Abstract
To substantiate a common genetic background of ciprofloxacin-resistant Enterococcus faecium, 32 ciprofloxacin-resistant (Cipr) and 31 ciprofloxacin-susceptible (Cips) isolates from outbreaks, clinical infections, surveillances, and animals from 10 different countries were genotyped by multilocus sequence typing. Additionally, susceptibilities to ampicillin and vancomycin and the presence of esp were determined and the quinolone resistance-determining regions of parC, gyrA, parB, and gyrE were sequenced. High-level Cipr (MIC ≥ 64 μg/ml) due to point mutations in the quinolone resistance-determining region was unique to a distinct hospital-adapted genetic complex in E. faecium, previously designated CC17. Low-level Cipr (MIC = 4 μg/ml) in non-CC17 strains is not attributable to point mutations in any subunit of the topoisomerase genes, and the mechanism of resistance remains unclear. Acquisition of mutations in parC and gyrA, leading to high-level Cipr, is, in addition to ampicillin resistance and the presence of a putative pathogenicity island, another cumulative step in hospital adaptation of CC17.
Over the last two decades, enterococci have become increasingly important as nosocomial pathogens (3). These organisms are intrinsically resistant to a large number of antimicrobials and have the ability to easily acquire new resistance traits (40, 50). The emergence of nosocomial infections caused by ampicillin-, high-level aminoglycoside-, and glycopeptide-resistant Enterococcus faecium has caused clinical concern due to intra- and interhospital spread and limited therapeutic options (40, 50). Glycopeptide-resistant enterococci (vancomycin-resistant enterococci [VRE]) are nowadays endemic to the United States, with ∼30% of enterococcal infections caused by VRE (38, 43); in Europe, the epidemiology of VRE is now changing from a near absence of VRE in hospital-acquired infections at the turn of the century to a situation in which nosocomial epidemics and infections are increasingly reported (3, 13, 17). We recently described a hospital-adapted genetic subtype of E. faecium associated with epidemics and clinical infections, which has spread globally (20, 32, 62, 65). This subpopulation belongs to a distinct genetic lineage labeled clonal complex 17 (CC17) (62) and is associated with the presence of the variant esp gene as part of a pathogenicity island (31) and resistance to ampicillin. Recently, resistance to ciprofloxacin appeared to be associated with ampicillin resistance in genotypically related E. faecium isolates from Norway (25, 55, 56) and Spain (7).
To substantiate a common genetic background of quinolone-resistant E. faecium and association with the hospital-adapted CC17, we studied the genetic relatedness of 32 ciprofloxacin-resistant and 31 ciprofloxacin-susceptible E. faecium isolates from various human and animal origins by multilocus sequence typing (MLST) and determined susceptibility to vancomycin and ampicillin and the presence of esp. Finally, we sequenced the quinolone resistance-determining regions (QRDR) of parC and gyrA to identify mutations involved in ciprofloxacin resistance.
(This study was presented in part at the 44th Interscience Conference on Antimicrobial Agents and Chemotherapy, Washington, D.C., November 2004.)
MATERIALS AND METHODS
Bacterial isolates and growth conditions.
Sixty-three isolates of E. faecium (32 ciprofloxacin resistant and 31 ciprofloxacin susceptible) were collected from 10 different countries from nosocomial epidemics (n = 13; United Kingdom, n = 1; The Netherlands, n = 5; United States, n = 7), clinical infections (n = 27; Austria, n = 2; Germany, n = 4; Spain, n = 3; France, n = 2; United Kingdom, n = 2; Israel, n = 1; Italy, n = 3; The Netherlands, n = 6; Portugal, n = 3; United States, n = 7) (e.g., from blood, urine, or wounds), surveillance for colonization among hospitalized patients and in the community (France, n = 3; The Netherlands, n = 8) (all fecal samples), and animals (The Netherlands, n = 12) (2, 4, 10, 24, 32, 54, 65). Strains were considered epidemic as defined before (65). Epidemic isolates were recovered from clinical sites, blood, and urine, as well as from feces. Only one representative isolate from each outbreak was used for analysis. Bacterial strains were grown according to standard growth conditions on tryptic soy agar sheep blood plates and Todd Hewitt broth. DNA was extracted according to previous defined methods (64).
Identification and susceptibility testing.
Enterococcal species were confirmed by multiplex PCR, as described by Dutka-Malen et al. (11). Ciprofloxacin, vancomycin, and ampicillin susceptibilities were determined by standard agar dilution methods, according to the CLSI (formerly NCCLS) guidelines (42), and according to CLSI guidelines, MICs of ≥4 μg/ml of ciprofloxacin, ≥16 μg/ml of ampicillin, and ≥8 μg/ml of vancomycin were considered resistant. High-level ciprofloxacin resistance was defined in this study by a MIC of ≥64 μg/ml, low-level ciprofloxacin resistance was defined in this study by a MIC of 4 μg/ml.
esp PCR.
Seventy strains were screened for esp by PCR, with two different primer sets (esp 11 [5′-TTGCTAATGCTAGTCCACGACC-3′] and esp 12 [5′-GCG TCAA CAC TTG CAT TGC CGA A-3′] and the combination 14F [5′-AGA TTT CAT CTT TGA TTC TTG G-3′] and 12R [5′-AAT TGA TTC TTT AGC ATC TGG-3′]). PCR conditions included an initial denaturation at 95°C for 15 min for activation of the HotStarTaq DNA polymerase (QIAGEN GmbH, Hilden, Germany), followed by 30 cycles of 94°C for 30 s, 52°C for 30 s, and 72°C for 1 min, followed by an extension at 72°C for 7 min. Reactions were performed in 25-μl volumes by using the HotStarTaq master mix (QIAGEN GmbH). Strains negative by PCR were checked for the presence of the esp gene by Southern hybridization, as described previously (65). For this check, we generated an esp-specific probe (956 bp) using primers esp 11 and esp 12 (see previous explanation).
Sequencing the QRDRs of parC, gyrA, parE, and gyrB.
The QRDRs of the E. faecium parC and gyrA genes were amplified and sequenced (12). The primers used were previously designed by el Amin et al. (12) for gyrA (gyrA-1F [5′-CGG GAT GAA CGA ATT GGG TGT GA-3′] and gyrA-1R [5′-AAT TTT ACT CAT ACG TGC TTC GG-3′]) and Brisse et al. (5) for parC (parC-A [5′-TTC CCG TGC ATT TCG ATC AGT ACT TC-3′] and parC-C [5′-CGT ATG ACA AAG GAT TCG GTA AAT C-3′]). The QRDRs of parE and gyrB were determined by aligning the Escherichia coli ParE and GyrB QRDR sequence to the E. faecium DO genome in GenBank. With forward and reverse primers (parE-F [5′-GTC CGT AAA GCA ATC AAA G-3′] and parE-R [5′-CTT TAT ATA AAG GCG GTA ACG-3′], gyrB-F [5′-TGA AAT TCT TGC TGG AAA A C-3′] and gyrB-R [5′-CAA CAA TAG GAC GCA TGT AAC-3′]), the parE and gyrB genes of 44/63 and 63/63 isolates, respectively, were amplified. All PCR conditions were similar to those for esp (see above). PCR products were purified with a PCR purification kit (QIAGEN, Hilden, Germany) and sequenced with PCR forward and reverse primers, an ABI PRISM Big Dye cycle sequencing ready reaction kit (Perkin-Elmer, Applied Biosystems, Foster City, Calif.), and an ABI PRISM 3700 DNA sequencer (Perkin-Elmer). Alignments of amino acids were made with Megalign software (DNAstar). Sequences with no mutations were defined as being identical to the reference partial EMBL sequence (gyrA, accession no. AF060881; parC, accession no. AB017811) or the E. faecium DO sequence determined by E. coli QRDR sequence alignment (gyrB and parE).
MLST.
MLST and computer analysis of allelic profiles of E. faecium isolates to determine the genetic relatedness of isolates was performed as described previously (20) with the use of updated primer sequences (available at http://efaecium.mlst.net/misc/info.asp). Different sequences of a given locus were assigned allele numbers, and different allelic profiles were assigned sequence types (STs) by interrogating the E. faecium MLST database, which is available on the MLST website (http://www.mlst.net). Cluster analysis of allelic profiles was performed by using a categorical coefficient and a graphic method called a minimum spanning tree with Bionumerics software (version 4.0; Applied Maths, Sint-Martens-Latem, Belgium), as previously described by Schouls et al. (49). The criteria to first link types that have the highest number of single-locus variants, taken from the eBURST criteria (14), were chosen. The stringent criterion of a maximum one-allele difference was used to create complexes.
RESULTS
Genotyping and population modeling.
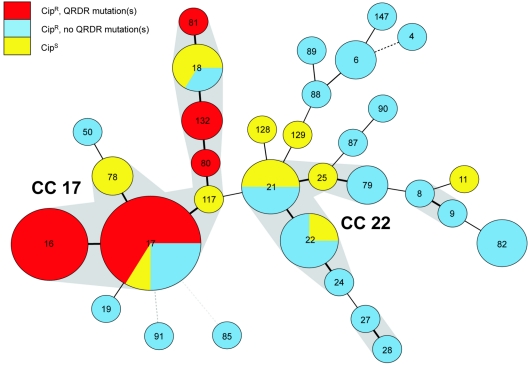
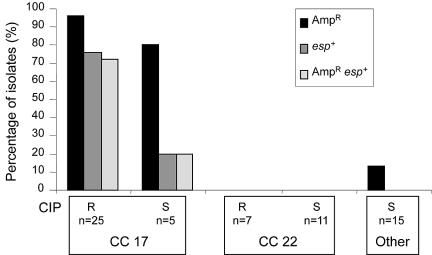
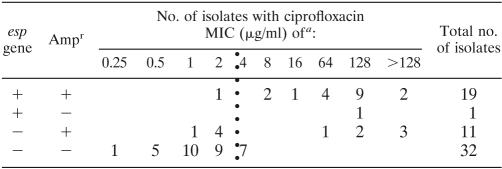
Genotyping of the 63 E. faecium isolates by MLST revealed 32 different STs, of which 10 STs (18 isolates) were found exclusively among Cipr strains and 18 STs (22 isolates) were found only among Cips strains. Only 4 STs (23 isolates) were found among both Cipr and Cips strains, strongly suggesting that the majority of Cipr and Cips strains are genetically unrelated. A minimum spanning tree based on allelic profiles, using the most stringent definition of complexes of ≥6 identical alleles, resolved 2 dominant clonal complexes (CC), CC17 and CC22, 2 minor complexes, and 11 singletons (Fig. 1). Almost all isolates (28/30, 93%) belonging to the previously described hospital-adapted CC17 were Cipr. In the other dominant complex, CC22 (n = 18), 7 isolates (39%) were Cipr, whereas all isolates belonging to minor complexes and singletons, defined in this study as “other genotype” (n = 15), were Cips (Table 1). According to the source of isolation, all epidemic isolates (100%), 61% of clinical infectious isolates, 17% of animal isolates, 9% of clinical surveillance isolates, and none of the community isolates were Cipr (data not shown). Ampicillin resistance (Ampr) was almost exclusively confined (28/30 isolates, 93%) to CC17, and 20 CC17 isolates (67%) carried the variant esp gene. Vancomycin resistance (Vanr), though, was distributed among all major and minor complexes (Table 1; Fig. 2). Within CC17, Cipr was associated with the presence of esp and Ampr; 56% of Cipr isolates were esp-positive Ampr, compared to 3% of Cips isolates (chi-square test, P < 0.01) (Table 1). Ampr was correlated to high-level Cipr (chi-square test, P < 0.01) and with a concomitant presence of esp (chi-square test, P < 0.01) (Table 2).
FIG. 1.
Population structure of 63 Cipr and Cips E. faecium isolates in CC. Clusters of STs are displayed in a minimum spanning tree. Each circle in the tree represents a different ST, and the ST is indicated by the number in the circle. One ST can contain multiple strains; circle size corresponds to the number of strains. Two CCs are identified (17 and 22) as well as 2 minor complexes and a couple of singletons. The CC and minor complexes are surrounded by gray shading. Heavy lines connecting two STs denote STs differing at a single locus, thin lines connect double-locus variants, and dotted lines connect STs differing at more than 2 loci. The colors indicate susceptibility to ciprofloxacin. If different resistance types occur simultaneously in one ST, pie charts are used to indicate distribution.
TABLE 1.
Origin, percentage of isolates resistant to shown antibiotics and positive for esp, and detailed Cipr range among 63 E. faecium isolates of CC17, CC22 and other genotypes
| CC (n) | Sourcea
|
% of isolates
|
Ciprofloxacin MIC (μg/ml)
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Out | Inf | Surv | Anim | Vanr | Ampr | esp+ | Cipr | MIC50 | MIC90 | Range | |
| 17 (30) | 13 | 15 | 2 | 0 | 77 | 93 | 67 | 83 | 128 | 256 | 1-256 |
| 22 (18) | 0 | 6 | 6 | 6 | 83 | 0 | 0 | 39 | 2 | 4 | 0.5-4 |
| Other (15) | 0 | 6 | 3 | 6 | 93 | 13 | 0 | 0 | 1 | 2 | 0.25-2 |
Out, outbreak; Inf, infectious; Surv, surveillance; Anim, animal.
FIG. 2.
Cipr in association with the presence and absence of esp and Ampr among E. faecium isolates (n = 63) of CC17, CC22, and others (genotypes not belonging to CC17 or CC22). Percentages of Ampr, presence of the esp gene, and Ampr in combination with the esp gene are shown by bars for Cipr and Cips isolates in CC17, Cipr and Cips isolates in CC22, and Cips isolates in other genotypes (no Cipr isolates found). For Cipr and Cips in each CC, the number of isolates is given. R, resistant, S, susceptible.
TABLE 2.
MIC of ciprofloxacin in association with presence and absence of the esp gene and ampicillin resistance
Vertical dotted line divides resistant and nonresistant E. faecium isolates.
Sequence analysis of the QRDRs of parC, gyrA, parE, and gyrB.
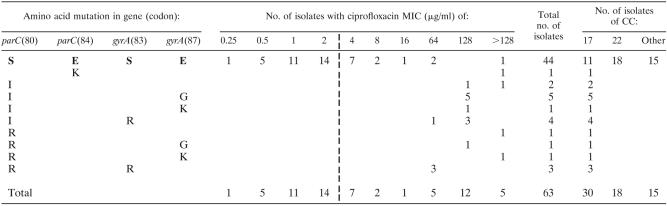
Ten different single and double amino acid changes were detected in ParC and GyrA of 19 of 32 Cipr isolates of E. faecium (Table 3). No amino acid changes in ParC or GyrA were found in the remaining 13 Cipr isolates or in the 31 Cips isolates. Nineteen isolates had mutations in parC, leading to an amino acid change at codon 80 (Ser to Ile [n = 12] or Ser to Arg [n = 6]) or codon 84 (Glu to Lys [n = 1]). In 15 of the 19 isolates with a mutation in parC, a secondary mutation was found in gyrA leading to an amino acid change at codon 83 (Ser to Arg [n = 7]) or codon 87 (Glu to Gly [n = 6] or Glu to Lys [n = 2]). In conclusion, among Cipr isolates, 4 contained amino acid changes only in ParC and 15 had mutations in both parC codon 80 and gyrA. Subsequently, the QRDRs of the gyrB and parE genes were sequenced of 11 Cipr isolates that did not contain mutations in gyrA and parC, 10 Cipr isolates containing QRDR mutations, and 23 Cips isolates. No amino acid substitutions were found in the topoisomerases (data not shown).
TABLE 3.
parC and gyrA mutations in 63 CIPr and CIPs E. faecium isolates corresponding to ciprofloxacin MIC and CCa
S, serine; E, glutamic acid; R, arginine; K, lysine; I, isoleucine; G, glutamine. Boldface type indicates wild-type amino acids; mutations are shown below. Dashed vertical line divides resistant and nonresistant E. faecium isolates.
Isolates with amino acid changes in ParC and GyrA had higher MICs (MIC50 = 128 μg/ml; MIC90 > 128) than isolates without mutations (MIC50 = 2 μg/ml; MIC90 = 8) (Student's t test, P < 0.01). No correlation was found between the number of substitutions in ParC and GyrA and the level of ciprofloxacin resistance (Table 3). Although some silent mutations were identified in ParE and GyrB, these were not associated with higher MICs (data not shown). Mutations in parC and gyrA associated with high-level Cipr were only found in isolates confined to CC17.
DISCUSSION
High-level ciprofloxacin resistance (MIC ≥ 64 μg/ml) in E. faecium is associated with amino acid changes in topoisomerase IV and DNA gyrase and confined to a single clonal complex, previously identified as a global hospital-adapted subpopulation of E. faecium (CC17). CC17 is further characterized by ampicillin resistance and a pathogenicity island, including the variant esp gene (20, 31, 32, 65) (Fig. 1). Cipr isolates belonging to other genetic backgrounds are only low-level resistant (MIC = 4 μg/ml) and do not carry these point mutations. These findings further disclose the cumulative adaptive mechanisms that have resulted in the evolution of a hospital-adapted genetic complex in E. faecium.
We have previously described the global spread of CC17 E. faecium, which is characterized by ampicillin resistance, although pandemic spread of VRE was instrumental for its identification. The strong association with quinolone resistance strongly suggests that this clonal complex has previously emerged as ampicillin-resistant but vancomycin-sensitive in Scandinavian countries. Torell et al. (55) described a clonal subset of Swedish vancomycin-susceptible Ampr E. faecium strains with high-level ciprofloxacin resistance and mutations in the QRDR genotyped by biochemical fingerprinting (PhenePlate). In addition, Jureen et al. (25), showed a correlation between ampicillin and ciprofloxacin resistance among genetically highly similar Norwegian isolates that also carried the purK-1 allele, indicative for CC17. More specifically, Mohn et al. (38) identified fluoroquinolone prescription as a risk factor for fecal carriage with an outbreak of ampicillin-resistant E. faecium in a Norwegian hospital. However, Fortún et al. (15) found that Spanish patients with bacteremia from ampicillin-resistant E. faecium were only likely to have received quinolones, but there is no significant association. van der Steen et al. identified previously administered ciprofloxacin as a risk factor for colonization with an outbreak-associated, vancomycin-resistant, ampicillin-resistant E. faecium isolate in a Dutch hospital (57). A mouse model nevertheless showed that ciprofloxacin administration does not promote high-level vancomycin-resistant, ampicillin-resistant E. faecium colonization (9).
Linkage of ampicillin and quinolone resistance in CC17 illustrates that specific clones have different likelihoods of acquiring resistance traits, which depends on the availability of resistance genes in the local gene pool and intrinsic capacities of bacterial strains to acquire foreign DNA or to accumulate mutations. It is interesting, in this respect, that E. faecium isolates that harbored amino acid substitutions in the DNA mismatch repair proteins MutS and MutL belong to CC17 (61), which may suggest that CC17 isolates are more prone to undergo mutations in, e.g., parC or gyrA, conferring high-level resistance to quinolones. Clonal dissemination of quinolone resistance was also reported among methicillin-resistant Staphylococcus aureus, Neisseria gonorrhoeae, Salmonella enterica serotype Typhimurium strain DT 104, Streptococcus pneumoniae, and Yersinia enterocolitica (28, 46, 48).
In gram-negative bacteria, such as Escherichia coli, Neisseria gonorrhoeae, and Pseudomonas aeruginosa, gyrA is the primary target in mutation-mediated quinolone resistance. In these species, low-level ciprofloxacin resistance was reported in strains with mutations only in gyrA, while a higher level of resistance was discerned in strains carrying mutations in both gyrA and parC (1, 19, 30, 41). Conversely, in gram-positive bacteria, such as Staphylococcus aureus and Streptococcus pneumoniae (16, 52), parC appears to be the primary target, with mutations resulting in low-level resistance, which changes to high-level resistance with subsequent gyrA mutations. The latter phenomenon has also been proposed for Enterococcus faecalis and E. faecium from mutant analysis (5, 12, 27, 29, 53). Our finding that all ciprofloxacin-resistant isolates carried mutations in parC and only 15 isolates carried additional mutations in gyrA, confirms parC as the main target in mutation-mediated ciprofloxacin resistance in enterococci. In contrast, the only enzymatic study in E. faecalis showed that, for levofloxacin and ciprofloxacin, gyrase is the primary target, possibly explained by higher in vivo lethality of the quinolone-gyrase-DNA complex (44).
In various pathogens, including E. faecalis, a clear correlation was found between the number of mutations in gyrA and parC and the level of ciprofloxacin resistance (27). For E. faecium, Brisse et al. described higher MICs for isolates that acquired an amino acid change in gyrA in addition to an already existing mutation in parC, even though this was of only limited impact compared to S. aureus or S. pneumoniae and not consistently true for all double mutants (5). We could not confirm this correlation, since the mean MIC for isolates with only one mutation in parC was not significantly lower than that for isolates with two mutations in parC and gyrA. It is unclear whether additional mutations, without increased ciprofloxacin resistance, have other selective advantages. Selection pressure of mutants by quinolones other than ciprofloxacin could be attributable to the observed inconsistency, as already suggested for the accumulation of mutations in both subunits of both topoisomerases in levofloxacin-resistant S. pneumoniae (6).
Several other resistance mechanisms may be responsible for (low-level) ciprofloxacin resistance in CC22 and CC17 isolates that do not contain amino acid changes in the QRDRs of ParC and GyrA. In Staphylococcus aureus, Streptococcus pneumoniae, and E. coli, mutations in parE and/or gyrB have been associated with fluoroquinolone resistance. It is possible that mutations in these genes account for the low-level quinolone resistance as observed in this study, though Grohs et al. reported one high-level Cipr E. faecalis isolate (with substitutions in ParC and GyrA) with an additional ParE substitution in codon 453 (Pro to Ser) which did not affect the MIC of ciprofloxacin (and other quinolones) significantly (18). No amino acid substitutions in parE and gyrB genes were found in the E. faecium isolates in this study. Silent mutations in parE in some highly Cipr strains were identified, but this does not explain increased MICs and possibly reflects higher mutation frequencies in certain subtypes of E. faecium.
Resistance by active efflux of quinolones or increased expression of endogenous efflux pumps are alternative quinolone resistance mechanisms reported for S. pneumoniae (45), S. aureus (26), and E. faecalis (22, 33, 34). Davis et al. reported the presence of 34 potential multidrug resistance-encoding genes in E. faecalis by a bioinformatics approach (8). Three E. faecalis multidrug efflux pumps have been characterized, EmeA, EfrAB, and Lsa, of which only the first two confer resistance to quinolones (22, 33, 34, 51). Therefore, it seems highly likely that efflux pumps exist in E. faecium as well. So far, only active efflux of norfloxacin has been reported for E. faecium strain ATCC 19434 (35), but the efflux pump hasn't been identified yet. A novel quinolone resistance mechanism includes a Klebsiella pneumoniae multidrug resistance plasmid-mediated qnr gene that could be transferred horizontally (37) and has ever since been described in clinical isolates of K. pneumoniae in the United States (47, 58), E. coli, Citrobacter freundii, and Enterobacter sp. in Europe and China (23, 36, 59), and Providencia stuartii in Egypt (60). However, until now, the qnr locus has been described only in gram negative bacteria. Microarray hybridization failed to support the presence of a qnr probe from Klebsiella in 100 E. faecium isolates (different from the strains used in this study; data not shown). Different mechanisms leading to (low-level) ciprofloxacin resistance in E. faecium, therefore, remain to be identified.
Acknowledgments
This work was supported by a ZonMW MD clinical research trainee grant 2903322 from The Netherlands Organization for Health Research and Development.
We thank Emile Spalburg for technical assistance in determining MICs.
REFERENCES
- 1.Belland, R. J., S. G. Morrison, C. Ison, and W. H. Huang. 1994. Neisseria gonorrhoeae acquires mutations in analogous regions of gyrA and parC in fluoroquinolone-resistant isolates. Mol. Microbiol. 14:371-380. [DOI] [PubMed] [Google Scholar]
- 2.Bonten, M. J. M., M. K. Hayden, C. Nathan, T. W. Rice, and R. A. Weinstein. 1998. Stability of vancomycin-resistant enterococcal genotypes isolated from long-term-colonized patients. J. Infect. Dis. 177:378-382. [DOI] [PubMed] [Google Scholar]
- 3.Bonten, M. J. M., R. J. L. Willems, and R. A. Weinstein. 2001. Vancomycin-resistant enterococci: why are they here, and where do they come from? Lancet Infect. Dis. 1:314-325. [DOI] [PubMed] [Google Scholar]
- 4.Bonten, M. J. M, M. K. Hayden, C. Nathan, J. van Voorhis, M. Matushek, S. Slaughter, T. Rice, and R. A. Weinstein. 1996. Epidemiology of colonisation of patients and environment with vancomycin-resistant enterococci. Lancet 348:1615-1619. [DOI] [PubMed] [Google Scholar]
- 5.Brisse, S., A. C. Fluit, U. Wagner, P. Heisig, D. Milatovic, J. Verhoef, S. Scheuring, K. Kohrer, and F. J. Schmitz. 1999. Association of alterations in ParC and GyrA proteins with resistance of clinical isolates of Enterococcus faecium to nine different fluoroquinolones. Antimicrob. Agents Chemother. 43:2513-2516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Canton, R., M. Morosini, M. C. Enright, and I. Morrissey. 2003. Worldwide incidence, molecular epidemiology and mutations implicated in fluoroquinolone-resistant Streptococcus pneumoniae: data from the global PROTECT surveillance proGramme. J. Antimicrob. Chemother. 52:944-952. [DOI] [PubMed] [Google Scholar]
- 7.Coque, T. M., R. J. L. Willems, J. Fortún, J. Top, S. Diz, E. Loza, R. Cantón, and F. Baquero. 2005. Population structure of Enterococcus faecium causing bacteremia in Spain: setting the scene for a future increase of vancomycin-resistance? Antimicrob. Agents Chemother. 49:2673-2700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Davis, D. R., J. B. McAlpine, C. J. Pazoles, M. K. Talbot, E. A. Alder, C. White, B. M. Jonas, B. E. Murray, G. M. Weinstock, and B. L. Rogers. 2001. Enterococcus faecalis multi-drug resistance transporters: application for antibiotic discovery. J. Mol. Microbiol. Biotechnol. 3:179-184. [PubMed] [Google Scholar]
- 9.Donskey, C. J., J. A. Hanrahan, R. A. Hutton, and L. B. Rice. 2000. Effect of parenteral antibiotic administration on establishment of colonization with vancomycin-resistant Enterococcus faecium in the mouse gastrointestinal tract. J. Infect. Dis. 181:1830-1833. [DOI] [PubMed] [Google Scholar]
- 10.Dunne, W. M., and W. Wang. 1997. Clonal dissemination and colony morphotype variation of vancomycin-resistant Enterococcus faecium isolates in metropolitan Detroit, Michigan. J. Clin. Microbiol. 35:388-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dutka-Malen, S., S. Evers, and P. Courvalin. 1995. Detection of glycopeptide resistance genotypes and identification to the species level of clinically relevant enterococci by PCR. J. Clin. Microbiol. 33:24-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.el Amin, N., S. Jalal, and B. Wretlind. 1999. Alterations in GyrA and ParC associated with fluoroquinolone resistance in Enterococcus faecium. Antimicrob. Agents Chemother. 43:947-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.European Antimicrobial Resistance Surveillance System (EARSS). 28 January 2005. EARSS annual report 2003. [Online.] http://www.earss.rivm.nl.
- 14.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fortún, J., T. M. Coque, P. Martín-Dávila, L. Moreno, R. Cantón, E. Loza, F. Baquero, and S. Moreno. 2002. Risk factors associated with ampicillin resistance in patients with bacteriaemia caused by Enterococcus faecium. J. Antimicrob. Chemother. 50:1003-1009. [DOI] [PubMed] [Google Scholar]
- 16.Fukuda, H., and K. Hiramatsu. 1999. Primary target of fluoroquinolones in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 43:410-412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goossens, H., D. Jabes, R. Rossi, C. Lammens, G. Privitera, and P. Courvalin. 2003. European survey of vancomycin-resistant enterococci in at-risk hospital wards and in-vitro susceptibility testing of ramoplanin against these isolates. J. Antimicrob. Chemother. 51(Suppl. 3):iii5-12. [DOI] [PubMed] [Google Scholar]
- 18.Grohs, P., S. Houssaye, A. Aubert, L. Gutmann, and E. Varon. 2003. In vitro activities of garenoxacin (BMS-284756) against Streptococcus pneumoniae, viridans group streptococci, and Enterococcus faecalis compared to those of six other quinolones. Antimicrob. Agents Chemother. 47:3542-3547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heisig, P., H. Schedletzky, and H. Falkenstein-Paul. 1993. Mutations in the gyrA gene of a highly fluoroquinolone-resistant clinical isolate of Escherichia coli. Antimicrob. Agents Chemother. 37:696-701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Homan, W. L., D. Tribe, S. Poznanski, M. Li, G. Hogg, E. Spalburg, J. D. van Embden, and R. J. Willems. 2002. Multilocus sequence typing scheme for Enterococcus faecium. J. Clin. Microbiol. 40:1963-1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reference deleted.
- 22.Jonas, B. M., B. E. Murray, and G. M. Weinstock. 2001. Characterization of emeA, a norA homolog and multidrug resistance efflux pump in Enterococcus faecalis. Antimicrob. Agents Chemother. 45:3574-3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jonas, D., K. Biehler, D. Hartung, B. Spitzmuller, and F. D. Daschner. 2005. Plasmid-mediated quinolone resistance in isolates obtained in German intensive care units. Antimicrob. Agents Chemother. 49:773-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jordens, J. Z., J. Bates, and D. T. Griffiths. 1994. Faecal carriage and nosocomial spread of vancomycin-resistant Enterococcus faecium. J. Antimicrob. Chemother. 34:515-528. [DOI] [PubMed] [Google Scholar]
- 25.Jureen, R., J. Top, S. C. Mohn, S. Harthug, N. Langeland, and R. J. L. Willems. 2003. Molecular characterization of ampicillin-resistant Enterococcus faecium isolates from hospitalized patients in Norway. J. Clin. Microbiol. 41:2330-2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaatz, G. W., and S. M. Seo. 1995. Inducible NorA-mediated multidrug resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 39:2650-2655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kanematsu, E., T. Deguchi, M. Yasuda, T. Kawamura, Y. Nishino, and Y. Kawada. 1998. Alterations in the GyrA subunit of DNA gyrase and the ParC subunit of DNA topoisomerase IV associated with quinolone resistance in Enterococcus faecalis. Antimicrob. Agents Chemother. 42:433-435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Klugman, K. P. 2003. The role of clonality in the global spread of fluoroquinolone-resistant bacteria. Clin. Infect. Dis. 36:783-785. [DOI] [PubMed] [Google Scholar]
- 29.Korten, V., W. M. Huang, and B. Murray. 1994. Analysis by PCR and direct DNA sequencing of gyrA mutations associated with fluoroquinolone resistance in Enterococcus faecalis. Antimicrob. Agents Chemother. 38:2091-2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumagai, Y., J. Kato, K. Hoshino, T. Akasaka, K. Sato, and H. Ikeda. 1996. Quinolone-resistant mutants of Escherichia coli DNA topoisomerase IV parC gene. Antimicrob. Agents Chemother. 40:710-714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Leavis, H., J. Top, K. Borgen, M. Bonten, J. van Embden, and R. J. L. Willems. 2004. A novel enterococcal putative pathogenicity island linked to the esp virulence gene of Enterococcus faecium associated with epidemicity. J. Bacteriol. 186:672-682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leavis, H. L., R. J. L. Willems, J. Top, E. Spalburg, E. M. Mascini, A. C. Fluit, A. Hoepelman, A. J. de Neeling, and M. J. Bonten. 2003. Epidemic and nonepidemic multidrug-resistant Enterococcus faecium. Emerg. Infect. Dis. 9:1109-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, E.-W., M. N. Huda, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. Functional cloning and expression of emeA, and characterization of EmeA, a multidrug efflux pump from Enterococcus faecalis. Biol. Pharm. Bull. 26:266-270. [DOI] [PubMed] [Google Scholar]
- 34.Lee, E.-W., M. N. Huda, T. Kuroda, T. Mizushima, and T. Tsuchiya. 2003. EfrAB, an ABC multidrug efflux pump in Enterococcus faecalis. Antimicrob. Agents Chemother. 47:3733-3738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lynch, C., P. Courvalin, and N. Hiroshi. 1997. Active efflux of antimicrobial agents in wild-type strains of enterococci. Antomicrob. Agents Chemother. 41:869-871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mammeri, H., M. van de Loo, L. Poirel, L. Martinez-Martinez, and P. Nordmann. 2005. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother. 49:71-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martínez-Martínez, L., A. Pascual, and G. A. Jacoby. 1998. Quinolone resistance from a transferable plasmid. Lancet 351:797-799. [DOI] [PubMed] [Google Scholar]
- 38.Mohn, S. C., S. Harthug, and N. Langeland. 2000. Outbreak of ampicillin-resistant Enterococcus faecium—risk factor for faecal colonization. APMIS 108:296-302. [DOI] [PubMed] [Google Scholar]
- 39.Reference deleted.
- 40.Murray, B. E. 2000. Vancomycin-resistant enterococcal infections. N. Engl. J. Med. 342:710-721. [DOI] [PubMed] [Google Scholar]
- 41.Nakano, M., T. Deguchi, T. Kawamura, M. Yasuda, M. Minura, Y. Okano, and Y. Kawada. 1997. Mutations in the gyrA and parC genes in fluoroquinolone-resistant clinical isolates of Pseudomonas aeruginosa. Antimicrob. Agents Chemother. 41:2289-2291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.National Committee for Clinical Laboratory Standards. 2004. Methods for dilution antimicrobial susceptibility tests for bacteria that grow aerobically. Approved standards M7-A6 and M100S14. NCCLS, Wayne, Pa.
- 43.National Nosocomial Infections Surveillance System. 2000. National Nosocomial Infections Surveillance (NNIS) System report data summary from January 1992-April 2000, issued June 2000. Am. J. Infect. Control 28:429-448. [DOI] [PubMed] [Google Scholar]
- 44.Onodera, Y., J. Okuda, M. Tanaka, and K. Sato. 2002. Inhibitory activities of quinolones against DNA gyrase and topoisomerase IV of Enterococcus faecalis. Antimicrob. Agents Chemother. 46:1800-1804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piddock, L. J., M. M. Johnson, S. Simjee, and L. Pumbwe. 2002. Expression of efflux pump gene pmrA in fluoroquinolone-resistant and -susceptible clinical isolates of Streptococcus pneumoniae. Antimicrob. Agents Chemother. 46:808-812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pletz, M. W. R., L. McGee, J. Jorgensen, B. Beall, R. R. Facklam, C. G. Whitney, and K. P. Klugman. 2004. Levofloxacin-resistant invasive Streptococcus pneumoniae in the United States: evidence for clonal spread and the impact of conjugate pneumococcal vaccine. Antimicrob. Agents Chemother. 48:3491-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rodríguez-Martínez, J. M., A. Pascual, I. Gracía, and L. Martínez-Martínez. 2003. Detection of the plasmid-mediated quinolone resistance determinant qnr among clinical isolates of Klebsiella pneumoniae producing AmpC-type β-lactamase. J. Antimicrob. Chemother. 52:703-706. [DOI] [PubMed] [Google Scholar]
- 48.Sanchez-Cespedes, J., M. M. Navia, R. Martinez, B. Orden, R. Millan, J. Ruiz, and J. Vila. 2003. Clonal dissemination of Yersinia enterocolitica strains with various susceptibilities to nalidixic acid. J. Clin. Microbiol. 41:1769-1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Schouls, L. M., H. G. J. van der Heide, L. Vauterin, P. Vauterin, and F. Mooi. 2004. Multiple-locus variable-number tandem repeat analysis of Dutch Bordetella pertussis strains rapid genetic changes with clonal expansion during late 1990s. J. Bacteriol. 186:5496-5505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shepard, B. J., and M. S. Gilmore. 2002. Antibiotic-resistant enterococci: the mechanisms and dynamics of drug introduction and resistance. Microbes Infect. 4:215-224. [DOI] [PubMed] [Google Scholar]
- 51.Singh, K. V., G. M. Weinstock, and B. E. Murray. 2002. An Enterococcus faecalis ABC homologue (Lsa) is required for the resistance of this species to clindamycin and quinopristin-dalfopristin. Antimicrob. Agents Chemother. 46:1845-1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tanaka, M., Y. Onodera, Y. Uchida, K. Sato, and I. Hayakawa. 1997. Inhibitory activities of quinolones against DNA gyrase and topoisomerase IV purified from Staphylococcus aureus. Antimicrob. Agents Chemother. 41:2362-2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tankovic, J., F. Mahjoubi, P. Courvalin, J. Duval, and R. Leclercq. 1996. Development of fluoroquinolone resistance in Enterococcus faecalis and role of mutations in the DNA gyrase gyrA gene. Antimicrob. Agents Chemother. 40:2558-2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Timmers, G. J., W. C. van der Zwet, I. M. Simoons-Smit, P. H. Savelkoul, H. H. Meester, C. M. vandenbroucke-Grauls, and P. C. Huijgens. 2002. Outbreak of vancomycin-resistant Enterococcus faecium in a haematology unit: risk factor assessment and successful control of the epidemic. Br. J. Haematol. 116:826-833. [DOI] [PubMed] [Google Scholar]
- 55.Torell, E., I. Kühn, B. Olsson-Liljequist, S. Hæggman, B.-M. Hoffman, C. Lindahl and L. G. Burman. 2003. Clonality among ampicillin-resistant Entercoccus faecium isolates in Sweden and relationships with ciprofloxacin resistance. Clin. Microbiol. Infect. 9:1011-1019. [DOI] [PubMed] [Google Scholar]
- 56.Torell, E., O. Cars, B. Olsson-Liljequist, B.-M. Hoffman, J. Lindbäck, and L. G. Burman. 1999. Near absence of vancomycin-resistant enterococci but high carriage rates of quinolone-resistant ampicillin-resistant enterococci among hospitalized patients and nonhospitalized individuals in Sweden. J. Clin. Microbiol. 37:3509-3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.van der Steen, L. F., M. J. M. Bonten, E. van Kregten, J. J. C. Charssema-Poot, R. Willems, and C. A. Gaillard. 2000. An outbreak of vancomycin-resistant Enterococcus faecium in a nephrology ward. Ned. Tijdschr. Geneesk. 144:2568-2571. [PubMed] [Google Scholar]
- 58.Wang, M., D. F. Sahm, G. A. Jacoby, and D. C. Hooper. 2004. Emerging plasmid-mediated quinolone resistance associated with the qnr gene in Klebsiella pneumoniae clinical isolates in the United States. Antimicrob. Agents Chemother. 48:1295-1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wiegand, I., M. Khalaf, M. Al-Agamy, and B. Wiedemann. 2004. First detection of the transferable quinolone resistance determinant in clinical Providencia stuartii strains in Egypt. Clin. Micobiol. Infect. 10(Suppl. 3):64. [Google Scholar]
- 61.Willems, R. J., J. Top, D. J. Smith, D. I. Roper, S. E. North, and N. Woodford. 2003. Mutations in the DNA mismatch repair proteins MutS and MutL of oxazolidinone-resistant or -susceptible Enterococcus faecium. Antimicrob. Agents Chemother. 47:3061-3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Willems, R. J. L., J. Top, M. van Santen, D. A. Robinson, T. M. Coque, F. Baquero, H. Grundmann, and M. J. M. Bonten. 2005. Global spread of vancomycin-resistant Enterococcus faecium from distinct nosocomial genetic complex. Emerg. Infect. Dis. 11:821-828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Reference deleted.
- 64.Willems, R. J. L., J. Top, N. van den Braak, A. van Belkum, D. J. Mevius, G. Hendriks, M. van Santen-Verheuvel, and J. D. van Embden. 1999. Molecular diversity and evolutionary relationships of Tn1546-like elements in enterococci from humans and animals. Antimicrob. Agents Chemother. 43:483-491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Willems, R. J. L., W. Homan, J. Top, M. van Santen-Verheuvel, D. Tribe, X. Manzioros, C. Gaillard, C. M. Vandenbroucke-Grauls, E. M. Mascini, E. van Kregten, J. D. van Embden, and M. J. Bonten. 2001. Variant esp gene as a marker of a distinct genetic lineage of vancomycin-resistant Enterococcus faecium spreading in hospitals. Lancet 357:853-855. [DOI] [PubMed] [Google Scholar]