Abstract
The Mycobacterium avium species consists of a group of organisms that are genetically related but phenotypically diverse, with certain variants presenting clear differences in terms of their host association and disease manifestations. The ability to distinguish between these subtypes is of relevance for accurate diagnosis and for control programs. Using a comparative genomics approach, we have uncovered large sequence polymorphisms that are, respectively, absent from bird-type M. avium isolates and from cattle types and sheep types of M. avium subsp. paratuberculosis. By evaluating the distribution of these genomic polymorphisms across a panel of strains, we were able to assign unique genomic signatures to these host-associated variants. We propose a simple PCR-based strategy based on these polymorphisms that can rapidly type M. avium isolates into these subgroups.
Mycobacterium avium organisms responsible for Johne's disease, avian tuberculosis, and opportunistic infections in humans have been classified as belonging to one species based on numerical taxonomy (27), DNA-DNA hybridization (20), and 16S rRNA sequencing (29). This shared designation has placed emphasis on the genetic relatedness of these organisms. Yet, there is ample evidence that members of this group exhibit phenotypic differences in laboratory characteristics and have widely varying propensities to cause disease in different hosts. In agreement with this diversity, M. avium organisms differ according to the presence of genetic elements, such as insertion elements (11, 12, 14), and also vary extensively at certain genomic regions (18, 22, 23).
Of the M. avium organisms, M. avium subsp. paratuberculosis presents a clear diagnostic concern, as it is a serious pathogen of cattle and other ruminants and a potential zoonotic agent (1, 2, 4, 5, 13, 21). Traditionally identified through phenotypic assays, and more recently by the presence of the insertion element IS900, M. avium subsp. paratuberculosis can now be specifically characterized by the deletion of a large sequence called LSPA 8 that is present in other M. avium organisms (22). Whole-genome comparisons of a small number of M. avium subsp. paratuberculosis isolates have shown relatively few genomic differences among strains (18, 23), suggesting genomic features specific to tested strains should lend themselves to robust diagnostic algorithms. One source of variability described within M. avium subsp. paratuberculosis is the existence of two host-associated types: the more prevalent cattle (C) strains, also known as type II strains, and the rarer sheep (S) strains, also called type I strains. These types have been distinguished through molecular fingerprints and PCR assays exploiting the variable presence of the mobile insertion elements IS900 and IS1311 (7, 8, 25). The ability to both detect and differentiate between these types of strains is of obvious importance for both accurate diagnosis and to guide control programs.
Among M. avium members other than M. avium subsp. paratuberculosis, phenotypic and genetic studies suggest the existence of two principal groups of organisms. Certain strains are characterized by the presence of the insertion element IS901 and a particular molecular fingerprint based on the closely related insertion sequences IS1311 and IS1245 (6, 14) referred to in some papers in which low-stringency hybridization was used as based solely on IS1245 (16, 19). These strains are associated with severe disease in domestic and wild birds and are thought to cluster into a unique group, called M. avium subsp. avium (16, 19). The genetic distinction between this group and the subspecies known as M. avium subsp. silvaticum is not yet clear. In contrast, the remaining M. avium strains manifest a diverse range of IS1245/IS1311 patterns and are pathogens of pigs and opportunistic pathogens of susceptible humans. These observations suggest that nonparatuberculosis M. avium organisms might be divided into two epidemiologically relevant variants, those called M. avium subsp. avium or bird-type M. avium and the rest, recently named M. avium subsp. hominissuis (16).
The goal of our study was to determine if these different variants of M. avium could be structured into distinct genomic profiles. Specifically, we hypothesized that cattle and sheep strains of M. avium subsp. paratuberculosis may be associated with unique genomic signatures, that bird-type M. avium strains would constitute a separate cluster of organisms with their own genomic profile, and that PCR-based testing for these regions would provide a robust method of identifying these M. avium variants.
MATERIALS AND METHODS
Bacterial isolates.
We assembled a panel of M. avium isolates, representative of each of the subspecies, selected on the basis that they were isolated from different hosts and from different geographic provenance. The isolates had been cultured with standard mycobacterial media with or without mycobactin J supplementation, and DNA extraction was performed according to standard methods (30).
All isolates were initially identified as M. avium based on 16S rRNA sequencing and were further subspeciated based on mycobactin J dependence and presence of IS900 (M. avium subsp. paratuberculosis; n = 21), initial mycobactin J dependence and presence of IS901 (M. avium subsp. silvaticum; n = 3), or mycobactin J independence and presence of IS1245/IS1311 (M. avium subsp. avium or M. avium subsp. hominissuis; n = 23), for a total of 47 isolates.
The M. avium subsp. paratuberculosis isolates were further characterized as C type (n = 11) or as S type (n = 10) by restriction fragment length polymorphism (RFLP) with IS900 (6) and additionally by PCR testing with primers able to distinguish between these two types (7). Strains were carefully chosen to ensure that they originated from different geographic locations and, further, had nonidentical S or C IS900 fingerprints. The M. avium subsp. avium and M. avium subsp. hominissuis isolates were characterized as bird type (M. avium subsp. avium; n = 13) or as multiband, hominissuis type (M. avium subsp. hominissuis; n = 10) by RFLP with IS1245/IS1311 (16).
Microarray-based genomic comparisons.
Comparative genomic studies were performed on a whole-genome DNA microarray composed of 70-bp oligonucleotide probes, printed in duplicate on microarray slides (Sigmascreen; Sigma) using a microarray robot (Chipwriter model SDDC2; Virtek). The array is representative of 98% of the open reading frames (ORFs) for the genome of M. avium subsp. paratuberculosis strain K10 and 93% of the predicted ORFs for the genome of M. avium strain 104. DNA from M. avium 104 (the reference sequenced strain, a clinical isolate from a human AIDS patient in the United States) served as the comparator DNA in cohybridization experiments with the following test isolates: two M. avium subsp. avium strains characterized as bird type by RFLP (strains R13 and D71076), two M. avium subsp. silvaticum strains (ATCC 49884 and 9800851), two M. avium subsp. paratuberculosis strains of the cattle type (sequenced strain K10, synonymous with ATCC BAA-968, and strain 17, a clinical isolate from a bison), and two M. avium subsp. paratuberculosis strains of the sheep type (strains LN20 and 4857). Fluorescent labeling of the DNA samples and hybridizations were performed according to previously described methods (23), and arrays were scanned using a confocal scanner (ScanArray Lite; Packard BioScience, Massachusetts). Fluorescence intensities were quantified using a software package (ScanArray Express version 2; Packard BioScience, Massachusetts). The fluorescence ratio for each spot was determined; this ratio was log10 transformed and then normalized with respect to the mean value of the fluorescence ratios. A Z value [Z = (log10 of the fluorescence ratio) − (mean/standard deviation)] was calculated for each spot, and spots with a Z score of greater than 2 were flagged for study. We screened for regions of three or more contiguous ORFs which hybridized with DNA from M. avium strain 104 but not with test isolates. Regions that were thus identified as divergent in the test isolates were further analyzed using a PCR approach. We first verified that these sequences were missing from test isolates using primers designed towards a sequence internal to these regions. In a second step, we performed PCR using primers designed towards the flanking regions, such that an amplicon would be obtained only if the region were missing. The resulting amplicons were sequenced in a core sequencing facility (McGill University and Génome Québec Innovation Centre) on a 3730XL DNA Analyzer system, using ABI dye terminator chemistry. Resulting sequences were aligned to the genomes of M. avium 104 and M. avium subsp. paratuberculosis K10 to identify the exact site at which the sequence polymorphism occurs.
Testing for the distribution of specific LSPs across a panel of isolates.
Each isolate in our panel was first tested for the presence or absence of LSPA 8, a large sequence previously shown to be specifically missing from M. avium subsp. paratuberculosis (22). This was done to ensure that isolates designated as M. avium subsp. paratuberculosis as per conventional methods (phenotypic characterization and presence of IS900) conformed to a uniform genomic definition of M. avium subsp. paratuberculosis. In a subsequent step, isolates were tested for the presence or absence of the large sequences identified by microarray as potentially associated with distinct groups of organisms. This was done with a multiplex PCR approach using a set of three primers: two primers (forward and reverse) designed towards the flanking regions (bridging primers) of the large sequence polymorphism (LSP) and a third primer designed to recognize a sequence internal to the LSP (internal primer). The primers were designed using Primer 3 software (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi), and primers were designed such that the resulting PCR products would be of different sizes depending on the presence or absence of the LSP under study. Primer sequences and predicted amplicon lengths are provided in Table 1.
TABLE 1.
Primers used for testing of LSPs
| Name of LSP | Primer 1 sequence (forward) | Primer 2 sequence (reverse) | Primer 3 sequence (reverse) | Product size (bp)a
|
|
|---|---|---|---|---|---|
| LSP present | LSP absent | ||||
| LSPA 8 | CCAGGTCGAAGAGGTGCTC | CCTATACCGCCAACGACATC | GTGCTGCCGTCCAGGTAG | 222 | 399 |
| LSPA 17 | CTGGAGTACTTCCACGACCA | GTCCAGGAAGAACCGGAAC | GCACTCGAATTCACGAAATG | 202 | 398 |
| LSPA 20 | GGCGTTACAGAATTGCCTTG | GCTCGAAGTTGGAGATCAGG | GTACGTGGTGACCAATGTCG | 197 | 306 |
| LSPA 4-II | TAGAAGGTGCGGGAAAGTTG | GTCTATCTGGCGGTGCTCTC | GTCGAAGCAGCGTTGATTGT | 540 | 462 |
| LSPA 18 | GACGTCCTGATCATCGGTTC | GTCCACCGAGGAGGTCAG | 1,967 | ||
| LSPA 18 | GTAGTAGTCGTCGCCCAGCA | GTGGGATCACGACCTGGAC | 403 | ||
For all but one LSP (LSPA 18), PCR tests were done using the three primers in a multiplex PCR; expected product sizes are listed in the last two columns. Primers 1 and 3 are the bridging primers, and primer 2 is the internal primer. For LSPA 18, PCR testing was performed using two primers only, such that a product was obtained with one set of (bridging) primers when the region was missing and one set of (internal) primers when the region was present.
PCRs.
PCRs were performed in 50-μl volumes, using 5 μl (equivalent to 5 ng) of DNA. For all but one set of reaction mixtures, we used 1 U Taq polymerase (MBI Fermentas), 5 μl of 10× PCR buffer (MBI Fermentas), 2.5 mM MgCl2, 5 μl acetamide 50% (wt/vol), 0.2 mM deoxynucleoside triphosphates (dNTPs), and 0.5 μM of each primer. In one instance where the expected product was close to 2 kb (LSPA 18), we were unable to design a reliable three-primer PCR because of the presence of an insertion element and a sequence inversion; therefore, we did separate PCRs for the presence or the absence of this genomic polymorphism. In this case, to amplify the 2-kb product, we used 3.75 U Taq polymerase and 5 μl of 10× buffer 3 (Expand Long template PCR system; Roche Diagnostics Corporation, Indianapolis, IN), 5 μl acetamide 50% (wt/vol), 0.4 mM lithium-stabilized dNTPs (dNTP set Li-salt solution; Roche Diagnostics Corporation), and 0.5 μM of each primer. For all reactions, PCR amplification consisted of an initial denaturation step at 94°C for 5 min followed by 35 cycles of denaturation at 94°C for 45 s, with annealing at 60°C (LSPA 8) or 55°C (other LSPs) for 45 s, elongation at 72°C for 2 min, and a final elongation step at 72°C for 10 min. PCR products were separated by electrophoresis in 1.5% (wt/vol) agarose gels containing ethidium bromide. When the amplicon indicated that the LSP was missing, this PCR product was subjected to sequencing to ensure that the exact same polymorphism was detected in each isolate.
RESULTS
From analysis of DNA microarray-based cohybridization experiments, we identified three large sequences that were polymorphic among the four strains of M. avium subsp. paratuberculosis studied by microarray. Of these, two sequences, LSPA 4-II and LSPA 18, were present in both of the S strains but missing from the two C strains. Another sequence, LSPA 20, was present in the C strains but missing from the S strains. We also identified one large sequence, LSPA 17, that was missing from the M. avium subsp. avium bird-type strains and M. avium subsp. silvaticum but present in M. avium 104 and the M. avium subsp. paratuberculosis strains studied by microarray. These LSPs and their distribution across a panel of isolates are described in greater detail below.
Description of sequences polymorphic in strains of M. avium subsp. paratuberculosis. (i) LSPA 4-II.
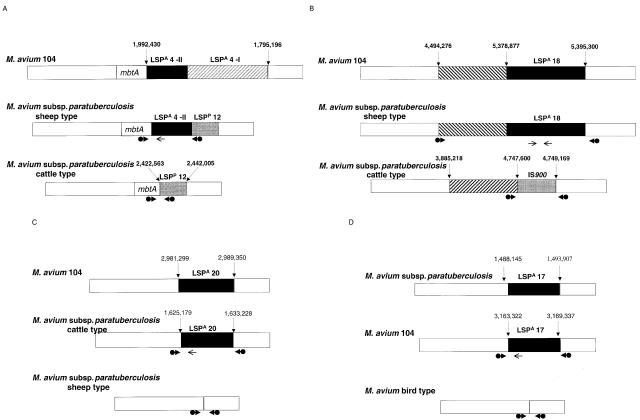
From previous genomic comparisons of members of the M. avium complex, we had identified LSP 4, a large sequence present in M. avium 104 but missing in M. avium subsp. paratuberculosis K10 (23). In the former, this 197-kb sequence is located within the mycobactin synthesis operon between mbtA and mbtJ. In M. avium subsp. paratuberculosis K10, LSP 4 is replaced by a different, 19-kb sequence called LSPP 12 (MAP 2179 to MAP 2197), determined by PCR to be highly specific to M. avium subsp. paratuberculosis isolates (22). Microarray data for C and S strains of M. avium subsp. paratuberculosis indicated that LSPP 12 was present in both but that a 26-kb segment of the M. avium LSP 4 element was present only in the S strains. These results suggested that 171 kb of LSP 4 was present in M. avium 104 but absent from S strains and that a further 26-kb segment was deleted from C strains; we named these two polymorphisms at the same locus LSPA 4-I and LSPA 4-II, respectively (Fig. 1A).
FIG. 1.
Schematic representation of large sequence polymorphisms. LSPA 4-II and LSPA 18 are specifically absent from M. avium subsp. paratuberculosis cattle type, LSPA 20 is absent from M. avium subsp. paratuberculosis sheep type, and LSPA 17 is absent from M. avium subsp. avium bird type. Coordinates on the genome are given as base pairs, starting from the first nucleotide of the start codon of dnaA in M. avium 104 and M. avium subsp. paratuberculosis K10, respectively. White boxes represent homologous sequences across M. avium 104, M. avium subsp. paratuberculosis sheep type, and M. avium subsp. paratuberculosis cattle type. (A) LSPA 4-II is depicted by the black box, LSPA 4-I is depicted by the striped box, and LSPP 12 is indicated by the gray box. (B) The striped box represents a large sequence that is conserved but inverted in M. avium subsp. paratuberculosis cattle type, and the black box represents LSPA 18. (C) The black box represents LSPA 20. (D) The black box represents LSPA 17. Thick arrows represent primers flanking the LSP (bridging primers); a PCR product is obtained if the region is missing. Thin arrows represent primers targeting a sequence that is within the LSP (internal primers); a PCR product is obtained if the region is present.
(ii) LSPA 18.
LSPA 18 is a 16-kb sequence present in M. avium subsp. avium and S strains of M. avium subsp. paratuberculosis that was absent in C strains of M. avium subsp. paratuberculosis. This sequence is immediately adjacent to an 800-kb sequence that is conserved in M. avium subsp. paratuberculosis K10 (C type) but inverted in relation to M. avium 104 and S strains of M. avium subsp. paratuberculosis. In M. avium subsp. paratuberculosis K10, this sequence is replaced by an IS900 element, MAP4281 (Fig. 1B).
(iii) LSPA 20.
LSPA 20, an 8-kb sequence spanning MAP1490 to MAP1484c, was absent from S strains of M. avium subsp. paratuberculosis. This sequence is predicted to encode proteins involved in metabolism, notably includes genes annotated as putatively encoding pyruvate dehydrogenases, and is highly conserved in other mycobacteria, including Mycobacterium tuberculosis. Sequence analysis of S strains indicated that MAP1490 and MAP1484c were both truncated compared to the annotated ORF in C strains, with the polymorphism occurring at position 1633228 of the M. avium subsp. paratuberculosis K10 genome, suggesting that the loss of LSPA 20 is a deletion event that occurred selectively in S strains of M. avium subsp. paratuberculosis (Fig. 1C).
(iv) Distribution of large sequence polymorphisms among strains of M. avium subsp. paratuberculosis: LSPA 8.
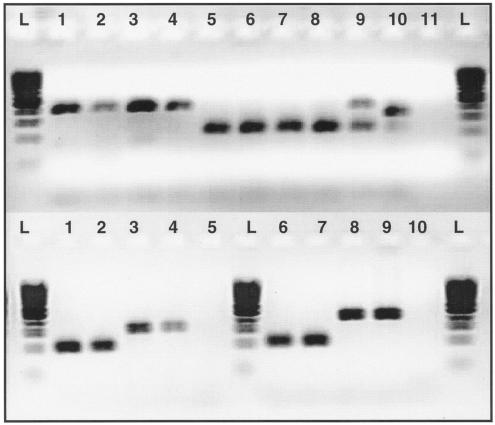
All 21 strains of M. avium subsp. paratuberculosis isolates in this study lacked the LSPA 8 sequence, while the sequence was detected as present in all non-M. avium subsp. paratuberculosis isolates, as shown by a multiplex PCR approach (Fig. 2). In a previous report, we noted that in a small minority of M. avium subsp. paratuberculosis isolates, the absence of LSPA 8 could not be demonstrated but that intervening sequence could not be amplified for these samples (22). In the present study, the absence of this region was 100% sensitive for M. avium subsp. paratuberculosis, suggesting a now-resolved technical limitation in our previous report (Table 2).
FIG. 2.
Detection of subspecies and subtypes of M. avium using PCR for large sequences polymorphic among strains of M. avium. On the top panel, 11 samples were tested for LSPA 8 using a three-primer PCR. Lanes: L, 100-bp ladder; 1 to 4, M. avium subsp. paratuberculosis strains; 5 and 6, nonparatuberculosis strains of M. avium; 9, mixed sample (M. avium 104 and M. avium subsp. paratuberculosis K10); 10, M. intracellulare ATCC 13950 strain; 11, water. In the bottom panel, four samples were tested for LSPA 20 and four samples are tested for LSPA 17 using three-primer PCRs. Lanes: L, 100-bp ladder; 1 and 2, M. avium subsp. paratuberculosis C type; 3 and 4, M. avium subsp. paratuberculosis S type; 5, water; 6 and 7, M. avium subsp. hominissuis; 8 and 9, M. avium subsp. avium bird type; 10, water.
TABLE 2.
PCR testing for presence or absence of LSPs across a panel of M. avium isolatesa
| Isolate | Isolate identity | Strain typeb | Host | Origin of isolate | IS900 | IS901 | LSPA 8 | LSPA 17 | LSPA 20 | LSPA 4-II | LSPA 18 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| K10 | M. avium subsp. paratuberculosis | C | Cow | USA | + | − | − | + | + | − | − |
| 17 | M. avium subsp. paratuberculosis | C | Bison | Canada | + | − | − | + | + | − | − |
| 989 | M. avium subsp. paratuberculosis | C | Cow | New Zealand | + | − | − | + | + | − | − |
| 6024 | M. avium subsp. paratuberculosis | C | Cow | New Zealand | + | − | − | + | + | − | − |
| 316 | M. avium subsp. paratuberculosis | C | Cow | United Kingdom | + | − | − | + | + | − | − |
| 6770B | M. avium subsp. paratuberculosis | C | Deer | New Zealand | + | − | − | + | + | − | − |
| 6354 | M. avium subsp. paratuberculosis | C | Deer | New Zealand | + | − | − | + | + | − | − |
| 1515 | M. avium subsp. paratuberculosis | C | Human | USA (ATCC 43015) | + | − | − | + | + | − | − |
| TMC 1613 | M. avium subsp. paratuberculosis | C | Cow | USA | + | − | − | + | + | − | − |
| 1518 | M. avium subsp. paratuberculosis | C | Cow | USA (ATCC 19698) | + | − | − | + | + | − | − |
| 7296 | M. avium subsp. paratuberculosis | C | Deer | New Zealand | + | − | − | + | + | − | − |
| LN 20 | M. avium subsp. paratuberculosis | S | Pig | Canada | + | − | − | + | − | + | + |
| 4857 | M. avium subsp. paratuberculosis | S | Sheep | New Zealand | + | − | − | + | − | + | + |
| 4873 | M. avium subsp. paratuberculosis | S | Sheep | New Zealand | + | − | − | + | − | + | + |
| 6758 | M. avium subsp. paratuberculosis | S | Sheep | New Zealand | + | − | − | + | − | + | + |
| 506c | M. avium subsp. paratuberculosis | S | Sheep | South Africa | + | − | − | + | − | + | + |
| 575A | M. avium subsp. paratuberculosis | S | Sheep | South Africa | + | − | − | + | − | + | + |
| 85/14 | M. avium subsp. paratuberculosis | S | Sheep | Canada | + | − | − | + | − | + | + |
| P465 | M. avium subsp. paratuberculosis | S | Sheep | Iceland | + | − | − | + | − | + | + |
| 6282 | M. avium subsp. paratuberculosis | S | Deer | New Zealand | + | − | − | + | − | + | + |
| 3579 | M. avium subsp. paratuberculosis | S | Deer | New Zealand | + | − | − | + | − | + | + |
| 9801574 | M. avium subsp. silvaticum | B | Bird (penguin) | Belgium | − | + | + | − | + | + | + |
| 9800851 | M. avium subsp. silvaticum | B | Bird (wood pigeon) | Belgium | − | + | + | − | + | + | + |
| 49884 | M. avium subsp. silvaticum | B | Bird (wood pigeon) | France (ATCC 49884) | − | + | + | − | + | + | + |
| R13 | M. avium subsp. avium | B | Bird | The Netherlands | − | + | + | − | + | + | + |
| 160-74 | M. avium subsp. avium | B | Bird (goose) | The Netherlands | − | + | + | − | + | + | + |
| D71076 | M. avium subsp. avium | B | Bird (owl) | The Netherlands | − | + | + | − | + | + | + |
| 9501266 | M. avium subsp. avium | B | Bird (chicken) | Argentina | − | + | + | − | + | + | + |
| 34211 | M. avium subsp. avium | B | Bird (spoonbill) | The Netherlands | − | + | + | − | + | + | + |
| 726 | M. avium subsp. avium | B | Bird (pigeon) | The Netherlands | − | + | + | − | + | + | + |
| 1262 | M. avium subsp. avium | B | Bird (owl) | The Netherlands | − | + | + | − | + | + | + |
| 238 | M. avium subsp. avium | B | Bird (black swan) | The Netherlands | − | + | + | − | + | + | + |
| 469 | M. avium subsp. avium | B | Bird (parrot) | The Netherlands | − | + | + | − | + | + | + |
| 33599/16 | M. avium subsp. avium | B | Bird (peacock) | The Netherlands | − | + | + | − | + | + | + |
| Wag 205 | M. avium subsp. avium | B | Cow | New Zealand | − | + | + | − | + | + | + |
| Wag 206 | M. avium subsp. avium | B | Cow | New Zealand | − | + | + | − | + | + | + |
| Strain 18 | M. avium subsp. avium | B | Cow | USA | − | + | + | − | + | + | + |
| 9602012 | M. avium subsp. hominissuis | HS | Human | The Netherlands | − | − | + | − | + | + | + |
| Wag 207 | M. avium subsp. hominissuis | HS | Deer | New Zealand | − | − | + | + | + | + | + |
| Wag 208 | M. avium subsp. hominissuis | HS | Deer | New Zealand | − | − | + | + | + | + | + |
| 104 | M. avium subsp. hominissuis | HS | Human | USA | − | − | + | + | + | + | + |
| 9601596 | M. avium subsp. hominissuis | HS | Human | The Netherlands | − | − | + | + | + | + | + |
| 9601288 | M. avium subsp. hominissuis | HS | Human | The Netherlands | − | − | + | + | + | + | + |
| ATCC700897 | M. avium subsp. hominissuis | HS | Human | USA (ATCC700897) | − | − | + | + | + | + | + |
| 9601544 | M. avium subsp. hominissuis | HS | Human | The Netherlands | − | − | + | + | + | + | + |
| 9601034 | M. avium subsp. hominissuis | HS | Human | The Netherlands | − | − | + | + | + | + | + |
| 9700642 | M. avium subsp. hominissuis | HS | Pig | The Netherlands | − | − | + | + | + | + | + |
+, sequence present; −, sequence absent. Results are based on the presence or absence of PCR products of different sizes.
Strain type abbreviations: C, cattle (type II); S, sheep (type I); B, bird; HS, hominissuis.
LSPs absent from C strains.
All 11 C strains of M. avium subsp. paratuberculosis lacked LSPA 4-II, and sequencing of their PCR products revealed identical sequences with a truncated mbtA gene (MAP2178). In contrast, LSPA 4-II amplified as present in S strains of M. avium subsp. paratuberculosis and all nonparatuberculosis M. avium strains studied. All C strains of M. avium subsp. paratuberculosis also lacked LSPA 18. With primers flanking the LSPA 18 sequence and under PCR conditions that were optimized to amplify a 2-kb product across an IS900 element, we were able to successfully obtain a PCR product in all 11 C strains of M. avium subsp. paratuberculosis and in none of the S strains. We sequenced these amplicons and confirmed they were identical to the sequence of M. avium subsp. paratuberculosis K10. In contrast, the presence of LSPA 18 was demonstrated in all other strains by PCR using primers internal to the sequence.
LSP absent from S strains.
All S strains studied lacked LSPA 20, which was present in all C strains, as demonstrated by a multiplex PCR approach. Sequences obtained from all 10 S strains were identical, with the polymorphism occurring at the exact same site and confirming the truncation of the ORFs of each end of this sequence.
Sequence polymorphic in M. avium strains isolated from birds: LSPA 17.
Analysis of the cohybridization experiments of M. avium 104 with two strains of M. avium subsp. avium characterized as bird type and two isolates labeled M. avium subsp. silvaticum revealed the consistent absence of a 6-kb sequence called LSPA 17, spanning MAP1375c to MAP1381c. From in silico analyses of the genomes of M. avium 104 and M. avium subsp. paratuberculosis K10, we determined that this region is conserved between these genomes (sequence identity of 98%), although a 253-bp portion situated in the middle of this sequence is missing from the latter. This polymorphism occurs at the same junction site in M. avium subsp. avium as well as in strains called M. avium subsp. silvaticum, corresponding to position 1493907 of the M. avium subsp. paratuberculosis strain K10 genome (Fig. 1D). LSPA 17 contains several genes with homology to those encoding short-chain dehydrogenases, in addition to a transcriptional regulator of the LysR family. Of note, LSPA 17 is not syntenous with the genome of the M. tuberculosis complex and does not appear to be conserved in other mycobacteria.
From testing for the presence or absence of this region using a multiplex PCR assay, we noted that the 12 M. avium subsp. avium isolates of the bird type and the 3 isolates designated as M. avium subsp. silvaticum all lacked LSPA 17. This region was detected as present in all M. avium subsp. paratuberculosis isolates and in the majority of M. avium subsp. hominissuis type isolates. Exceptionally, we determined that one M. avium subsp. hominissuis isolate, obtained from a patient with AIDS, also lacked this sequence. We further characterized this isolate by amplification and sequencing of the 3′ end of the hsp65 gene, a genotyping method that we recently applied towards classification of M. avium organisms (28). The M. avium subsp. hominissuis isolate that lacked LSPA 17 was shown to differ from the bird-type hsp65 sequevar (code 4) by only one SNP and belonged to the sequevar called code 3. Consistent with other strains belonging to this sequevar, the isolate did not possess IS901, an insertion element generally found to be present in bird strains of M. avium. To determine if the absence of LSPA 17 might be a feature of code 3 strains, we tested five other isolates belonging to this sequevar and noted that these isolates also lacked LSPA 17. These data suggest that the absence of LSPA 17 is a shared feature of code 3 strains and bird strains and is therefore sensitive for M. avium subsp. avium but not perfectly specific.
DISCUSSION
M. avium organisms present many phenotypic differences between and within subspecies, leading to concerted efforts to understand their genomic diversity. In this work, we uncovered large regions of genomic differences between phenotypically and genetically distinct subsets of M. avium subsp. paratuberculosis and nonparatuberculosis M. avium isolates. The recognition of these sequence differences facilitates their use in determining which M. avium subsets are associated with human and veterinary diseases. As has been done with polymorphisms of the M. tuberculosis complex (3, 15), we have adopted a three-primer PCR strategy that is practical and immediately applicable in diagnostic and reference laboratories (17, 26).
M. avium subsp. avium strains (generally associated with severe disease in birds) consistently lack a large sequence, LSPA 17, although the absence of this region was not strictly restricted to this cluster. Isolates previously designated as M. avium subsp. silvaticum also lacked this sequence and could not specifically be identified with a particular genomic profile. This is in agreement with our recent typing scheme for M. avium isolates, in which bird-type M. avium and M. avium subsp. silvaticum could not be distinguished (28). In that hsp65-based study, we noted that some IS901-negative strains associated with disseminated disease in AIDS patients had a closely related sequevar which differed from that of bird-type M. avium by just one SNP. Both these groups, termed hsp65 code 3 and code 4, lacked LSPA 17, suggesting that this LSP event preceded the SNP that distinguishes these two lineages.
Our findings also show that S and C strains of M. avium subsp. paratuberculosis have undergone distinct evolutionary paths. One sequence, LSPA 20, appears to represent a genomic deletion specific to S strains of M. avium subsp. paratuberculosis. Conversely, two other sequences, LSPA 18 and LSPA 4-II, were absent from all C strains of M. avium subsp. paratuberculosis tested but present in S strains, likely indicating deletions characteristic of the C lineage of M. avium subsp. paratuberculosis. Because these represent complex genetic events, the genomic evidence points to S strains as being closer to the M. avium subsp. paratuberculosis ancestor and C strains having a more derivative status (Fig. 1). Our data for the regions missing from C strains of M. avium subsp. paratuberculosis are in agreement with and expand upon findings from a representational difference analysis-based study that identified three loci missing from type II (C type) strains (9). The 233-bp locus they identified as pig-RDA10 (AY266300) forms part of the 16-kb region we called LSPA 18. The 197-bp locus pig-RDA20 (AY266301) is located within the 26-kb segment we called LSPA 4-II. While the third locus they describe (AY266302) also forms part of a larger segment, microarray-based comparisons indicated that this segment was also variably missing from nonparatuberculosis M. avium isolates (unpublished observations); therefore, the loss of this region does not appear to be specifically associated with M. avium subsp. paratuberculosis.
In this work we focused only on regions consistently associated with distinct M. avium subgroups, as these would have the greatest applicability for diagnostic laboratories. We have not confirmed genomic differences proposed by microarrays that distinguish among closely related M. avium organisms and, thus, it is possible that other large sequences are missing from selected groups of strains. For instance, in previous work we identified an LSP (called LSP 11) which was missing in the S strain tested by microarray (23). Further analysis revealed that this region was missing from only a subset of S strains, indicating that even within subgroups or clusters of strains significant genomic variability may exist and may be of value for molecular epidemiologic applications.
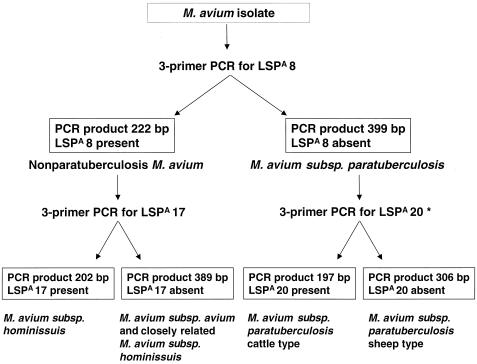
For any typing method, the optimal utility depends on the question being asked and the available technologies. In comparison to sequencing of the 3′ region of hps65 (28), this LSP-based PCR method is of lower resolution but greater simplicity (Fig. 3). Reassuringly, the two methods provide consistent results, in that lineages defined by hsp65 sequencing were branded by a shared LSP profile. This PCR-based method was not able to reliably distinguish the Mycobacterium intracellulare species; testing for the presence or absence of LSPA 8 on a small number of M. intracellulare strains gave ambiguous results (Fig. 2). The little sequence information that is currently available for this species and our hsp65 sequencing results provide evidence that the M. intracellulare species is clearly separate from the M. avium group and, further, that there is genetic diversity among the former. Therefore, testing using this LSP-based PCR method should be reserved for isolates determined to belong to the M. avium species by AccuProbe or alternate methods.
FIG. 3.
Diagnostic algorithm for PCR-based identification and typing of an M. avium isolate. *, testing for the presence or absence of LSPA 4-II or LSPA 18 are other alternatives for typing M. avium subsp. paratuberculosis isolates into cattle or sheep types.
Unlike hsp65-based sequencing, PCR for LSPs is restricted to testing for the currently described genomic variations. However, with the addition of reactions to test for newly described polymorphic regions, this modality will be readily applicable to testing for other variants of M. avium and can ultimately be packaged in the form of a deligotype platform (10). An advantage of PCR-based testing is the capacity to detect mixed infections (Fig. 2), a recognized concern with M. avium disease (24), in which case the sequencing of hsp65 may only return the result for the predominant clone. Finally, in settings where sequencing is not readily available or large volumes of isolates are to be screened, PCR with the primers we have described can provide an immediate gel-based indication of which M. avium variant is present and stimulate additional testing by other methods as indicated.
Acknowledgments
This work was supported by a grant from the Natural Science and Engineering Research Council (grant number GEN2282399). M.S. is funded by the Fonds de la Recherche en Sante du Quebec. M.B. is a New Investigator of the Canadian Institutes of Health Research.
None of the authors have a conflict of interest or any commercial association that may pose a conflict of interest.
We thank Fiona McIntosh for her assistance with microarray experiments and Murray E. Hines III for providing DNA for “strain 18,” formerly identified as “MAP 18.”
REFERENCES
- 1.Autschbach, F., S. Eisold, U. Hinz, S. Zinser, M. Linnebacher, T. Giese, T. Loffler, M. W. Buchler, and J. Schmidt. 2005. High prevalence of Mycobacterium avium subspecies paratuberculosis IS900 DNA in gut tissues from individuals with Crohn's disease. Gut 54:944-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Behr, M. A., M. Semret, A. Poon, and E. Schurr. 2004. Crohn's disease, mycobacteria, and NOD2. Lancet Infect. Dis. 4:136-137. [DOI] [PubMed] [Google Scholar]
- 3.Behr, M. A., M. A. Wilson, W. P. Gill, H. Salamon, G. K. Schoolnik, S. Rane, and P. M. Small. 1999. Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520-1523. [DOI] [PubMed] [Google Scholar]
- 4.Bull, T. J., E. J. McMinn, K. Sidi-Boumedine, A. Skull, D. Durkin, P. Neild, G. Rhodes, R. Pickup, and J. Hermon-Taylor. 2003. Detection and verification of Mycobacterium avium subsp. paratuberculosis in fresh ileocolonic mucosal biopsy specimens from individuals with and without Crohn's disease. J. Clin. Microbiol. 41:2915-2923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chacon, O., L. E. Bermudez, and R. G. Barletta. 2004. Johne's disease, inflammatory bowel disease, and Mycobacterium paratuberculosis. Annu. Rev. Microbiol. 58:329-363. [DOI] [PubMed] [Google Scholar]
- 6.Collins, D. M., S. Cavaignac, and G. W. de Lisle. 1997. Use of four DNA insertion sequences to characterize strains of the Mycobacterium avium complex isolated from animals. Mol. Cell Probes. 11:373-380. [DOI] [PubMed] [Google Scholar]
- 7.Collins, D. M., M. De Zoete, and S. M. Cavaignac. 2002. Mycobacterium avium subsp. paratuberculosis strains from cattle and sheep can be distinguished by a PCR test based on a novel DNA sequence difference. J. Clin. Microbiol. 40:4760-4762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Collins, D. M., D. M. Gabric, and G. W. de Lisle. 1990. Identification of two groups of Mycobacterium paratuberculosis strains by restriction endonuclease analysis and DNA hybridization. J. Clin. Microbiol. 28:1591-1596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dohmann, K., B. Strommenger, K. Stevenson, L. de Juan, J. Stratmann, V. Kapur, T. J. Bull, and G. F. Gerlach. 2003. Characterization of genetic differences between Mycobacterium avium subsp. paratuberculosis type I and type II isolates. J. Clin. Microbiol. 41:5215-5223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goguet de la Salmoniere, Y. O., C. C. Kim, A. G. Tsolaki, A. S. Pym, M. S. Siegrist, and P. M. Small. 2004. High-throughput method for detecting genomic-deletion polymorphisms. J. Clin. Microbiol. 42:2913-2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Green, E. P., M. L. Tizard, M. T. Moss, J. Thompson, D. J. Winterbourne, J. J. McFadden, and J. Hermon-Taylor. 1989. Sequence and characteristics of IS900, an insertion element identified in a human Crohn's disease isolate of Mycobacterium paratuberculosis. Nucleic Acids Res. 17:9063-9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guerrero, C., C. Bernasconi, D. Burki, T. Bodmer, and A. Telenti. 1995. A novel insertion element from Mycobacterium avium, IS1245, is a specific target for analysis of strain relatedness. J. Clin. Microbiol. 33:304-307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Harris, N. B., and R. G. Barletta. 2001. Mycobacterium avium subsp. paratuberculosis in veterinary medicine. Clin. Microbiol. Rev. 14:489-512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johansen, T. B., B. Djonne, M. R. Jensen, and I. Olsen. 2005. Distribution of IS1311 and IS1245 in Mycobacterium avium subspecies revisited. J. Clin. Microbiol. 43:2500-2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mahairas, G. G., P. J. Sabo, M. J. Hickey, D. C. Singh, and C. K. Stover. 1996. Molecular analysis of genetic differences between Mycobacterium bovis BCG and virulent M. bovis. J. Bacteriol. 178:1274-1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mijs, W., P. de Haas, R. Rossau, L. T. Van der, L. Rigouts, F. Portaels, and D. van Soolingen. 2002. Molecular evidence to support a proposal to reserve the designation Mycobacterium avium subsp. avium for bird-type isolates and “M. avium subsp. hominissuis” for the human/porcine type of M. avium. Int. J. Syst. Evol. Microbiol. 52:1505-1518. [DOI] [PubMed] [Google Scholar]
- 17.Parsons, L. M., R. Brosch, S. T. Cole, A. Somoskovi, A. Loder, G. Bretzel, D. van Soolingen, Y. M. Hale, and M. Salfinger. 2002. Rapid and simple approach for identification of Mycobacterium tuberculosis complex isolates by PCR-based genomic deletion analysis. J. Clin. Microbiol. 40:2339-2345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paustian, M. L., V. Kapur, and J. P. Bannantine. 2005. Comparative genomic hybridizations reveal genetic regions within the Mycobacterium avium complex that are divergent from Mycobacterium avium subsp. paratuberculosis isolates. J. Bacteriol. 187:2406-2415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pavlik, I., P. Svastova, J. Bartl, L. Dvorska, and I. Rychlik. 2000. Relationship between IS901 in the Mycobacterium avium complex strains isolated from birds, animals, humans, and the environment and virulence for poultry. Clin. Diagn. Lab. Immunol. 7:212-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saxegaard, F., and I. Baess. 1988. Relationship between Mycobacterium avium, Mycobacterium paratuberculosis and “wood pigeon mycobacteria.” Determinations by DNA-DNA hybridization. APMIS 96:37-42. [PubMed] [Google Scholar]
- 21.Sechi, L. A., A. M. Scanu, P. Molicotti, S. Cannas, M. Mura, G. Dettori, G. Fadda, and S. Zanetti. 2005. Detection and isolation of Mycobacterium avium subspecies paratuberculosis from intestinal mucosal biopsies of patients with and without Crohn's disease in Sardinia. Am. J. Gastroenterol. 100:1529-1536. [DOI] [PubMed] [Google Scholar]
- 22.Semret, M., D. C. Alexander, C. Y. Turenne, P. de Haas, P. Overduin, D. van Soolingen, D. Cousins, and M. A. Behr. 2005. Genomic polymorphisms for Mycobacterium avium subsp. paratuberculosis diagnostics. J. Clin. Microbiol. 43:3704-3712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Semret, M., G. Zhai, S. Mostowy, C. Cleto, D. Alexander, G. Cangelosi, D. Cousins, D. M. Collins, D. van Soolingen, and M. A. Behr. 2004. Extensive genomic polymorphism within Mycobacterium avium. J. Bacteriol. 186:6332-6334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Slutsky, A. M., R. D. Arbeit, T. W. Barber, J. Rich, C. F. von Reyn, W. Pieciak, M. A. Barlow, and J. N. Maslow. 1994. Polyclonal infections due to Mycobacterium avium complex in patients with AIDS detected by pulsed-field gel electrophoresis of sequential clinical isolates. J. Clin. Microbiol. 32:1773-1778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stevenson, K., V. M. Hughes, L. de Juan, N. F. Inglis, F. Wright, and J. M. Sharp. 2002. Molecular characterization of pigmented and nonpigmented isolates of Mycobacterium avium subsp. paratuberculosis. J. Clin. Microbiol. 40:1798-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Talbot, E. A., D. L. Williams, and R. Frothingham. 1997. PCR identification of Mycobacterium bovis BCG. J. Clin. Microbiol. 35:566-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Thorel, M. F., M. Krichevsky, and V. V. Levy-Frebault. 1990. Numerical taxonomy of mycobactin-dependent mycobacteria, emended description of Mycobacterium avium, and description of Mycobacterium avium subsp. avium subsp. nov., Mycobacterium avium subsp. paratuberculosis subsp. nov., and Mycobacterium avium subsp. silvaticum subsp. nov. Int. J. Syst. Bacteriol. 40:254-260. [DOI] [PubMed] [Google Scholar]
- 28.Turenne, C. Y., M. Semret, D. Cousins, D. M. Collins, and M. A. Behr. 2006. Sequencing of hsp65 distinguishes among subsets of the Mycobacterium avium complex. J. Clin. Microbiol. 44:433-440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van der Giessen, J. W., A. Eger, J. Haagsma, and B. A. van der Zeijst. 1993. Rapid detection and identification of Mycobacterium avium by amplification of 16S rRNA sequences. J. Clin. Microbiol. 31:2509-2512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.van Soolingen, D., J. Bauer, V. Ritacco, S. C. Leao, I. Pavlik, V. Vincent, N. Rastogi, A. Gori, T. Bodmer, C. Garzelli, and M. J. Garcia. 1998. IS1245 restriction fragment length polymorphism typing of Mycobacterium avium isolates: proposal for standardization. J. Clin. Microbiol. 36:3051-3054. [DOI] [PMC free article] [PubMed] [Google Scholar]