an ELISA with the 12 SU5-total, 11 SU5-variable, SG6393-SU5-consensus, and TM3 peptides.
As shown in Table 3, 77 sera (95%) showed a positive reaction with at least one SU5-total peptide. Fifty-one (63%) reacted to at least one of the shorter SU5-variable peptides encompassing the highly variable region of SU5. In contrast, only four sera (5%) reacted to the SG6393-SU5-consensus peptide encompassing the constant region of SU5. Two of these sera were from animals infected with an MVV-like virus, and two were from animals infected with a CAEV-like virus. These results provide additional evidence that the constant region of SU5 does not contain an immunodominant epitope, as indicated by the competition experiments described in the previous paragraph. Finally, the TM3 peptide detected 51 sera (63%), confirming that this highly conserved peptide, in spite of its immunodominance and conservation, has only limited sensitivity.
TABLE 3.
Serological results of 81 Gag PCR-positive animalsa
| Test | No. (%) positive | No. negative | No. indeterminate |
|---|---|---|---|
| IB | 81 (100) | 0 | 0 |
| Che | 69 (85) | 7 | 5 |
| SU5-total | 77 (95) | 4 | NAb |
| SU5-2PK | 51 (63) | 30 | NA |
| SU5-cons | 4 (5) | 77 | NA |
| TM3 | 51 (63) | 30 | NA |
This group comprises nine sheep and 72 goats. Tests: IB, immunoblot assay; Che, Chekit ELISA. The cutoff for all of the ELISAs was set at an OD of 0.3.
NA, not applicable.
Seroepidemiology.
Potentially, the serological reactivity to the SU5 peptides, and in particular to the short SU5-variable peptides encompassing the most variable portion of the SU5 region, may provide information on the phylogenetic classification of the infecting virus.
To test this hypothesis, the panel of 62 sera from animals for which we had access to sequence information on the infecting virus was tested on all of the SU5-total and SU5-variable peptides (Table 4). With one exception, all sera showed a positive reaction to more than one SU5-total peptide (96%).
TABLE 4.
Serological results of 62 Gag PCR-positive animalsa for which gag sequence information is available
| Test | No. (%) positive | No. negative | No. indeterminate |
|---|---|---|---|
| IB | 62 (100) | 0 | 0 |
| Che | 56 (90) | 3 | 3 |
| SU5-total | 61 (98) | 1 | NAb |
| SU5-2PK | 43 (69) | 19 | NA |
| SU5-cons | 4 (6.5) | 58 | NA |
| TM3 | 41 (66) | 21 | NA |
This group includes eight sheep and 54 goats. Tests: IB, immunoblot assay; Che, Chekit ELISA. The cutoff for all of the ELISAs was set at an OD of 0.3.
NA, not applicable.
Thirty-three (77%) of 43 sera obtained from animals infected with a virus clustering in the CAEV-like group showed the strongest reactivity with the 615-SU5-total peptide. Of the 19 sera obtained from animals infected with a virus clustering in the MVV-like group, 8 (42%) showed the strongest binding to the 1163-SU5-total peptide and 7 (37%) showed the strongest binding to the SG6393-SU5-total peptide. These peptides were derived from the SU5 sequences of two highly related Swiss field strains detected in both goat and sheep populations (reference 33 and next paragraph).
In summary, the 615-, 1163-, and SG6393-SU5-total peptides represent the dominating virus types circulating in the animals that provided our serum panel. With the SU5-variable peptides, spanning the most variable part of SU5, 43 sera were positive (69%). All but one serum showed the strongest reactivity with peptides belonging to the same phylogenetic group as the infecting virus, that is, the MVV-like or the CAEV-like group.
The SU5-variable peptides were less sensitive when applied to sera of animals infected with an MVV-like virus (8 negative results of 19, 42.1%) than by testing sera from animals infected with a CAEV-like virus strain (11 negative results of 43, 25.6%). Nine of the 11 positive sera from animals infected with MVV-like viruses showed the highest reaction to the 1163-SU5-variable peptides, which confirms the dominance of this strain in this population. In contrast, the reaction pattern of the sera from animals infected with CAEV-like viruses was more diverse. As for the SU5-total peptides, the overall reaction was dominated by two peptides. Nine sera reacted to the 615-SU5-variable peptide, and 11 reacted to the 4663-SU5-variable peptide. Three sera in this panel were from animals experimentally infected with the CAEV-CO strain. All three sera showed the strongest reaction with the CAEV-CO-SU5-variable peptide, confirming that these peptides can provide information on the phylogenetic origin of the infecting virus.
Application to a field diagnostic case.
To test the suitability of this novel SU5-ELISA in a diagnostic setup, we applied it to a complex diagnostic case presented to our laboratory. In a flock of six goats that had been seronegative for years, all of a sudden five animals seroconverted. As these animals did not have any contact with other goats, a diagnostic mistake was suspected. The investigation of this case yielded the following results.
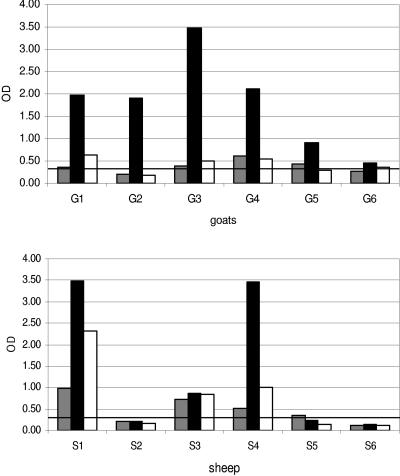
The WB performed in our laboratory confirmed the five positive results and one negative result obtained with the Chekit ELISA. All six goat sera were positive in an SU5-total ELISA. All six sera reacted strongly to the 1163-SU5-total peptide. Upon enquiry, the farmer disclosed that he was keeping six sheep on the same farm. Five sheep sera were negative in the Chekit ELISA, as well as by WB. The sixth serum was also negative by WB but gave an indeterminate result in the Chekit ELISA. In contrast, four sera were positive in the SU5-total ELISA and, analogously to the goat sera, showed the strongest reaction to the 1163-SU5-total peptide. The only exception was the serum of one sheep reacting only to the 1355-SU5-total peptide, with an OD close to the cutoff value (Fig. 6).
FIG. 6.
Serological results of the diagnostic-case sera tested in an SU5-total ELISA. The peptides used were 1355-SU5-total (dark gray), 1163-SU5-total (black), and 615-SU5-total (white). The OD at 405 nm was measured at 30 min after addition of the chromophore. The black line indicates the cutoff value, which was set at an OD of 0.3. G, goat; S, sheep.
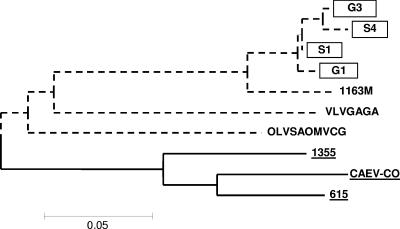
To obtain information on the viruses infecting these animals, we amplified by PCR and sequenced a fragment encompassing the SU5 region of these viruses. Positive amplifications were obtained starting from DNA isolated directly from alveolar macrophages of one goat, from milk cells of a second goat cocultivated with GSM cells for two passages, and finally, from the PBMC of two SU5-seropositive sheep. The sequences obtained from goat and sheep cells were phylogenetically closely related, clustering in a group close to the 1163-SU5 sequence derived from a Swiss isolate (Fig. 7). This confirmed the sensitivity of the SU5 ELISA and the applicability of this test for the serotyping of unknown field strains.
FIG. 7.
Phylogenetic analysis of the SU5 sequences obtained from the diagnostic-case isolates. The SU5 regions of the SRLV strains infecting goats 1 and 3 (G1 and G3) and sheep 1 and 4 (S1 and S4) were amplified by PCR and sequenced. The MVV-like group is shown by dash lines, and the CAEV-like group is shown by solid lines. Novel sequences are boxed.
DISCUSSION
SRLV infections in goats and sheep are usually diagnosed by serological analysis (4). Slow seroconversions and low antibody titers in particular goats and sheep complicate the detection of infected animals (4). In this respect, the Env glycoprotein may be a promising antigen to improve the sensitivity of these assays. Goats and sheep seroconvert rapidly upon experimental infection with SRLV, and antibody directed to Env dominates the humoral immune response in both naturally and experimentally infected animals (3, 10, 13, 17). Serological tests with Env proteins, and based on the competitive inhibition of monoclonal antibody binding to a single epitope, have been developed (5, 8, 9). Tests based on purified Env proteins could be developed as well, but the high variability of this antigen, inducing a type-specific antibody response, may require the use of different antigens covering the broad spectrum of circulating SRLV strains. Such antigen preparations may be prohibitively expensive if based on recombinant antigens but may become affordable if based on short synthetic peptides. Previously, we showed that SU5, a B-cell epitope located in the carboxy-terminal region of Env, is a prominent target of the antibody response and a potential candidate for developing novel serological tools for SRLV diagnostics (2). To expand the panel of SU5 peptides available, which comprised only three sequences derived from field isolates, we amplified by PCR and sequenced the genomic region encoding the SU5 region of 18 SRLV isolates. To avoid the sampling of minor variants, for every amplified sequence several clones were sequenced and the corresponding SU5 peptides were synthesized according to the consensus sequences.
In order to use these peptides in an ELISA, we had to define a cutoff. In the absence of a “gold standard” for SRLV diagnostics, we analyzed a panel of 208 goat sera that did not show any of the bands that characterize a positive WB reaction (37). We aimed at developing a test with a specificity of ≥99%, and the 615-SU5-total ELISA reached this specificity at an OD of 0.3. The cutoff was set at an OD of 0.3 for all peptides on the assumption that, due to the similar lengths and amino acid patterns, the background should be similar for all peptides. Indeed, the overall background was between an OD of 0.1 and an OD of 0.2 for all of the peptides.
With sera of animals experimentally infected with a field isolate or with the CAEV-CO molecular clone, we monitored the kinetics of the antibody response to SU5 and the maturation of its avidity. The goat infected by blood transfusion, most likely due to the low-dose infection, showed relatively slow seroconversion, analogous to that observed in naturally infected animals. This goat seroconverted with similar kinetics by WB and in an SU5 ELISA and only 6 weeks later in a Chekit ELISA. All animals infected with a high dose of CAEV-CO seroconverted much more rapidly, and their kinetics of seroconversion in the SU5 assay perfectly paralleled the results obtained with the sensitive WB. All seven of the goats analyzed showed the typical two-peak kinetics of seroconversion. A lower antibody peak was detected shortly after infection, followed by decreasing reactivity and a strong rise to a second peak, leading to a stable antibody response, which constantly remained well above the positive cutoff of our test. In some animals, the antibody levels between the two peaks crossed the negative threshold of the assay. This implies that false-negative results may occur during the interval between the two peaks and that only negative results based on at least two samples taken at 6- to 12-month intervals may be considered to be true negatives. The two-peak kinetics of seroconversion observed in all seven of the animals analyzed may be due to depletion of the anti-SU5 antibody by an excess of antigen. This, however, is unlikely, because the viral load of the six experimentally infected animals, as monitored by virus isolation from PBMC, was particularly low during this phase of the experiment (data not shown). Therefore, the decrease in antibody titers following the first peak most likely results from the low antigenic burden as a consequence of the efficient control of the virus by the immune system. This immune response, however, cannot eliminate the virus, which induces efficient maturation of the avidity of anti-SU5 antibodies. This was demonstrated by analyzing several sequential sera, all showing a rise in avidity over time, to values close to 100% late in infection. The avidity scores are generally higher than those observed in horses infected with equine infectious anemia virus (6), possibly as a consequence of the striking immunodominance of the SU5 Env fragment.
Antibody avidity maturation has been used to distinguish primary from established infections or to detect passively acquired high-avidity maternal antibodies in several infections such as those with HIV, Epstein-Barr virus, rubella virus, or hepatitis C virus and may be applied as well in the context of SRLV diagnostics (7, 21, 26, 36).
The different patterns of reaction to the SU5-total peptides, varying from strictly type specific to broadly reacting, suggested that several epitopes may be present in this short stretch of amino acids. In order to map these antibody-binding sites, we performed competition experiments with several peptides. As shown in Fig. 4, the reaction of sera A and B to the 1163-SU5-total peptide can be blocked by the addition of soluble 1163-SU5-total peptide, demonstrating its specificity. The type-specific reaction of serum A was blocked by adding soluble 1163-SU5-variable peptide, spanning the variable region of SU5, but not with peptides corresponding to other strains such as 615 and Zn, thus confirming the presence of a type-specific B-cell epitope in this region. In contrast, broadly reacting serum B was blocked by the homologous 1163-SU5-total peptide but not by the 1163-SU5-variable peptide, spanning the variable region of the SU5 region, suggesting that the epitope recognized by this serum may be located in the constant region of SU5. We excluded this possibility by showing that a peptide spanning the constant region of SU5 (SG6393-SU5-consensus), as well as several SU5-total peptides derived from the SU5 sequences of other SRLV strains that encompass the common constant region, failed to block the binding of serum B to the 1163-SU5-total peptide.
Cross-reacting antibodies to the SU5-CO-total and SU5-1163-total peptides were obtained from the serum of a goat infected with the CAEV-CO strain by affinity purification on an 1163-SU5-total peptide column. These antibodies were used to demonstrate that an immunodominant epitope is located at the junction between the variable and constant regions. These cross-reacting antibodies have a very high avidity for the specific SU5 sequence of the infecting virus (CAEV-CO) and an intermediate avidity for the heterologous 1163-SU5-total peptide used on the affinity column. Additionally, we showed that by depleting these affinity-purified antibodies from their CO-SU5-total specific component, a background reactivity with very low avidity to the 1163-SU5-total peptide remains, which probably represents non-SRLV-specific, cross-reacting antibodies. The presence of these unspecific antibodies with very low avidity should be taken into account when using SU5 peptides for diagnostic purposes.
Taken together, these results confirm that broadly reacting sera do not bind exclusively to the constant region of SU5 but make contact with amino acids comprised in both the constant and the variable regions. Presumably, the amino acid composition of the variable region determines the avidity of the reaction. Therefore, by disrupting the binding of low-avidity antibody with 8 M urea washes, it will be possible to obtain precise phylogenetic information on the infecting virus also with broadly reacting sera.
These results also confirm the fine mapping of this immunogenic region performed by two independent laboratories by the Pepscan technique (34, 35). The successful use of synthetic peptides corresponding to a single epitope for diagnostic purposes strictly depends on the immunodominance of the epitope chosen. An immunodominant epitope should induce an antibody response in the majority of the infected animals, independently of their genetic background and the phylogenetic origin of the infecting virus. To test if this applies to the SU5 epitope, we analyzed sera from animals positive for both the virus genome and antiviral antibody, as demonstrated by a positive result in a Gag-PCR assay and WB, respectively.
As shown in Table 3, SU5-total peptides detected 77 of 81 WB-positive animals, proving the immunodominance of this region. The four potentially false-negative sera may be explained by the peculiar kinetics of the anti-SU5 antibody discussed above or may reflect the fact that these animals were infected with viruses phylogenetically distant from our SU5 sequences. To test this hypothesis, we restricted the analysis to 62 sera of animals from which the amplified PCR fragments were sequenced, permitting a phylogenetic analysis of the infecting virus. Only two sera eluded detection by the SU5-total ELISA. Animal ZS3794 was experimentally infected with virus strain 4663, and the negative result must be considered a true false negative, possibly due to the biphasic kinetics of seroconversion described above. Unfortunately, a second serum sample from this animal taken at a later time point was not available. Genomic DNA isolated from PBMC of the second seronegative animal was available, and we have PCR amplified and sequenced the SU5 region of the SRLV infecting this animal. The SU5-total peptide based on the consensus sequence of strain 3151, which contains an unusual mutation of a well-conserved proline (P) into a glutamine (Q) at position 13 (Fig. 1), detected the homologous serum in an ELISA, confirming the immunodominance of this epitope. These results indicate that the SU5-total ELISA is an efficient test that detected 61 of the 62 infected animals in spite of the phylogenetic diversity of the viruses involved. At the same time, however, the result obtained with goat 3151 implies that while interpreting diagnostic results based on SU5-total peptides one must always consider the rare possibility that a single animal or a flock may be infected with a virus variant which is not yet covered by the panel of SU5 peptides available.
In an attempt to simplify the use of the SU5-total ELISA, we explored the possibility of coating the ELISA plates with a mixture of SU5 peptides. Unfortunately, this approach markedly reduced the sensitivity of our assay and was abandoned (data not shown). The reasons for the conspicuous loss of sensitivity are not known and may involve unfavorable interactions between the peptides or competition for binding to the ELISA plates.
The panel of sera obtained from PCR-positive animals was also tested on the SU5-variable peptides encompassing the most variable region of this epitope. As expected, the use of these peptides caused a loss of sensitivity but increased the capacity of the assay to provide phylogenetic information on the infecting virus. The correlation between serological results and the corresponding Gag sequences was excellent. In all but one case, this ELISA predicted the phylogenetic origin (CAEV or MVV) of the infecting virus, confirming analogous results obtained with HIV (1, 27). The sole exception was a potentially false-positive serum with reactivity close to the negative cutoff of our assay.
The assay based on the SU5-variable peptides appeared to be more sensitive in detecting animals infected with CAEV strains than in detecting MVV strains, possibly indicating that our peptide panel does not yet cover some of the MVV strains circulating in Switzerland.
This ELISA, based exclusively on the variable region of SU5, revealed a clear hierarchy of reactivity to different peptides. The majority of the animals reacting with SU5 peptides derived from CAEV strains were infected with virus strains close to prototypic Swiss sequence 615. By testing several field sera from our diagnostic serum bank, we confirmed that before the start of the eradication campaign, the 615 virus had been the most prevalent strain circulating in the Swiss goat population (data not shown). Equally, the 1163 and SG6393 strains, which are phylogenetically closely related, appeared to dominate the MVV-like strains circulating in this population.
This novel SU5 assay was tested in a diagnostic setup based on an episode of seroconversions in a flock of goats that had been certified SRLV free for years.
In this context, the SU5 assay completely matched our expectations. The seropositivity of the goat sera as detected in the Chekit ELISA and by WB was confirmed. Moreover, the SU5 ELISA pointed to a virus closely related to the 1163 strain as the infecting agent. This was confirmed by PCR amplification and sequencing of SU5 sequences obtained from both goats and sheep of this flock. The SG6393 strain infecting these animals was shown to be phylogenetically very close, but not identical, to the 1163 strain, therefore excluding potential PCR contamination. The SU5 assay performed even better with the sheep sera, where the Chekit ELISA and the WB failed to detect any seropositive animals. As mentioned above, the accuracy of the SU5 ELISA results was demonstrated by PCR amplification and sequencing of the SU5 region of the virus infecting these animals.
In conclusion, we sampled and sequenced PCR amplicons encompassing the SU5 region of several Swiss-SRLV field isolates. The serological analysis of the corresponding SU5 peptides confirmed the immunodominance of the humoral immune response to these epitopes and showed rapid seroconversions accompanied by striking maturation of the avidity of the antibody directed to SU5. This paves the way for the use of these synthetic peptides as an effective tool complementing the serological tests currently used for SRLV diagnosis. The modular use of long SU5-total peptides containing the constant region of SU5 or shorter SU5-variable peptides restricted to the most variable part of SU5 permits both the application of these novel ELISAs as a broad screening tool and as a rapid test to obtain phylogenetic information on the infecting virus.
Acknowledgments
We thank Ruth Parham for valuable linguistic help. We are indebted to C. Shah, J. Böni, and J. Schüpbach of the Swiss National Center for Retroviruses in Zürich, Switzerland, for providing the panel of purified genomic DNA of animals with defined SRLV PCR status.
This work was supported by Swiss Federal Office for Education and Science (BBW) grant 95.0614 (FP5/COWDANADIAVAC/ICA4-CT-2000-30026) and by Swiss Federal Veterinary Office grant 1.01.19.
REFERENCES
- 1.Barin, F., Y. Lahbabi, L. Buzelay, B. Lejeune, A. Baillou-Beaufils, F. Denis, C. Mathiot, S. M'Boup, V. Vithayasai, U. Dietrich, and A. Goudeau. 1996. Diversity of antibody binding to V3 peptides representing consensus sequences of HIV type 1 genotypes A to E: an approach for HIV type 1 serological subtyping. AIDS Res. Hum. Retrovir. 12:1279-1289. [DOI] [PubMed] [Google Scholar]
- 2.Bertoni, G., C. Hertig, M. L. Zahno, H. R. Vogt, S. Dufour, P. Cordano, E. Peterhans, W. P. Cheevers, P. Sonigo, and G. Pancino. 2000. B-cell epitopes of the envelope glycoprotein of caprine arthritis-encephalitis virus and antibody response in infected goats. J. Gen. Virol. 81:2929-2940. [DOI] [PubMed] [Google Scholar]
- 3.Bertoni, G., M.-L. Zahno, R. Zanoni, H. R. Vogt, E. Peterhans, G. Ruff, W. P. Cheevers, P. Sonigo, and G. Pancino. 1994. Antibody reactivity to the immunodominant epitopes of the caprine arthritis-encephalitis virus gp38 transmembrane protein associates with the development of arthritis. J. Virol. 68:7139-7147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.de Andres, D., D. Klein, N. J. Watt, E. Berriatua, S. Torsteinsdottir, B. A. Blacklaws, and G. D. Harkiss. 2005. Diagnostic tests for small ruminant lentiviruses. Vet. Microbiol. 107:49-62. [DOI] [PubMed] [Google Scholar]
- 5.Fevereiro, M., S. Barros, and T. Fagulha. 1999. Development of a monoclonal antibody blocking-ELISA for detection of antibodies against maedi-Visna virus. J. Virol. Methods 81:101-108. [DOI] [PubMed] [Google Scholar]
- 6.Hammond, S. A., S. J. Cook, D. L. Lichtenstein, C. J. Issel, and R. C. Montelaro. 1997. Maturation of the cellular and humoral immune responses to persistent infection in horses by equine infectious anemia virus is a complex and lengthy process. J. Virol. 71:3840-3852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hedman, K., J. Hietala, A. Tiilikainen, A. L. Hartikainen-Sorri, K. Raiha, J. Suni, P. Vaananen, and M. Pietilainen. 1989. Maturation of immunoglobulin G avidity after rubella vaccination studied by an enzyme linked immunosorbent assay (avidity-ELISA) and by haemolysis typing. J. Med. Virol. 27:293-298. [DOI] [PubMed] [Google Scholar]
- 8.Herrmann, L. M., W. P. Cheevers, K. L. Marshall, T. C. McGuire, M. M. Hutton, G. S. Lewis, and D. P. Knowles. 2003. Detection of serum antibodies to ovine progressive pneumonia virus in sheep by using a caprine arthritis-encephalitis virus competitive-inhibition enzyme-linked immunosorbent assay. Clin. Diagn. Lab. Immunol. 10:862-865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herrmann, L. M., W. P. Cheevers, T. C. McGuire, D. S. Adams, M. M. Hutton, W. G. Gavin, and D. P. Knowles. 2003. Competitive-inhibition enzyme-linked immunosorbent assay for detection of serum antibodies to caprine arthritis-encephalitis virus: diagnostic tool for successful eradication. Clin. Diagn. Lab. Immunol. 10:267-271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Herrmann, L. M., T. C. McGuire, I. Hotzel, G. S. Lewis, and D. P. Knowles. 2005. Surface envelope glycoprotein is B-lymphocyte immunodominant in sheep naturally infected with ovine progressive pneumonia virus. Clin. Diagn. Lab. Immunol. 12:797-800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Houwers, D. J., A. H. Visscher, and P. R. Defize. 1989. Importance of ewe/lamb relationship and breed in the epidemiology of maedi-visna virus infections. Res. Vet. Sci. 46:5-8. [PubMed] [Google Scholar]
- 12.Johnson, G. C., A. F. Barbet, P. Klevjer-Anderson, and T. C. McGuire. 1983. Preferential immune response to virion surface glycoproteins by caprine arthritis-encephalitis virus-infected goats. Infect. Immun. 41:657-665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Knowles, D. P., W. P. Cheevers, T. C. McGuire, T. A. Stem, and J. R. Gorham. 1990. Severity of arthritis is predicted by antibody response to Gp135 in chronic infection with caprine arthritis-encephalitis virus. J. Virol. 64:2396-2398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Krieg, A., and E. Peterhans. 1990. Die caprine Arthritis-Encephalitis in der Schweiz: epidemiologische und klinische Untersuchungen. Schweiz. Arch. Tierheilkd. 132:345-352. [PubMed] [Google Scholar]
- 15.Kühn, N. 1998. Retrospektive Analyse der Bekämpfung der caprinen Arthritis-Enzephalitis in der Schweiz. PhD. thesis. Faculty of Veterinary Medicine, University of Bern, Bern, Switzerland.
- 16.Leroux, C., J. Chastang, T. Greenland, and J. F. Mornex. 1997. Genomic heterogeneity of small ruminant lentiviruses: existence of heterogeneous populations in sheep and of the same lentiviral genotypes in sheep and goats. Arch. Virol. 142:1125-1137. [DOI] [PubMed] [Google Scholar]
- 17.McGuire, T. C., D. P. Knowles, Jr., W. C. Davis, A. L. Brassfield, T. A. Stem, and W. P. Cheevers. 1992. Transmembrane protein oligomers of caprine arthritis-encephalitis lentivirus are immunodominant in goats with progressive arthritis. J. Virol. 66:3247-3250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McGuire, T. C., K. I. O'Rourke, D. P. Knowles, and W. P. Cheevers. 1990. Caprine arthritis encephalitis lentivirus transmission and disease. Curr. Top. Microbiol. Immunol. 160:61-76. [DOI] [PubMed] [Google Scholar]
- 19.Narayan, O., D. Sheffer, D. E. Griffin, J. Clements, and J. Hess. 1984. Lack of neutralizing antibodies to caprine arthritis-encephalitis lentivirus can be overcome by immunization with inactivated mycobacterium tuberculosis. J. Virol. 49:349-355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pancino, G., H. Ellerbrok, M. Sitbon, and P. Sonigo. 1994. Conserved framework of envelope glycoproteins among lentiviruses. Curr. Top. Microbiol. Immunol. 188:77-105. [DOI] [PubMed] [Google Scholar]
- 21.Parekh, B. S., M. S. Kennedy, T. Dobbs, C. P. Pau, R. Byers, T. Green, D. J. Hu, S. Vanichseni, N. L. Young, K. Choopanya, T. D. Mastro, and J. S. McDougal. 2002. Quantitative detection of increasing HIV type 1 antibodies after seroconversion: a simple assay for detecting recent HIV infection and estimating incidence. AIDS Res. Hum. Retrovir. 18:295-307. [DOI] [PubMed] [Google Scholar]
- 22.Pepin, M., C. Vitu, P. Russo, J. F. Mornex, and E. Peterhans. 1998. Maedi-visna virus infection in sheep: a review. Vet. Res. 29:341-367. [PubMed] [Google Scholar]
- 23.Peterhans, E., T. Greenland, J. Badiola, G. Harkiss, G. Bertoni, B. Amorena, M. Eliaszewicz, R. A. Juste, R. Krassnig, J. P. Lafont, P. Lenihan, G. Petursson, G. Pritchard, J. Thorley, C. Vitu, J. F. Mornex, and M. Pepin. 2004. Routes of transmission and consequences of small ruminant lentiviruses (SRLVs) infection and eradication schemes. Vet. Res. 35:257-274. [DOI] [PubMed] [Google Scholar]
- 24.Pisoni, G., A. Quasso, and P. Moroni. 2005. Phylogenetic analysis of small-ruminant lentivirus subtype B1 in mixed flocks: evidence for natural transmission from goats to sheep. Virology 339:147-152. [DOI] [PubMed] [Google Scholar]
- 25.Plantier, J. C., F. Damond, M. Lasky, J. L. Sankale, C. Apetrei, M. Peeters, L. Buzelay, S. M'Boup, P. Kanki, E. Delaporte, F. Simon, and F. Barin. 1999. V3 serotyping of HIV-1 infection: correlation with genotyping and limitations. J. Acquir. Immune Defic. Syndr. Hum. Retrovirol. 20:432-441. [DOI] [PubMed] [Google Scholar]
- 26.Robertson, P., S. Beynon, R. Whybin, C. Brennan, U. Vollmer-Conna, I. Hickie, and A. Lloyd. 2003. Measurement of EBV-IgG anti-VCA avidity aids the early and reliable diagnosis of primary EBV infection. J. Med. Virol. 70:617-623. [DOI] [PubMed] [Google Scholar]
- 27.Robey, W. G., P. L. Nara, C. M. Poore, M. Popovic, M. F. McLane, F. Barin, M. Essex, and P. J. Fischinger. 1987. Rapid assessment of relationships among HIV isolates by oligopeptide analyses of external envelope glycoproteins. AIDS Res. Hum. Retrovir. 3:401-408. [DOI] [PubMed] [Google Scholar]
- 28.Ruff, G., and S. Lazary. 1988. Evidence for linkage between the caprine leucocyte antigen (CLA) system and susceptibility to CAE virus-induced arthritis in goats. Immunogenetics 28:303-309. [DOI] [PubMed] [Google Scholar]
- 29.Ruff, G., J. G. Regli, and S. Lazary. 1993. Occurrence of caprine leucocyte class I and II antigens in Saanen goats affected by caprine arthritis (CAE). Eur. J. Immunogenet. 20:285-288. [DOI] [PubMed] [Google Scholar]
- 30.Saitou, N., and M. Nei. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4:406-425. [DOI] [PubMed] [Google Scholar]
- 31.Saltarelli, M., G. Querat, D. A. M. Konings, R. Vigne, and J. E. Clements. 1990. Nucleotide sequence and transcriptional analysis of molecular clones of CAEV which generate infectious virus. Virology 179:347-364. [DOI] [PubMed] [Google Scholar]
- 32.Shah, C., J. Boni, J. B. Huder, H. R. Vogt, J. Muhlherr, R. Zanoni, R. Miserez, H. Lutz, and J. Schupbach. 2004. Phylogenetic analysis and reclassification of caprine and ovine lentiviruses based on 104 new isolates: evidence for regular sheep-to-goat transmission and worldwide propagation through livestock trade. Virology 319:12-26. [DOI] [PubMed] [Google Scholar]
- 33.Shah, C., J. B. Huder, J. Boni, M. Schonmann, J. Muhlherr, H. Lutz, and J. Schupbach. 2004. Direct evidence for natural transmission of small-ruminant lentiviruses of subtype A4 from goats to sheep and vice versa. J. Virol. 78:7518-7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trujillo, J. D., N. M. Kumpula-McWhirter, K. J. Hotzel, M. Gonzalez, and W. P. Cheevers. 2004. Glycosylation of immunodominant linear epitopes in the carboxy-terminal region of the caprine arthritis-encephalitis virus surface envelope enhances vaccine-induced type-specific and cross-reactive neutralizing antibody responses. J. Virol. 78:9190-9202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Valas, S., C. Benoit, G. Baudry, G. Perrin, and R. Z. Mamoun. 2000. Variability and immunogenicity of caprine arthritis-encephalitis virus surface glycoprotein. J. Virol. 74:6178-6185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ward, K. N., W. Dhaliwal, K. L. Ashworth, E. J. Clutterbuck, and C. G. Teo. 1994. Measurement of antibody avidity for hepatitis C virus distinguishes primary antibody responses from passively acquired antibody. J. Med. Virol. 43:367-372. [DOI] [PubMed] [Google Scholar]
- 37.Zanoni, R., A. Krieg, and E. Peterhans. 1989. Detection of antibodies to caprine arthritis-encephalitis virus by protein G enzyme-linked immunosorbent assay and immunoblotting. J. Clin. Microbiol. 27:580-582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zanoni, R. G., H. R. Vogt, B. Pohl, J. Bottcher, W. Bommeli, and E. Peterhans. 1994. An ELISA based on whole virus for the detection of antibodies to small-ruminant lentiviruses. Zentbl. Veterinarmed. B 41:662-669. [DOI] [PubMed] [Google Scholar]
- 39.Zink, M. C., J. A. Yager, and J. D. Myers. 1990. Pathogenesis of caprine arthritis encephalitis virus—cellular localization of viral transcripts in tissues of infected goats. Am. J. Pathol. 136:843-854. [PMC free article] [PubMed] [Google Scholar]