Abstract
Monitoring of serotypes and their clonal associations is critical as pneumococci adapt to the selective pressures exerted by the pneumococcal seven-valent conjugate vaccine (PCV7). We genotyped 1,476 invasive isolates from the Active Bacterial Core surveillance (705 [89.8%] of the isolates were obtained from children <5 years of age, and 771 [18.4%] of the isolates were obtained from individuals >5 years of age) in 2001 and 2002 (after the introduction of PCV7). The data were compared to the results for 1,168 invasive isolates (855 [83.9%] of the isolates were from children <5 years of age) collected in 1999. Among children <5 years of age, the incidence of invasive disease due to non-PCV7 serogroups together with serogroup 19A increased (P < 0.001). Eighty-three clonal sets, representing 177 multilocus sequence types (STs), were compiled from the 3-year isolate set. Among the non-PCV7 serogroups, newly emerging clones were uncommon; and a significant expansion of already established clones occurred for serotypes 3 (ST180), 7F (ST191), 15BCF (ST199), 19A (ST199), 22F (ST433), 33F (ST662), and 38 (ST393). However, additional minor clonal types within serotypes 1, 6A, 6B, 7C, 9N, 10A, 12F, 14, 15B/C, 17F, 19A, 19F, 20, 22F, and 33F that were absent in 1999 were found during 2001 and 2002. Although 23 clonal sets exhibited multiple serotypes, for most serotypes there were either no changes or modest changes in clonal compositions since the introduction of PCV7. The only example of an identical ST shared between non-PCV7 and PCV7 or PCV7-related serotypes was ST199; however, ST199 was prevalent within serotypes 15B/C and 19A before and after PCV7 introduction. Continued genotypic surveillance is warranted, since certain clones not targeted by PCV7 are expanding, and their emergence as significant pathogens could occur with maintained vaccine pressure.
The effects of the pneumococcal seven-valent conjugate vaccine (PCV7) upon the genetic structure of the pneumococcal population and on serotype-genotype associations are still unknown, since PCV7 was only licensed in 2000. Although PCV7 has been very effective in decreasing the prevalence of PCV7 and PCV7-related (PCV7R) serotypes among pediatric isolates, there are reports of serotype replacement, representing increases of serotypes not targeted by PCV7 (16, 19, 23, 31, 33). Recently, in a country that does not use PCV7, multiresistant strains of nonvaccine serotypes have been characterized and shown to be probable capsular switch variants of highly successful clones of PCV7 serotypes (29). Such findings are important, since variants not targeted by vaccines have the potential to become highly successful pathogens within the pediatric population. Genetic characterization of replacement strains and monitoring of their prevalence over time within specific geographic areas will determine the relative impacts of newly emerging strains, of strains arising by capsular switching, and of already established strains. With time, active surveillance may detect all three categories of replacement strains that may increase as a result of intense PCV7 pressure. These issues can be addressed only with accurate molecular surveillance over extended periods.
Previously, we used pulsed-field gel electrophoresis (PFGE) of chromosomal SmaI digests combined with multilocus sequence typing (MLST) to describe the clonal structure within the majority of serotypes commonly observed among isolates causing invasive disease immediately prior to the introduction of PCV7 (12). That work provided the baseline information of PFGE profiles associated with MLST identifiers for future comparisons. The present study was undertaken with the objectives of continuing the active molecular surveillance of invasive pneumococcal isolates collected during each year of Active Bacterial Core surveillance (ABCs) (1) subsequent to PCV7 implementation and comparing these results to the baseline information obtained during previous years. Here we present the findings of the genotypic surveillance of selected pneumococcal isolates collected in the 2 years immediately following PCV7 introduction. Declines in the numbers of invasive pneumococcal disease cases due to the clonal types targeted by PCV7 serotypes compared to the numbers of cases in 1999 and increases in the numbers of invasive pneumococcal cases due to strains within clonal types not targeted by the vaccine are shown.
MATERIALS AND METHODS
Year 1999 isolates.
The year 1999 isolates were described previously (12).
Year 2001 isolates.
The year 2001 isolates were obtained through ABCs (1), an active population-based laboratory based surveillance program that is part of the Centers for Disease Control and Prevention (CDC) Emerging Infections Program. Surveillance areas included California (San Francisco County and children <5 years of age in Alameda and Contra Costa Counties), Colorado (the 5-county Denver area), Connecticut, Georgia (the 20-county Atlanta area), Maryland (the 6-county Baltimore area), Minnesota (the 7-county Twin Cities area), New York (the 7-county Rochester area and the 8-county Albany area), Oregon (the 3-county Portland area), and Tennessee (11 urban counties). These surveillance areas represented a total population of 22,479,308 people. For year 2001, all available sterile-site isolates from individuals less than 5 years of age (pediatric isolates [PIs]) were analyzed. Additionally, selected sterile-site isolates of serotypes from individuals ≥5 years of age (nonpediatric isolates [NPIs]) were analyzed as representatives of certain serotypes common or represented in adults but not in children or isolates from adults with decreased antibiotic susceptibilities. A total of 630 isolates with PCV7 or PCV7-related serotypes (PCV7-related serotypes are serotypes within the same serogroup as a PCV7 serotype) and 496 isolates with serotypes unrelated to those included in PCV7 were analyzed. These included a small number of isolates added to the study due to unusual resistance profiles, such as fluoroquinolone resistance, or as a result of an initial serotyping error.
Year 2002 isolates.
The surveillance areas for the year 2002 isolates were the same as those for the 2001 isolates, except that Minnesota was not included. For year 2002, we evaluated all available isolates from individuals <5 years of age that were not PCV7 serotypes. We also included a selection of penicillin-nonsusceptible non-PCV7 isolates from individuals ≥5 years of age. A small number of isolates with PCV7 serotypes were also included due to unusual resistance profiles, such as fluoroquinolone resistance, or as a result of an initial serotyping error. The year 2002 isolates included 190 isolates with non-PCV7-related serotypes, 150 isolates with PCV7-related serotypes, and 37 isolates with PCV7 serotypes.
Serotype and antibiotic susceptibility testing.
The isolates were serotyped by latex agglutination, and the results were confirmed by positive Quellung reactions. The antibiotic susceptibility patterns were determined by use of PML Microbiologicals (Wilsonville, Oreg.) custom broth dilution MIC panels.
Serotype classification.
The seven PCV7 serotypes are 4, 6B, 9V, 14, 18C, 19F, and 23F. In this study, serotypes considered to be PCV7R include serotypes not included in PCV7 but of the same serogroups (serogroups 6A, 9A, 9N, 18B, 18F, 19A, and 23A). PCV7-nonrelated (PCV7NR) serotypes include all serotypes not of the PCV7 serogroups.
Isolates used for calculation of rate changes.
Isolates used for calculation of rate changes were from surveillance areas under continuing surveillance from 1999 to 2002. These surveillance areas included California (San Francisco County only), Connecticut state, Georgia (the 20-county Atlanta metro area), Maryland (the 7-county Baltimore area), Minnesota (the 7-county Twin Cities area), New York (the 15-county Albany and Rochester areas), Oregon (the 3-county Portland area), and Tennessee (4 counties). The surveillance population for 1999 was 17,677,681 (1,211,130 individuals <5 years of age and 16,466,551 individuals ≥5 years of age). The surveillance population for 2002 was 18,904,366 (1,280,890 individuals <5 years of age and 17,623,476 individuals ≥5 years of age).
Genotyping.
PFGE and MLST were used to delineate clonal sets; and both procedures were carried out as described previously (7, 12), with the inclusion of all 26 currently recognized Pneumococcal Molecular Epidemiology Network (PMEN) clones (27). Sequence type (ST) identifiers from the pneumococcal multilocus sequence typing site (28) or new STs were determined for representative isolates of PFGE clusters in which two or more isolates within the same serotype shared PFGE patterns with ≥80% similarity, as determined by use of a Dice coefficient of 1.5. All instances of unusual serotype-genotype associations were reexamined by serotyping and/or genotyping. Isolates of different serotypes that shared nearly identical PFGE patterns were examined by MLST before they were assigned to a clonal set. Single PFGE outliers within a serotype were screened against the entire PFGE database consisting of isolates of all serotypes. All instances of strong PFGE similarity between isolates of different serotypes were examined by MLST.
CCs.
STs were divided into sets by using eBURST (11), which is available at the MLST database (28). eBURST sets have the requirement in which all STs must be a single-locus variant (SLV) of at least one other ST within the group and are illustrated by solid vertical lines in Fig. 1. Founder STs were defined as described previously (11) as being the ST within the eBURST set that has the greatest number of single-locus variants (STs in color in Fig. 1). For this work clonal complexes (CCs) consisted of eBURST sets or eBURST sets plus related STs that shared five of seven allelic identities with at least one ST within the set (double locus variants [DLVs]) and, additionally, that shared at least four allelic identities to the majority of the other STs included within the set.
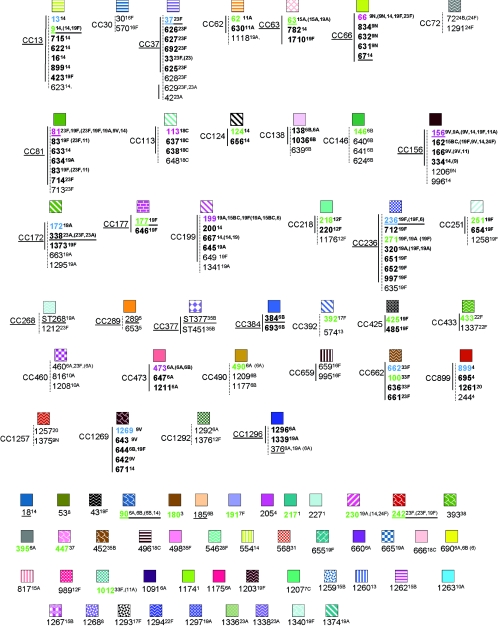
FIG. 1.
Eighty-three clonal sets representing the isolates genotyped and the STs encountered in 1999, 2001, and 2002. Solid lines connect STs within the same clonal group determined by eBURST analysis. STs within the same eBURST group are indicated in boldface. Closely related outliers within the same clonal set are connected to eBURST sets by dotted lines. Numbers in red indicate STs calculated as founders by using eBURST with the STs generated from this study and also calculated as founders by using all known STs (28). Numbers in green indicate STs calculated as founders solely by using all known STs. Numbers in blue indicate STs calculated as founders by using only the STs found in this study for the analysis. Serotypes in parentheses indicate different serotype associations listed at www.mlst.net for the indicated STs. When no parentheses are indicated, no differences in serotype associations for the STs were observed between this study and www.mlst.net.
Clonal sets.
The 83 total clonal sets referred to in this report refer to each of the genetically nonoverlapping CCs and STs that we have documented from ABCs (Fig. 1). All CCs and individual STs that include 1 of the 26 currently recognized internationally disseminated clones of PMEN (27) are underlined throughout this report. For example, clonal complex 37 (CC37) includes ST37 from PMEN clone Tennessee23F-4 and refers to the six-member clonal group shown in Fig. 1 plus the related STs 628, 629, and 42.
Assignment of clonal set designations by PFGE.
All isolates of the same serotype that shared ≥80% genetic relatedness within unweighted pair group method with arithmetic means-generated PFGE clusters (restricted to a single serotype) that included at least one isolate of a known ST were assigned within a clonal set (ST or clonal complex). The validity of this strategy was demonstrated previously (12). MLST was performed with all isolates of different serotypes that shared related PFGE profiles.
RESULTS
Invasive pneumococcal disease.
Analysis of pneumococcal isolates that caused invasive disease revealed that, with the exception of serotype 19A, the incidence of disease due to PCV7 serogroups decreased markedly during 2002 compared to the incidence in 1999 overall and in both age groups (Table 1). This was especially evident in 2002 for the pediatric (<5-year-old) age group, in which a nearly eightfold reduction in disease incidence due to serogroups contained in PCV7 (excluding serogroup 19A) was evident relative to the incidence in 1999 (P < 0.001). In contrast, the incidence of invasive disease in the pediatric population due to non-PCV7 serogroups (and serogroup 19A) increased 1.5-fold in 2002 relative to the incidence in 1999 (P < 0.001). Compared to the proportion of cases in 1999, the proportion of cases due to non-PCV7 serotypes 3, 7F, 15BCF, 19A, 22F, 33F, and 38 significantly increased in 2002 (P ≤ 0.002; Table 2).
TABLE 1.
Invasive pneumococcal disease incidence in 1999 and 2002a
| Serotype(s) | Age group (yr) | 1999
|
2002
|
Change in rate | P valuec | ||
|---|---|---|---|---|---|---|---|
| No. of cases | Rateb | No. of cases | Rate | ||||
| All | <5 | 1,167 | 96.36 | 308 | 24.05 | −72.31 | <0.001 |
| ≥5 | 3,141 | 19.08 | 2,418 | 13.72 | −5.36 | <0.001 | |
| All | 4,308 | 24.37 | 2,726 | 14.42 | −10.05 | <0.001 | |
| PCV7 serogroups except 19A | <5 | 1,064 | 87.85 | 143 | 11.16 | −76.69 | <0.001 |
| ≥5 | 2,067 | 12.55 | 1,233 | 7.00 | −5.55 | <0.001 | |
| All | 3,131 | 17.71 | 1,376 | 7.28 | −10.43 | <0.001 | |
| Non-PCV7 serogroups plus 19A | <5 | 103 | 8.50 | 165 | 12.88 | +4.38 | <0.001 |
| ≥5 | 1,067 | 6.48 | 1,185 | 6.72 | +0.24 | 0.4 | |
| All | 1,170 | 6.62 | 1,350 | 7.14 | +0.52 | 0.06 | |
The surveillance areas included California (San Francisco only), Connecticut (state), Georgia (the 20-county Atlanta metro area), Maryland (the 7-county Baltimore II area), Minnesota (the 7-county Twin Cities area), New York (the 15-county Albany and Rochester areas), Oregon (the 3-county Portland area), and Tennessee (4 counties).
Number of cases per 100,000 individuals.
P values are for 1999 versus 2002.
TABLE 2.
Incidence of invasive cases in group less than 5 years of age due to PCV7-nonrelated serotypes and serotype 19A in 2002 relative to incidence in 1999a
| Serotype | 1999
|
2002
|
P valued | ||
|---|---|---|---|---|---|
| Actual (projectedb) no. of cases | % of totalc | Actual (projected) no. of cases | % of total | ||
| 1 | 6 (7) | [0.6] | 3 | [1] | |
| 3 | 6 (7) | [0.6] | 8 (9) | [2.9] | 0.002 |
| 5 | 1 | [<0.1] | 0 | ||
| 7F | 8 (9) | [0.8] | 11 (13) | [4.2] | <0.001 |
| 10A | 1 | [<0.1] | 3 | [1] | |
| 11A | 1 | [<0.1] | 4 (5) | [1.6] | |
| 12F | 12 (14) | [1.2] | 7 (8) | [2.6] | 0.106 |
| 15B/15C/15F | 6 (7) | [0.6] | 26 (30) | [9.7] | <0.001 |
| 16F | 2 | [<0.1] | 2 | [0.6] | |
| 17F | 0 | 1 | [0.3] | ||
| 19A | 20 (23) | [2] | 41 (47) | [15.3] | <0.001 |
| 22F | 7 | [0.6] | 9 (10) | [3.2] | 0.002 |
| 24B/24F | 1 | [<0.1] | 0 | ||
| 33A | 0 | 1 | [0.3] | ||
| 33F | 8 (9) | [0.8] | 14 (16) | [5.2] | <0.001 |
| 35B | 2 | [<0.1] | 1 | [0.3] | |
| 35F | 0 | 1 | [0.3] | ||
| 38 | 5 (6) | [0.5] | 11 (13) | [4.2] | <0.001 |
| Nontypeable | 2 | [<0.1] | 0 | ||
| Total | 1,167 | 308 | |||
The surveillance areas are the same as those listed in footnote a of Table 1.
Projected number of cases in the group <5 years of age for the indicated surveillance areas since serotypes were not determined for 171 of the total (n = 1,167) year 1999 isolates and were not determined for 41 of the the total (n = 308) year 2002 isolates.
Percentage of total isolates recovered from this age group in the indicated year.
Comparison of proportions for serotypes which have ≥2% of total strains in at least 1 year by using the chi-square or the Fisher exact test.
New STs obtained in the study.
Overall, we identified 177 STs among ABCs isolates, including 39 new STs discovered during the year 2001 surveillance and the year 2002 surveillance. These included STs 715, 989, 1174 to 1177, 1207 to 1209, 1211, 1212, 1257 to 1263, 1267 to 1269, 1291 to 1297, 1336 to 1341, 1373, 1374, 1375, 1376, and 1710. Five of the 39 new STs (ST1175 [serotype 6A], ST1207 [serotype 7C], ST1262 [serotype 15BC], ST1293 [serotype 17F], and ST1374 [serotype 19A]), representing five different serotypes, shared four or fewer alleles with any other known ST. Of the 34 remaining new STs, 25 were closely similar (SLVs or DLVs) to STs with the same serotype, as listed in the MLST database (28). However, nine of these new STs (SLVs or DLVs) had serotype associations that were not described previously. Five of these nine new associations included STs associated with PCV7NR serotypes. The five serotype-ST associations 15BC-ST1259, 15BC-ST1267, 20-ST1261 (CC899), 10A-ST1263, and 12F-ST1376 (CC1292) potentially depict descendants of STs associated with PCV7 serotypes due to their relatedness with known STs from PCV7 serotype strains. This possibility seems likely for ST1261 (serotype 20), since it is the sole member of an eight-ST eBURST group (when all known STs at www.mlst.net are used) that is not associated with serotype 4 and is an SLV of the predicted founder ST, ST899 (Fig. 1).
The 21 new MLST alleles found within the year 2001 and the year 2002 isolates included aroE61, aroE62, gdh-87, gki-89, gki-90, gki-101, recP61, recP67, recP68, spi-93 to spi-95, xpt-112, xpt-130, xpt-137, xpt-140, ddl-149, ddl-151, and ddl-154. With one exception, all of the new alleles exhibited 99% or greater identity with the majority of the corresponding allele sequences in the MLST database. The highly divergent ddl-154 allele obtained from a penicillin-resistant type 19A isolate shared only 95% sequence identity with its closest match from penicillin-nonsusceptible isolates and only about 93% sequence identity with typical ddl alleles from penicillin-sensitive isolates. Consistent with previous observations (2, 8, 12), the various divergent ddl alleles encountered during this work were always associated with penicillin-nonsusceptible isolates.
Serotype-genotype associations of invasive pneumococcal isolates compared to those in the pneumococcal MLST database.
In general, STs were restricted to the same serotype and the results between our data set (Fig. 1) and the pneumococcal MLST database (28) were concordant. All instances of multiple serotype associations with single STs found in our study are shown in Fig. 1. For each ST, any differing serotype association patterns listed at the MLST database as of December 2004 are in parentheses (Fig. 1).
Clonal sets.
Eighty-three nonoverlapping clonal sets were documented from the 1999 to 2002 surveillance for invasive pneumococci (Fig. 1). Six of the founder STs for clonal complexes shown in Fig. 1 were calculated independently with eBURST by the use of our study-limited data set of 177 STs and were also determined to be founders, based on the data set containing all known STs in the database at mlst.net (1,510 STs as of 20 December 2004; these founder STs are indicated in red in Fig. 1). Twenty-three additional STs that were detected during our surveillance and that were predicted founders, based on the total ST data in the MLST database, are indicated in green in Fig. 1. Seven additional founders were predicted solely on the basis of the analysis of the STs found during this study (indicated in blue). The total of 36 “founder” STs generally represent STs that were most commonly found within the indicated clonal sets (Fig. 1), consistent with these particular strains being the originators of the clonal sets (11).
The relationships of the 83 clonal sets that were compiled by using study STs alone (the single STs that are listed but that are not highly related to others included in Fig. 1 are also considered clonal sets) are highly congruent with nonoverlapping eBURST groups and singlets (without DLVs within the study set) derived from all known STs (28). The exceptions to this involve 11 clonal sets that include CC138, CC172, CC226, CC156, CC1269, CC425, CC146, ST18, CC460, CC1292, and CC392 (Fig. 1). These 11 clonal sets represent a total of 13 nonoverlapping clonal sets at mlst.net, as follows: (i) STs comprising CC138, CC172, and CC226 are included within one large eBURST group comprised of 79 STs, where ST138 is the second most likely founder ST, based upon the numbers of SLVs. (ii) STs comprising CC156 and CC1269 are all included within a large eBURST group comprised of 73 STs, where ST156 is the predicted founder ST. (iii) ST146 is included within one eBURST group, while CC146 STs 640 and 641 are not linked as SLVs to any known STs. (iv) ST18 is included within CC13. (v) The three STs included in CC460 each represented small eBURST groups. (vi) CC1292 and CC392 both represent two distinct eBURST lineages when they are analyzed in the context of all known STs.
In summary, with the exceptions listed above, the 83 clonal sets derived from our study isolates closely correlated to the 86 clonal groups derived by eBURST analysis of the entire pneumococcal MLST database (28).
Genetic structures of individual serotypes.
PCV7 and PCV7R serotypes displayed an overall increased degree of genetic complexity (48 CCs and 124 STs) relative to that of the PCV7NR serotypes (37 CCs and 52 STs) (Fig. 1; see Table 4). This could be due in part to the fact that more PCV7 and PCV7R isolates than PCV7NR isolates (1,534 versus 1,012 total isolates) were analyzed from the 3 years. However, it is apparent that PCV7NR serotypes displayed a maximum of five clonal sets (within 15B/C), with types 1, 3, 7F, 10A, 11A, 12F, 15A, 15B/C, 22F, 33F, and 38 each primarily comprised of one distinct clonal set. In contrast PCV7 or PCV7R serotypes 6A, 6B, 14, 19A, 19F, and 23F each displayed 6 to 14 clonal types, with types 6A, 6B, 19F, and 23F each containing three major clonal sets that accounted for 5 to 35% of the isolates analyzed within their respective serotypes during each year (Fig. 2). A higher number of distinct clonal sets was most often observed within serotypes with a high percentage of penicillin-nonsusceptible isolates (including serotypes 6A, 6B, 14, 19A, 19F, and 23F) (Fig. 2). Two exceptions to this observation included serotype 9V, which primarily consisted of only two highly related clonal clusters or groups during all 3 years (CC156 and CC1269, which are included in a single eBURST cluster when all existing STs are considered), and serotype 35B, which consisted of a penicillin-nonsusceptible clonal group and a distinct penicillin-susceptible clone (CC377 and ST452, respectively) (2). PCV7 serotype 4 and PCV7 serogroup 18 (primarily serotype 18C) consisted entirely of penicillin-susceptible isolates.
TABLE 4.
Clonal complex distribution among invasive pneumococcal isolates in 1999, 2001, and 2002
| Serotype (clonal complex)a | No. of isolates within complex/no. of isolates analyzed (%) in the following years for the indicated age groups
|
|||||
|---|---|---|---|---|---|---|
| 1999
|
2001
|
2002b
|
||||
| <5 yr | ≥5 yr | <5 yr | ≥5 yr | <5 yr | ≥5 yr | |
| 1 (ST227) | 5/6 (83.3) | 15/15 (100) | 4/4 (100) | 28/34 (82.4) | 4/4 (100) | None analyzed |
| 1 (ST217) | 1/6 (16.7) | 0/15 | 0/4 | 1/34 (2.9) | 0/4 | None analyzed |
| 1 (ST1174) | 0/6 | 0/6 | 0/6 | 2/34 (5.9) | 0/4 | None analyzed |
| 3 (ST180) | 6/6 (100) | 48/50 (96.0) | 6/6 (100) | 68/70 (97.0) | 8/8 (100) | 11/11 (100) |
| 4 (CC899) | 53/71 (74.6) | None analyzed | 29/35 (82.9) | 18/34 (52.9) | None analyzed | 1/1 (100) |
| 4 (ST205) | 8/71 (11.3) | None analyzed | 6/35 (17.1) | 11/34 (32.4) | None analyzed | 0/1 (0) |
| 5 (ST653) | 100 (1) | None analyzed | 1/1 (100) | 1/1 (100) | No type 5s | None analyzed |
| 6A (CC1296) | 19/57 (33.3) | 0/1 | 10/38 (26.3) | 0/9 | 4/13 (30.7) | 14/41 (34.1) |
| 6A (CC460) | 13/57 (22.4) | 0/1 | 11/38 (29.4) | 3/9 (33.3) | 3/13 (23.1) | 0/41 |
| 6A (CC473) | 5/57 (8.8) | 0/1 | 7/38 (18.4) | 1/9 (11.1) | 1/13 (7.7) | 15/41 (36.6) |
| 6A (ST395) | 1/57 (1.8) | 1/1 (100) | 3/38 (7.9) | 0/9 | 3/13 (23.1) | 2/41 (4.9) |
| 6A (CC490) | None detectedb | 0/1 | 2/38 (5.3) | 0/9 | None detected | None detected |
| 6A (CC138) | None detected | 0/1 | 2/38 (5.3) | 0/9 | None detected | None detected |
| 6A (ST1091) | None detected | 0/1 | 1/38 (2.6) | 0/9 | None detected | None detected |
| 6A (ST1175) | None detected | 0/1 | 1/38 (2.6) | 0/9 | None detected | None detected |
| 6A (ST90) | 1/57 (1.8) | 0 | None detected | 0/9 | None detected | None detected |
| 6A (ST660) | 1/57 (1.8) | 0 | None detected | 0/9 | None detected | None detected |
| 6A (ST690) | None detected | 0 | None detected | 2/9 (22.2) | None detected | None detected |
| 6A (CC1292) | None detected | 0/1 | None detected | 3/9 (33.3) | None detected | 6/41 |
| 6B (CC138) | 13/103 (12.6) | 0/1 | 13/45 (28.9) | 2/15 (9.1) | None analyzed | 1/4 (25.0) |
| 6B (CC146) | 22/103 (21.4) | 0/1 | 11/45 (24.4) | 4/15 (28.6) | None analyzed | 0/4 |
| 6B (ST90) | 13/103 (12.6) | 0/1 | 13/45 (28.9) | 5/15 (28.6) | None analyzed | 0/4 |
| 6B (CC384) | 6/103 (5.8) | 0/1 | 5/45 (11.1) | 3/15 (20.0) | None analyzed | 0/4 |
| 6B (CC1269) | 3/103 (2.9) | 0/1 | None detected | 0/15 | None analyzed | 0/4 |
| 6B (CC490) | None detected | 0/1 | 2/45 (4.4) | 0/15 | None analyzed | 3/4 (75.0) |
| 6B (ST185) | 1/103 (1.0) | 0/1 | None detected | 0/15 | None analyzed | 0/4 |
| 6B (ST690) | None detected | 1/1 | None detected | 1/15 (6.7) | None analyzed | 0/4 |
| 7B (ST664) | 1/1 (100) | No type 7Bs | No type 7Bs | No type 7Bs | No type 7B | No type 7Bs |
| 7C (ST1207) | No type 7C | No type 7Cs | No type 7Cs | 1/1 (100) | 1/1 (100) | 1/1 (100) |
| 7F (ST191) | 7/7 (100) | 22/24 (91.7) | 13/15 (86.7) | 29/30 (96.7) | 5/6 (83.3) | 3/3 (100) |
| 7F (CC218) | 0/7 | 1/24 (4.2) | None detected | None detected | None detected | 0/3 |
| 8 (ST53) | No type 8 | None analyzed | 2/2 (100) | 1/10 (10.0) | No type 8 | 100 (1) |
| 8 (ST1268) | No type 8 | None analyzed | 0/2 | 9/10 (90.0) | No type 8 | 0/1 |
| 9A (CC156) | No type 9A | No type 9As | 3/3 (100) | No type 9As | No type 9A | No type 9As |
| 9N (CC66) | 2/2 (100) | 9/11 (81.8) | 3/3 (100) | 34/37 (91.9) | None analyzed | 3/5 (60.0) |
| 9N (CC156) | 0/2 | None detected | 0/3 | None detected | None analyzed | 1/5 (20.0) |
| 9N (CC1257) | 0/2 | None detected | 0/3 | None detected | None analyzed | 1/5 (20.0) |
| 9V (CC156) | 60/72 (83.3) | 21/26 (80.1) | 26/38 (68.4) | 28/42 (66.7) | 2/2 (100) | 7/7 (100) |
| 9V (CC1269) | 9/72 (12.5) | 3/26 (11.5) | 11/38 (28.9) | 14/42 (33.3) | 0/2 | 0/2 |
| 10A (CC460) | 1/1 (100) | None analyzed | 3/3 (100) | 2/5 (40) | 3/3 (100) | 2/2 (100) |
| 10A (ST1263) | 0/1 | None analyzed | 0/3 | 2/5 (40) | 0/3 | 0/2 |
| 11A (CC62) | 1/1 (100) | 22/24 (87.5) | 5/5 (100) | 41/41 (100) | 2/2 (100) | 9/9 (100) |
| 12F (CC218) | 11/11 (100) | 48/49 (97.8) | 17/17 (100) | 65/65 (100) | 4/6 (66.7) | 1/1 (100) |
| 12F (ST989) | 0/11 | None detected | 0/17 | 0/65 | 1/6 (16.7) | 0/1 |
| 12F (CC1292) | 0/11 | None detected | 0/17 | 0/65 | 1/6 (16.7) | 0/1 |
| 13 (CC392) | No type 13 | None analyzed | No type 13s | 2/2 (100) | No type 13 | 0/1 |
| 13 (ST1260) | No type 13 | None analyzed | No type 13s | 0/2 | No type 13 | 1/1 (100) |
| 14 (CC13) | 96/208 (46.2) | None analyzed | 48/91 (52.7) | 19/36 (52.7) | None analyzed | 3/3 (100) |
| 14 (CC124) | 78/208 (37.5) | None analyzed | 33/91 (30.0) | 9/36 (25.0) | None analyzed | 0/3 |
| 14 (CC63) | None detected | None detected | 6/91 (6.6) | 0/36 | None analyzed | 0/3 |
| 14 (CC1269) | 5/208 (2.4) | None analyzed | 2/91 (2.2) | 2/36 (5.9) | None analyzed | 0/3 |
| 14 (CC156) | None detected | None analyzed | 3/91 (3.3) | 1/36 (2.8) | None analyzed | 0/3 |
| 14 (CC66) | 4/208 (1.9) | None analyzed | 1/91 (1.1) | 1/36 (2.8) | None analyzed | 0/3 |
| 15A (CC63) | No type 15As | None analyzed | No type 15As | 3/3 (100) | No type 15As | 12/13 (92.3) |
| 15A (ST817) | No type 15As | None analyzed | No type 15As | 0/3 | No type 15As | 1/13 (7.7) |
| 15BC (CC199) | 6/6 (100) | None analyzed | 20/22 (90.9) | 3/3 (100) | 27/33 (81.8) | 5/5 (100) |
| 15BC (ST1262) | 0/6 | None analyzed | None detected | 0/3 | 2/33 (6.1) | 0/5 |
| 15BC (ST1267) | 0/6 | None analyzed | None detected | 0/3 | 2/33 (6.1) | 0/5 |
| 15BC (ST1259) | 0/6 | None analyzed | None detected | 0/3 | 1/33 (3.0) | 0/5 |
| 15BC (CC156) | 0/6 | None analyzed | None detected | 0/3 | 1/33 (3.0) | 0/5 |
| 16F (CC659) | 1/1 (100) | 0/2 | No type 16Fs | 7/7 (100) | 2/2 (100) | 1/3 (33.3) |
| 16F (CC30) | 0/1 | 2/2 (100) | No type 16s | 0/7 | 0/2 | 2/3 (66.7) |
| 17F (CC392) | No type 17F | None analyzed | 1/1 (100) | None analyzed | 1/1 (100) | None analyzed |
| 18BCF (CC113) | 74/93 (79.6) | None analyzed | 28/40 (68.3) | 9/11 (81.8) | 2/3 (66.7) | 2/2 (100) |
| 18BCF (ST496) | 8/93 (8.6) | None analyzed | 5/40 (12.5) | 1/11 (9.0) | 0/3 | 0/2 |
| 18BCF (ST666) | 1/93 (1.1) | None analyzed | 1/40 (2.4) | 1/11 (9.0) | 1/3 (33.3) | 0/2 |
| 18BCF (CC66) | 1/93 (1.1) | None analyzed | None detected | 0/11 | 0/2 | 0/2 |
| 19A (ST199) | 13/18 (72.2) | 46/64 (71.9) | 18/29 (62.1) | 10/10 (100) | 27/40 (67.5) | 32/45 (71.1) |
| 19A (CC172) | None detected | 2/64 (3.1) | 2/29 (6.9) | 0/10 | 0/40 | 1/45 (2.2) |
| 19A (ST665) | 1/18 (5.5) | None detected | 1/29 (3.4) | None detected | 0/40 | 0/45 |
| 19A (CC236) | None detected | None detected | 1/29 (3.4) | None detected | 3/40 (7.5) | 1/45 (2.2) |
| 19A (CC1296) | None detected | None detected | None detected | None detected | 4/40 (10.0) | 5/45 (11.1) |
| 19A (CC81) | 1/18 (5.5) | None detected | None detected | None detected | 1/40 (2.5) | 2/45 (4.4) |
| 19A (ST230) | None detected | None detected | 2/29 (6.9) | None detected | 0/40 | 0/45 |
| 19A (ST1297) | None detected | None detected | 1/29 (3.4) | None detected | 3/40 (7.5) | 2/45 (4.4) |
| 19A (CC62) | None detected | None detected | None detected | None detected | 2/40 (5.0) | 0/45 |
| 19A (ST1374) | None detected | None detected | None detected | 0/10 | 0/40 | 1/45 (2.2) |
| 19A (ST847) | None detected | None detected | None detected | 0/10 | 0/40 | 1/45 (2.2) |
| 19F (CC236) | 23/110 (20.9) | None examined | 10/47 (21.3) | 3/12 (25.0) | 1/2 (50) | 8/9 (88.9) |
| 19F (CC251) | 22/110 (20.0) | None examined | 19/47 (40.4) | 3/12 (25.0) | 0/2 | 0/9 |
| 19F (CC81) | 13/110 (11.8) | None examined | 3/47 (6.4) | 3/12 (25.0) | 0/2 | 0/9 |
| 19F (CC199) | 10/110 (9.1) | None examined | 1/47 (2.1) | 1/12 (9.1) | 0/2 | 0/9 |
| 19F (ST655) | 2/110 (1.8) | None examined | 0/47 | 0/12 | 0/2 | 0/9 |
| 19F (CC177) | 2/110 (1.8) | None examined | 4/47 (8.5) | 0/12 | 0/2 | 0/9 |
| 19F (CC1269) | 1/110 (0.9) | None examined | 2/47 (4.3) | 1/12 (9.1) | 0/2 | 0/9 |
| 19F (ST1340) | None detected | None examined | 0/47 | 0/12 | 1/2 (50) | 0/9 |
| 19F (CC13) | 1/110 (0.9) | None examined | 0/47 | 0/12 | 0/2 | 0/9 |
| 19F (ST43) | 1/110 (0.9) | None examined | 4/47 (8.5) | 0/12 | 0/2 | 0/9 |
| 19F (CC172) | None detected | None examined | 1/47 (2.1) | 0/12 | 0/2 | 0/9 |
| 19F (CC425) | 4/110 (3.6) | None examined | 3/47 (6.4) | 0/12 | 0/2 | 0/9 |
| 19F (CC63) | None detected | None examined | 0/47 | 1/12 (9.1) | 0/2 | 0/9 |
| 19F (ST1203) | None detected | None examined | 0/47 | 0/12 | 0/2 | 1/9 (11.1) |
| 20 (CC899) | No type 20 | None examined | 1/1 (100) | 4/6 (66.7) | No type 20 | None examined |
| 20 (CC1257) | No type 20 | None examined | 0/1 | 33.3 (2) | No type 20 | None examined |
| 22F (CC433) | 6/6 (100) | 30/31 (96.8) | 19/19 (100) | 90.2 (46) | 92.9 (13) | 100 (2) |
| 22F (ST1294) | 0/2 | None detected | 0/1 | None detected | 7.1 (1) | 0/2 |
| 23A (CC172) | 2/2 (100) | None examined | 0/1 | 0/1 | No type 23A | 5/6 (83.3) |
| 23A (ST1336) | 0/2 | None examined | 1/1 (100) | 0/1 | No type 23A | 0/6 |
| 23A (ST1338) | 0/2 | None examined | 0 | 2/4 (50.0) | No type 23A | 0/6 |
| 23A (CC37) | 0/2 | None examined | 0 | 2/4 (50.0) | No type 23A | 1/6 (16.7) |
| 23F (CC37) | 46/64 (71.9) | None examined | 26/37 (70.3) | 6/13 (46.2) | None examined | 0/5 |
| 23F (CC81) | 11/64 (17.2) | None examined | 5/37 (13.5) | 4/13 (30.7) | None examined | 4/5 (80.0) |
| 23F (ST242) | 4/64 (6.2) | None examined | 1/37 (2.7) | 2/13 (15.4) | None examined | 1/5 (20.0) |
| 23F (CC662) | 1/64 (1.6) | None examined | None detected | 0/13 | None examined | 0/5 |
| 23F (CC268) | None detected | None examined | 1/37 (2.7) | 1/13 (7.7) | None examined | 0/5 |
| 23F (CC460) | None detected | None examined | 2/37 (5.4) | 0/13 | None examined | 0/5 |
| 24BF (CC72) | No type 24BF | None examined | 1/1 (100) | None examined | None examined | 1/1 (100) |
| 28F (ST546) | No type 28F | No type 28F | No type 28F | No 28F isolates | No 28F isolates | 100 (1) |
| 31 (ST568) | No type 31 | 1/1 (100) | No type 31 | 2/2 (100) | 1/1 (100) | No type 31 |
| 33F (CC662) | 7/7 (100) | 1/1 (100) | 10/11 (90.9) | 7/8 (87.5) | 18/18 (100) | 1/1 (100) |
| 33F (ST1012) | 0/7 | 0/1 | 1/11 (9.1) | 1/8 (12.5) | 0/18 | 0/1 |
| 35B (CC377) | 2/2 (100) | 10/14 (71.4) | 2/2 (100) | 27/35 (77.1) | 100 (1) | 18/19 (94.7) |
| 35B (ST452) | 0/2 | 4/14 (28.6) | 0/2 | 8/35 (22.9) | 0/1 | 1/19 (5.3) |
| 35F (ST498) | No type 35F | None examined | 2/2 (100) | 2/2 (100) | None examined | None examined |
| 37 (ST447) | No type 37 | None examined | No type 37 | None examined | No type 37 | 1/2 (50.0) |
| 38 (ST393) | 5/5 (100) | None examined | 7/7 (100) | 1/1 (100) | 15/15 (100) | None examined |
Clonal complexes in boldface were found only among isolates from individuals ≥5 years of age. Up to 30% of the isolates within a given serotype had unique PFGE profiles and were not subjected to MLST (represented by gaps in the columns in Fig. 2). Such isolates could possibly belong to any of the clonal sets included or could represent additional clonal sets.
Year 2002 isolates were primarily selected from non-PCV7 serotypes.
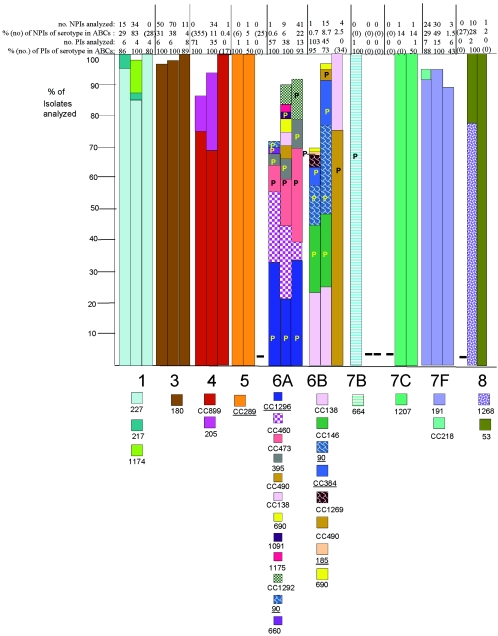
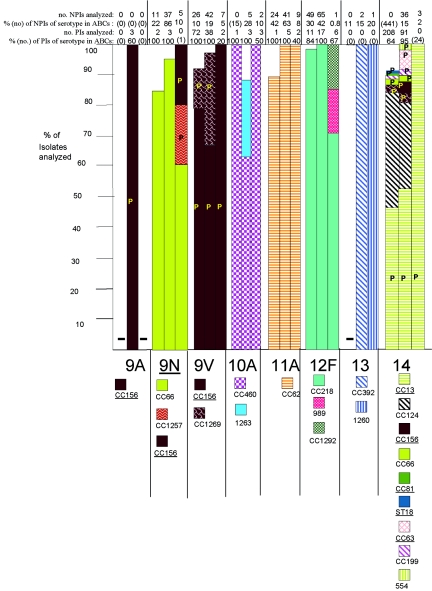
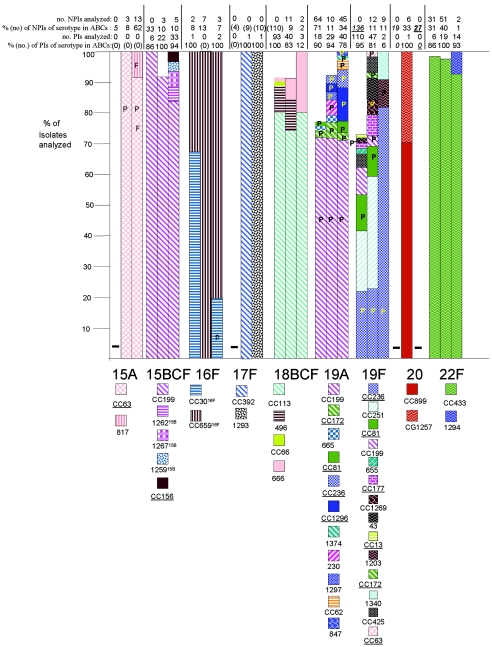
FIG.2.
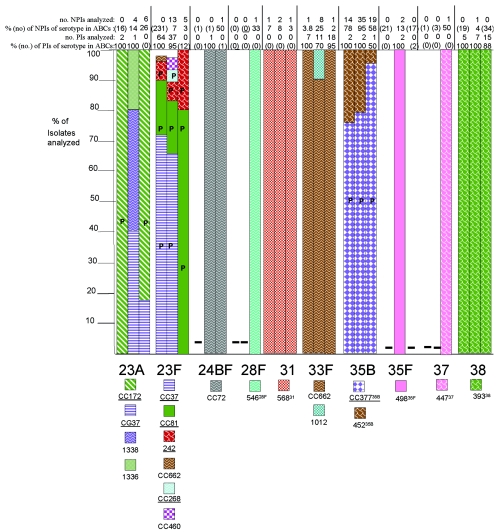
Clonal compositions of individual serotypes from invasive surveillance. The left, middle, and right columns represent isolates recovered in 1999, 2001, and 2002, respectively. The NPIs analyzed depict the numbers of isolates from individuals 5 years of age or older that were subjected to PFGE. The PIs analyzed depict the numbers of isolates from individuals less than 5 years of age that were subjected to PFGE. The percentages depict the proportions of invasive pneumococcal isolates within these serotypes that were subjected to genetic analysis. Where the numbers corresponding to these percentages are in parentheses, they refer to the total numbers of ABCs isolates within that age and serotype. The letter P indicates that at least 15% of the indicated clonal set within the indicated serotype were penicillin nonsusceptible. Serotypes in parentheses are shown when the serotype distributions listed in the global database (28) differed in any way from what we observed. Underlines indicate STs directly shared by PMEN clones (27) or clonal complexes containing these STs found among PMEN clones. Up to 30% of the isolates within a given serotype had unique PFGE profiles and were not subjected to MLST (represented by gaps in the columns). Such isolates could possibly belong to any of the clonal sets included or could represent additional clonal sets.
Serotype associations within clonal sets.
Sixty-three of the 83 clonal sets were represented solely by single serotypes. Forty-one clonal sets represented only PCV7 or PCV7R serotypes and were not inclusive of PCV7NR serotypes during the years of this study.
Although 22 of the 83 clonal sets shown in Fig. 1 were associated with multiple serotypes (25 of the clonal sets were associated with multiple serotypes, if the serotype distribution listed in the pneumococcal MLST database is also considered; see the serotypes in parentheses in Fig. 1), only 9 of these were associated in this study with both PCV7-PCV7R and PCV7NR serotypes. Ten clonal sets were associated with both PCV7R and PCV7NR serotypes if the serotype distribution listed in the pneumococcal MLST database is also taken into consideration (see ST230 in Fig. 1).
Of the nine clonal sets containing both PCV7-PCV7R and PCV7NR serotypes (CC62, CC63, CC156, CC199, CC899, CC662, CC460, CC1257, and CC1292), we observed only three instances in which a PCV7NR serotype strain shared a clonal complex where other members were associated only with PCV7 and PCV7R serotypes. One such instance was observed within CC156. Within CC156, which is comprised almost entirely of PCV7-related strains, we found a single ST162 serotype 15B pediatric isolate recovered during 2002. The second instance was within CC1292, where a DLV of ST1292, ST1376, was found in a single year 2002 type 12F pediatric isolate. The third instance was CC899, which accounted for the majority of serotype 4 isolates during 1999 and 2001. Five of seven year 2002 serotype 20 isolates, including the single pediatric isolate, were represented by ST1261, which is an SLV of ST899 associated with serotype 4.
We observed only 13 instances in which different serotypes were directly associated with an identical ST. Three of these instances (ST6329N,18C, ST21812F,7F, and ST906B,6A) were observed only in 1999 and involved only single instances of unusual associations. ST19915BC,19A,19F was the only ST shared between PCV7R-PCV7 serotype isolates and PCV7NR isolates and was the most common ST found within serotypes 19A and 15B/C during all 3 years of this study. The nine remaining instances involved only PCV7 or PCV7R serotypes (ST64419F,6B, ST8123F,19F, ST3766A,19A, ST1386A,6B, ST1569A,9V, ST27119F,19A, ST4606A,23F, ST62923F,23A, and ST6906A,6B). Of the ST-serotype combinations involved in these 13 instances, only ST19919A, ST37619A, and ST27119A represented serotypes that are not targeted by PCV7, as observed by increases of serotype 19A invasive disease cases from the <5-year-old age group (Table 2) (23). Notably, these three combinations are represented by a high percentage of penicillin-nonsusceptible isolates (Fig. 1) that are resistant to several other antibiotics (data not shown). ST271 is a single-locus variant of globally disseminated multiresistant clone Taiwan19F-14 (ST236) that is common within the United States (10) (Fig. 1 and 2 [see serotypes 19F and 19A]; see Table 4).
PMEN clones.
Isolates highly related to 17 of the 26 currently recognized PMEN clones were detected within our sample set (Table 3). From 1999 to 2002, large fractions of individual PCV7 or PCV7-related serotypes were comprised of isolates highly related to various PMEN clones primarily of the same serotypes (Tables 3 and 4; Fig. 2). Thirty-one percent of the isolates within the 1999 study set were highly related to individual PMEN clones listed in Table 3, whereas 28.2% of the isolates in the 2001 study set were highly related to individual PMEN clones. With the exception of CC289 from serotype 5, the isolates represented by these clones contained a high percentage of penicillin-nonsusceptible isolates (Fig. 2). Although the 2002 genotype surveillance focused primarily upon non-PCV7 serotypes, 26.5% of the isolates in this sample were within genetic sets representing PMEN clones. PCV7NR serotype 35B was comprised in large part of penicillin-nonsusceptible isolates highly related to PMEN clone Utah35B-24 (2) during the years 1999, 2001, and 2002 (Fig. 2; Table 4).
TABLE 3.
PMEN clones sharing genetic identity or close genetic relatedness to ABCs pneumococcal isolates recovered in 1999, 2001, and 2002
| PMEN clone (ST) | Serotype(s) of invasive pneumococcal isolates genetically related to PMEN clonea
|
||
|---|---|---|---|
| 1999 | 2001 | 2002 | |
| Spain23F-1 (ST81) | 19F, 23F, 14, 19A | 19F, 23F | 19A, 23F |
| Spain6B-2 (ST90) | 6B, 6A | 6B | |
| Spain9V-3 (ST156) | 9V, 6B, 14 | 9V, 14, 19F, 9A | 15B, 9N |
| Tennessee23F-4 (ST37) | 23F | 23F, 23A | 23A |
| Spain14-5 (ST18) | 14 | ||
| Hungary19A-6 (ST268) | 23F | ||
| South Africa6B-8 (ST185) | 6B | ||
| England14-9 (ST9) | 14, 19F | 14 | |
| Taiwan19F-14 (ST236) | 19F | 19F | 19A |
| Taiwan23F-15 (ST242) | 23F | 23F | 23F |
| Maryland6B-17 (ST384) | 6B | 6B | |
| Columbia5-19 (ST289) | 5 | 5 | |
| Portugal19F-21 (ST177) | 19F | 19F | |
| North Carolina6A-23 (ST376) | 6A | 6A | 6A, 19A |
| Utah35B-24 (ST377) | 35B | 35B | 35B |
| Sweden15A-25 (ST63) | 15A | 15A, 14 | |
| Colombia23F-26 (ST338) | 23A | 23A | |
A total of 1,168 invasive isolates were examined from year 1999, 1,126 invasive isolates were examined from year 2001, and 377 invasive isolates were examined from year 2001.
Associations of specific clonal types with patient age.
In general, when specific serotypes were amply represented by both PIs and NPIs, there were few examples of preferential clonal associations with certain age groups. Within this survey, 22 clonal set-serotype associations were found only within NPIs (Table 4, indicated in boldface). While the significance of these observations, if any, is as yet unknown, it is possible that these represent strains that are more commonly present among individuals 5 years of age or older. For example, CC1292 represented 9 isolates (18%) of the serotype 6A NPIs characterized from the 2001 and 2002 isolate sets, yet it was not found among 51 PIs that accounted for nearly all serotype 6A PIs from the same period. Similarly, CC1296 and ST1340 were not found in extensive surveys of both 1999 and 2001 serotype 19F PIs, yet they were found in a limited sampling of year 2002 NPIs. Certain genotypes were found only among NPIs, because the corresponding serotypes were not found among PIs (serotypes 13, 15A, 28F, and 37).
Resistance phenotypes.
In general, resistance to antibiotics was uncommon within serotypes which were represented primarily by penicillin-susceptible isolates. Resistance to erythromycin was common within most penicillin-nonsusceptible genetic sets specific to PCV7 or PCV7R serotypes and is not described here in detail. Occasional instances of resistance to one or more of the antibiotics co-trimoxazole, tetracycline, and chloramphenicol occurred within most serotypes and are not generally described here. During 2001 and 2002 fluoroquinolone resistance was seen only within serotypes 3 (three isolates), 4 (three isolates), 6B (four isolates), 9N (one isolate), 14 (five isolates), 15A (one isolate), 15B (one isolate), 18C (three isolates), 19A (two isolates), 19F (four isolates), 20 (one isolate), 22F (one isolate), 23A (one isolate), and 35B (one isolate), with most assigned to clonal sets common to their serotype (26; this work). Penicillin nonsusceptibility at frequencies of 15% or higher within specific genetic sets is referred to in Fig. 2 with the letter P and includes serotypes 6A, 6B, 7B, 9A, 9N, 9V, 14, 16F, 19A, 19F, 23A, 23F, and 35B.
Comparison of PCV7NR serotypes pre- and postvaccination.
We targeted all available PIs from PCV7NR serotypes and additionally included certain NPIs nonsusceptible to erythromycin, clindamycin, beta-lactam antibiotics, or fluoroquinolones. Below, the resultant data for each serotype from the 2001 and the 2002 data sets are combined and compared to the year 1999 data.
(i) Serotype 1.
A majority of the serotype 1 isolates in the present study were grouped within ST227, similar to our findings in the prevaccination period (12). ST227 included all PIs, including one that was intermediately penicillin resistant. Two minor STs (STs 217 and 1174) accounted for a total of five year 2001 NPIs. The data are in agreement with those from a recent report showing that ST227 is characteristic of a lineage (lineage A) of serotype 1 pneumococci that are apparently exclusive to Europe and North America (4). ST1174 shares only three loci with ST227; however, it shares six loci with one of the other STs within lineage A. ST217 is typical of lineage B serotype 1 pneumococci that are predominant within Africa and Israel (4) and is reported to represent a hypervirulent clonal complex (18).
(ii) Serotype 3.
As in the year 1999 surveillance, the majority of serotype 3 isolates (both PIs and NPIs) after vaccine introduction were within clonal set ST180. These included 13 antibiotic-resistant NPIs, including all 3 levofloxacin-resistant serotype 3 NPIs from 2001 and 2002 (26) and 10 erythromycin-resistant isolates (including the single PI and 9 of 22 NPIs from the 2 years).
(iii) Serotype 5.
The two serotype 5 isolates (one PI and one NPI, both of which were collected in 2001) had PFGE profiles highly similar to the profile from the single NPI described in the year 1999 surveillance, which was ST653. ST653 is a DLV of ST289, the tetracycline-resistant PMEN clone Colombia5-24 (27). Only the year 2001 NPI was resistant to antibiotics (tetracycline and co-trimoxazole).
(iv) Serotype 7B.
None of the isolates from 2001 to 2002 were serotype 7B, and so no comparison to the single penicillin-nonsusceptible ST664 PI recovered in 1999 could be made.
(v) Serotype 7C.
The three serotype 7C isolates genotyped shared highly similar PFGE profiles. These included one PI and a penicillin-nonsusceptible, tetracycline-resistant NPI with the novel ST ST1207 (which has only three allelic identities with the closest matches).
(vi) Serotype 7F.
Consistent with the year 1999 surveillance results, 94% of the year 2001 and 2002 isolates formed a single highly related PFGE cluster represented by individual ST191 isolates. ST191 represented 90% of the PIs and both year 2002 penicillin-nonsusceptible NPIs (one of which was also cefotaxime nonsusceptible, tetracycline resistant, chloramphenicol resistant, and co-trimoxazole resistant).
(vii) Serotype 8.
Nine of 10 serotype 8 NPIs collected in 2001 (all 10 were collected in Minnesota) shared very similar PFGE profiles, one of which was ST1268 (two new alleles), a DLV of ST404 from serotype 8 isolates causing meningitis in Brazil and from type 8 carriage isolates in the United Kingdom (28). Both PIs from 2001 and the single NPI from 2002 were highly related by PFGE and were represented by ST53, which has been associated with serotype 8 isolates causing invasive disease in Brazil, The Netherlands, Spain, and the United Kingdom.
(viii) Serotype 10A.
Due to the presence of STs 816 and 1208, which were shared among a PFGE cluster of highly related profiles, the majority of serotype 10A isolates (including all PIs) were assigned to CC460, which includes ST460, an ST commonly seen among serotype 6A isolates. Two of the year 2001 NPIs recovered in Minnesota were assigned to ST1263, which is an SLV of STs 877 and 1299 described from invasive serotype 6B and 9V strains from Korea and Brazil, respectively (28).
(ix) Serotype 11A.
The majority of the serotype 11A isolates tested during all 3 years had closely related PFGE patterns and were assigned to CC62 due to the occurrence of ST62 and its SLV, ST630. ST62 has been described from invasive serotype 11A isolates recently recovered in Spain and Italy (28). Six erythromycin-resistant isolates (five NPIs and one PI) were found within CC62 during 2001 and 2002.
(x) Serotype 12F.
Ninety-eight percent of the serotype 12F isolates tested clustered within CC218 due to STs 218, 220, and ST1176 (a DLV of ST218). Isolates not related to CC218 included two year 2002 PIs with the unique STs 989 and 1376. We recently observed ST989 within serotype 12F carriage isolates recovered in a village in Kenya (21). ST1376 is a DLV of ST1292, which is found among PCV7 antibiotic-resistant serotype 6A NPIs described in this study; however, this serotype 12F isolate was not antibiotic resistant. All erythromycin-resistant year 2001 serotype 12F isolates (three PIs and two NPIs) were in CC218. Although no erythromycin-resistant year 2002 isolates were genotyped, 14 erythromycin-resistant serotype 12F isolates (13 NPIs and 1 PI) were detected in 2002.
(xi) Serotype 13.
There were no serotype 13 PIs in our surveillance. The two year 2001 NPIs were ST574, which, together with its DLV, ST392 (from a serotype 17F strain), comprise CC392. A single tetracycline-resistant year 2002 NPI had a unique, new ST, ST1260.
(xii) Serotype 15A.
There were no serotype 15A PIs in our surveillance. The 15 intermediately penicillin-resistant NPIs (which were also resistant to combinations of clindamycin, erythromycin, and tetracycline) tested from the years 2001 and 2002 were determined by PFGE and MLST to be identical or highly related to ST63 from the PMEN clone Sweden15A-25 (27). The single levofloxacin-resistant isolate from 2001 and 2002 (a year 2002 NPI) was ST817, which is unrelated to ST63 and which was initially reported from a serogroup 15 conjunctivitis isolate in Scotland (28).
(xiii) Serotypes 15B and 15C.
The incidence of ABCs PIs with serotypes 15B and 15C incrementally increased in 2001 and 2002 relative to the incidence in 1999 (Fig. 2, Table 2). Serotypes 15B and 15C interconvert (32), which is consistent with the observation that ST199 was the most common ST found among U.S. invasive isolates of both serotypes during this surveillance (Fig. 2; Table 4). Six of 7 erythromycin-resistant isolates from 2001 and 2002 (three PIs and three NPIs) were within CC199. Although ST199 and ST199-related STs are also the most common STs found within serotype 19A, serotype 19A isolates were commonly penicillin nonsusceptible, while serotype 15B and 15C isolates were uniformly penicillin sensitive. Four additional clonal sets (ST1262, ST1267, ST1259, and CC156) were observed among six isolates collected during 2002. CC156, which primarily accounts for multiresistant serotype 9V isolates, was shared by a single antibiotic-sensitive ST162 type 15B PI recovered in 2002. ST162 has previously been associated with three PCV7 serotypes (28) and also with serotype 24F isolates (25).
(xiv) Serotype 16F.
Two clonal clusters accounted for the 12 serotype 16F isolates genotyped during 2001 and 2002. CC659 accounted for the majority of the isolates, including all three PIs from the 3 years. The three year 2002 serotype 16F CC659 NPIs were erythromycin resistant, accounting for all macrolide resistance within this serotype in 2002. Three isolates were genotyped as CC30, including a levofloxacin-resistant NPI from 1999 and the single penicillin-nonsusceptible NPI from the year 2002 (26).
(xv) Serotype 17F.
The single serotype 17F PIs from the years 2001 and 2002 were STs 392 and 1253, respectively. ST392 is a DLV of ST574 associated with serotype 13 isolates.
(xvi) Serotype 20.
Five year 2001 serotype 20 isolates, including the single PI and the single levofloxacin-resistant serotype 20 isolate in the 2001 and 2002 surveillance, were assigned to CC899 due to inclusion in a PFGE cluster represented by new ST1261. ST1261 was an SLV of ST899 commonly found among serotype 4 isolates. Two antibiotic-sensitive NPIs were found to share the unrelated ST1257. ST1257, with its DLV ST1375 from a single penicillin-nonsusceptible year 2002 serotype 9N NPI, form CC1257 (Fig. 1 and 2).
(xvii) Serotype 22F.
Most of the serotype 22F isolates were within CC433, including one of the two year 2002 erythromycin-resistant isolates (both NPIs) and the single levofloxacin-resistant 22F isolate (an NPI) recovered during 2001 and 2002 (26). A single year 2001 NPI with a divergent PFGE profile was determined to be a novel DLV of ST156 typically found among serotype 9V isolates and other PCV7 serotypes; however, since this NPI could not be regrown to verify its serotype, the ST was not submitted to the MLST database. A single year 2002 PI was determined to have ST1294, which shares four or fewer identical alleles with all known STs.
(xviii) Serotypes 24B and 24F.
Despite serotype differences (serotype 24F versus serotype 24B), both isolates typed were highly related and comprised CC72. The single year 2001 serotype 24B PI was ST72; and the single year 2002 serotype 24F NPI tested was ST1291, a new DLV of ST72.
(xix) Serotype 28F.
A single tetracycline-resistant year 2002 serotype 28F NPI was ST546, previously described from a serogroup 28 middle ear NPI recovered in 1994 in Finland (28).
(xx) Serotype 31.
The four serotype 31 NPIs genotyped from the 3 years shared highly similar PFGE profiles, and two of these NPIs were directly determined to have ST568, previously described from a serotype 31 cerebrospinal fluid isolate recovered in Scotland (28). The 1999 and 2002 NPIs chosen for analysis showed elevated fluoroquinolone MICs (ciprofloxacin MICs, 4 to 8 μg/ml; levofloxacin MICs, 2 μg/ml).
(xxi) Serotype 33F.
There were incremental increases in the numbers of pneumococcal serotype 33F PIs causing invasive disease in 2001 and 2002 compared to the numbers in 1999 (Table 2; Fig. 2). Erythromycin-resistant serotype 33F PIs increased with time, accounting for one year 1999 PI (14%), six year 2001 PIs (37.5%; four were genotyped), and eight year 2002 PIs (42%). All but two of these isolates (96%) tested from the 3 years, including all erythromycin-resistant isolates, were CC662, which is comprised of STs previously associated with type 33F invasive isolates. Two antibiotic-sensitive year 2001 isolates (one PI and one NPI) were found to be ST1012, previously reported from a year 1998 serotype 11A cerebrospinal fluid isolate recovered in Poland (28).
(xxii) Serotype 35B.
Two unrelated clonal sets comprise serotype 35B in the United States, one of which is penicillin nonsusceptible (CC377, which includes PMEN clone Utah35B-24) and is often resistant to additional classes of antibiotics (2). The true ratios of these clones during the 3 years are most accurately depicted by the frequency of penicillin nonsusceptibility. All six PIs recovered within the 3 years were penicillin resistant and on the basis of MLST and PFGE are within CC377 (Fig. 2; Table 4). Fifty-six of 74 (76%) year 2001 and 2002 isolates were penicillin nonsusceptible, and all 4 PIs were penicillin resistant. The two levofloxacin-resistant isolates from ABCs (1999 and 2001) were also penicillin nonsusceptible and within CC377 (26).
(xxiii) Serotype 35F.
One of the four year 2001 serotype 35F isolates was ST498 and had a PFGE profile highly similar to those of the other three isolates tested, which included two PIs. ST498 was previously associated with a serogroup 35 carriage isolate recovered in Finland in 1994 (28).
(xxiv) Serotype 38.
The numbers of invasive cases due to serotype 38 PIs incrementally increased in 2001 and 2002 relative to the numbers of cases in 1999 (Fig. 2; Table 2). All isolates examined from the 3 years (27 PIs and 1 NPI) were highly related on the basis of combined PFGE and MLST analysis. Four independent isolates were sequence typed as ST393. ST393 was previously associated with a type 38 carriage strain recovered during 1998 in the United Kingdom (28).
Pre- and postvaccination genetic structures of PCV7R serotypes. (i) Serotype 6A.
Serotype 6A displayed a total of 12 different clonal sets among the isolates examined from all 3 years, 4 of which (CC1296, CC460, CC473, and ST395) were evident among multiple isolates in the present study as well as among multiple isolates in 1999. Eight of the 12 clonal sets exhibited high levels of penicillin nonsusceptibility and resistance to other antibiotics. As in 1999, CC1296 (including ST376 of the PMEN clone North Carolina6A-23 [14, 27]) was common in 2001 and 2002.
The six distinct clonal sets CC490, CC138, ST690, ST1091, ST1175, and CC1292 accounted for 17 serotype 6A isolates from 2001 and 2002 and were not found among year 1999 isolates. Nine CC1292 NPIs (six penicillin-nonsusceptible isolates, including five isolates resistant to multiple antibiotics) that shared a high level of PFGE profile similarity, were recovered in 2001 and 2002 from four states. Two of these nine CC1292 NPIs were directly sequence typed as the newly discovered ST1292. ST1175, represented by a single isolate, was characterized by three new alleles and differed from its closest match at mlst.net by four loci. STs 138, 490, and 1091 have previously been seen among serotype 6A isolates from the United Kingdom, Finland, and Zambia, respectively (28) (note that ST138 is most commonly seen among serotype 6B isolates). ST690 has been associated with a serogroup 6 strain causing otitis media in the United States (28).
(ii) Serotype 9A.
As reported for the year 1999 surveillance (12), numerous (n = 29) year 2001 isolates were mistakenly originally serotyped as 9A when they were actually serotype 9V, were resistant to multiple antibiotics, and were members of CC156. Only 3 of 32 multiresistant year 2001 NPIs initially tested as serotype 9A were still found to be serotype 9A after they were retested. These three serotype 9A isolates were also designated CC156, since one was directly typed as ST156 and all three had PFGE profiles nearly indistinguishable from those of multiple independent ST156 serotype 9V isolates. Of nine year 2002 NPIs (all penicillin nonsusceptible) initially tested as serotype 9A, six were subsequently found to be serotype 9V (and CC156), two were serotype 6A (one was CC473 and one was CC1292), and one was serotype 19A (CC199).
(iii) Serotype 9N.
Serotype 9N was comprised primarily of antibiotic-sensitive CC66 isolates during 1999 (12), and similar results were obtained for the 2001 and 2002 surveillance (Fig. 2). Single erythromycin-nonsusceptible and penicillin-nonsusceptible CC66 year 2001 NPIs were observed. The single levofloxacin-resistant serotype 9N isolate within ABCs was a year 2002 representative of CC156 (ST1206, a DLV of ST156 [26]. A single penicillin-nonsusceptible CC1257 (ST1375) NPI was also observed among the five NPIs from 2002.
Four year 2002 isolates (three multiresistant NPIs and one antibiotic-sensitive PI) were initially erroneously labeled as serotype 9N and were found to be either serotype 9V (3 CC156 NPIs) or nonserotypeable.
(iv) Serotype 19A.
Unlike other PCV7 and PCV7R serotypes, there was an increase in the numbers of serotype 19A PIs in 2001 and 2002 relative to the numbers in 1999 (Fig. 1, Table 2). It is also evident that the numbers of serotype 19A isolates recovered from both age groups have increased incrementally each year in the period from 2001 to 2004 (23). Although about 70% of the 206 isolates analyzed in each of the 3 years shared CC199, 10 additional genetic sets were detected, 9 of which were comprised primarily of penicillin-nonsusceptible isolates with additional resistance determinants. Four of these nine multiply resistant clonal sets were comprised of isolates highly related to PMEN clones.
Multiresistant isolates highly related to PMEN clone Colombia23F-26 (CC172) were apparent during all 3 years. Isolates related to Spain23F-1 (CC81) were present in 1999 and 2002.
CC236, the clonal complex with Taiwan19F-14 (ST236) as the predicted founder, was detected as a single multiply resistant ST271 PI within the 2001 isolate set. CC236 accounted for an additional ST (ST320) and five multiply resistant isolates (three PIs and two NPIs) among the year 2002 isolate set. Three of these isolates had very high levels of resistance to beta-lactam antibiotics (MICs of 8 μg/ml for both penicillin and amoxicillin). It will be interesting to determine whether these multiresistant isolates carry both ermB and mefA determinants, as do other strains closely related to Taiwan19F-14 that were recently characterized in a global study of community-acquired respiratory tract infections (10).
CC1296, which contains ST376 from North Carolina6A-23, was found only among the year 2002 isolates, accounted for nine isolates (five NPIs and four PIs) that were resistant to penicillin and cefotaxime and that were also resistant to erythromycin and co-trimoxazole. ST376 and ST1339 (a DLV of ST376) represented this nine-isolate cluster.
(v) Serotype 23A.
Both serotype 23A PIs recovered from 1999 were penicillin nonsusceptible and were found to be ST338 (CC172), which is the genotype of PMEN clone Colombia23F-26. Of eight penicillin-intermediate year 2002 type 23A NPIs, we genotyped five and found that each isolate shared ST338 and/or highly similar PFGE profiles with Colombia23F-26. We suspect that the six penicillin-nonsusceptible NPIs among the 27 serotype 23A NPIs obtained during 2001 were also highly related to Colombia23F-26, but these isolates were not genotyped. We have subsequently encountered CC172 from PIs and NPIs recovered in 2003 (24).
The four penicillin-susceptible year 2001 isolates genotyped yielded three different genotypes, including the new ST1336 (a DLV of three other STs from serotype 23A isolates), ST1338 (a DLV of five other STs from serotype 23F isolates), and ST42. Three CC37 isolates (including two that were ST42, which is a DLV of ST37 [Tennessee23F-4] and which is associated with serotype 23A) were found in 2001 and 2002. Two of these three CC37 isolates were antibiotic resistant (one was resistant to clindamycin, erythromycin, co-trimoxazole, and tetracycline, while the other was resistant to levofloxacin). The levofloxacin-resistant isolate was ST629 and was originally found from an antibiotic-sensitive serotype 23F pediatric invasive isolate (11).
Genetic structures of PCV7 serotypes.
We targeted all available PIs from the 1999 ABCs (12) and the 2001 ABCs. Additionally, we added some NPIs for some serotypes, often targeting antibiotic-resistant isolates. Although PCV7 serotypes were not initially targeted from the 2002 data set, a small number of PCV7 serotype isolates that were originally mistakenly identified as non-PCV7 serotypes were included from the 2002 ABCs. Fluoroquinolone-resistant isolates were included from the 2002 ABCs.
(i) Serotype 4.
As in 1999, CC899 was the major genetic cluster, accounting for 68% of serotype 4 isolates in 2001 (Fig. 2; Table 2). It also included all four levofloxacin-resistant CC899 isolates (all NPIs) from the 2001 and 2002 ABCs (two of these were genotyped previously [26]). ST205 accounted for 17 (24.6%) of the year 2001 serotype 4 isolates and 11.3% of the year 1999 serotype 4 isolates that were genotyped (Fig. 2).
(ii) Serotype 6B.
The four most frequently occurring clonal sets were the same between the 1999 and 2001 isolate sets (CC138, CC146, ST90 [Spain6B-2], and CC384 [Maryland6B-17]), and three of these represented primarily multiresistant isolates (CC146, ST90, and CC384). Maryland6B-17 was the most recently characterized of these multiresistant clones and was originally discovered among invasive isolates recovered in the United States in 1997 (14). The five year 2001 and 2002 serotype 6B levofloxacin-resistant isolates were represented by ST138 (one NPI), CC146 (two NPIs) (26), ST690 (one NPI) (26), and ST90 (one NPI).
The remaining clonal set from year 2001 to 2002, CC490, was comprised of penicillin-nonsusceptible isolates with nearly identical PFGE profiles associated with the DLVs ST1177 and ST1209.
(iii) Serotype 9V.
Serotype 9V, unlike other PCV7 serotypes comprised of a large percentage of penicillin-nonsusceptible isolates, appeared to have a relatively simple genetic structure during all 3 years. Although two different clonal sets are shown for the years 1999 and 2001 because each had a distinct clonal group and founder ST defined by eBURST analysis of the study STs (Fig. 1), these STs all lie within the same complex when the stringency is relaxed to include each ST that shares five identical alleles with at least one other member ST. All STs of CC156 and CC1269 share at least four identical alleles with ST156. Additionally, all of the component STs of these two genetic sets are within the same large eBURST group (73 STs, with founder ST156) when eBURST is performed with all known STs.
The nine CC156 isolates shown for year 2002 were all multiply resistant and were originally erroneously typed as either serotype 9A (six NPIs) or serotype 9N (two PIs and one NPI).
(iv) Serotype 14.
Little difference between the genetic structure of the year 1999 PIs (208 isolates; 63.5% of the total serotype 14 PIs) and that in the 2001 data set (125 isolates, including 94.8% of total serotype 14 PIs) was seen. During both years, CC13 and CC124 accounted for the majority of the penicillin-nonsusceptible and the penicillin-susceptible isolates, respectively, and together accounted for more than 80% of the serotype 14 isolates. Although ST9 is predicted to be a more likely founder of this clonal complex when eBURST is extended to all known STs, ST13 was the most commonly encountered ST within CC13 and was the predicted founder by the use of only STs from this study (Fig. 1).
While most isolates grouped within CC13 and CG124 during 1999 and 2001, the rest of the serotype 14 isolates were comprised of penicillin-nonsusceptible isolates from eight minor clonal sets. Three of these clonal sets were found only among the year 2001 isolates, including CC63, CC156, and ST554. It is interesting that two of these clonal sets represent clonal groups that include PMEN clones of different serotypes, including Sweden15A-25 and Spain9V-3.
The five levofloxacin-resistant type 14 isolates in the 2001 and 2002 ABCs included four CC13 NPIs (26) and a single CC156 NPI. These included the only year 2002 isolates characterized (three CC13 NPIs).
(v) Serotype 18C(BF).
Ten isolates of serotype 18B or 18F are also included among the three data sets since all were found to be of CC113 or ST496, which are both commonly found within serotype 18C. These data suggest the possibility that serotypes 18C, 18B, and 18F interconvert.
The clonal distributions were similar within the 1999 and 2001 data sets. The five levofloxacin-resistant serogroup 18 ABCs isolates from 2001 and 2002 represented both CC113 (26) and ST496. Among the 2002 isolates was a PI that was resistant to clindamycin, erythromycin, co-trimoxazole, and tetracycline. This PI had a PFGE profile highly similar to that of the previously typed ST666 isolate from the year 1999 isolate set. The two other year 2002 PIs examined were serotype 18F and were within the major CC, CC113.
(vi) Serotype 19F.
Serotype 19F exhibited 14 distinct clonal sets (more than any other serotype), 7 of which were associated with penicillin-resistant isolates (Fig. 2). Clonal sets representing four PMEN clones were detected within the 1999 and the 2001 isolate sets, with CC236 (Taiwan19F-14) accounting for 20.9% of the PIs during 1999 and 21.3% of the PIs during 2001. CC236 consisted entirely of multiresistant isolates. Nine multiresistant year 2002 isolates (one PI and eight NPIs) containing both mef and erm macrolide resistance determinants were found to be within CC236 (Taiwan19F-14) (L. McGee, unpublished data), which is consistent with a recent report concerning this clonal complex (10). The proportion of CC81 (Spain23F-1) PIs, all of which were multiresistant, decreased from 11.8% in 1999 to 6.4% in 2001 (and CC81 was additionally found among the NPIs tested in 2001). CC177 (Portugal19F-21) was also detected among PIs during both years. A single ST423 isolate (a DLV of England14-9) was present only among the 1999 PIs. Two of the four year 2001 levofloxacin-resistant isolates (all NPIs) were CC81 and were directly typed as ST81 and its SLV, ST83 (26). There were no year 2002 levofloxacin-resistant serotype 19F isolates.
CC251 was the most common clonal complex in both 1999 (20% of the PIs) and 2001 (40.4% of the PIs and 18.2% of the NPIs examined). Other penicillin-susceptible clonal complexes represented during both 1999 and 2001 included CC177, CC425, and ST43.
Two additional year 2002 isolates originally mistakenly assigned to non-PCV7 serotypes were found to have the new unique ST (ST1340) and ST1203, which is associated with serotype 19F isolates causing multiple otitis media cases in Costa Rica from 2000 to 2003 (28).
(vii) Serotype 23F.
Serotype 23F was primarily represented by PMEN clones. Three multiresistant clonal complexes, CC37 (Tennessee23F-4), CC81 (Spain23F-1), and ST242 (Taiwan23F-15), were major constituents of serotype 23F in 1999 as well as in 2001. CC37 constituted 70 to 72% of PIs in 1999 and 2001, as well as 46% of the year 2001 NPIs genotyped. CC81 comprised 13.5 to 17.2% of the PIs from 1999 to 2001 (and four year 2001 NPIs). Three CC81 multiresistant NPIs, including one with ST713 (a DLV of ST81), were the sole levofloxacin-resistant serotype 23F isolates within the 2001 and 2002 ABCs (genotyping data for two of these isolates were shown previously [26]).
ST242 accounted for 6.1 to 6.3% of the isolates tested during 1999 and 2001. A double-locus variant of ST268 (Hungary19A-6), ST1212, was discovered in two multiresistant isolates (one PI and one NPI). Two penicillin-susceptible ST460 isolates were also detected among the year 2001 PIs.
Four year 2002 multiresistant NPIs were tested due to erroneous original serotyping results indicating novel associations of high-level beta-lactam resistance (penicillin MICs, 8 μg/ml or greater) with serotypes 23A and 23B. All four isolates were found to be serotype 23F; three of these were within CC81, and one was in ST242.
DISCUSSION
The introduction of PCV7 has resulted in a dramatic reduction in invasive pneumococcal infections caused by the serotypes targeted by the vaccine (33). However, the long-term effectiveness of the vaccine is not clearly known, in part due to the unknown extent that replacement disease—an increase in the incidence of non-PCV7 serotype disease—will become a factor. It is possible that PCV7 could have profound effects upon the prevalence and strain composition of individual serotypes that it does not target. The marked increase in the incidence of invasive disease due to serogroups not included in PCV7 (and serotype 19A) in 2002 relative to that in 1999 (P ≤ 0.001) provides ample rationale for continued monitoring of individual serotypes within this set (Table 1). It follows that certain individual non-PCV7 serotypes (serotypes 3, 7F, 15B/C, 19A, 22F, 33F, and 38) individually accounted for significantly higher proportions of invasive cases among children <5 years of age in 2002 relative to the proportions in 2001 (Table 2). These data are consistent with those from a recent report of the increase in the number of invasive pediatric cases due to serogroups 15 and 33 in the post-PCV7 era (16). The cumulative data indicate a need for continued monitoring of invasive pneumococci to detect changes in serotype-genotype associations and to determine the effects of the vaccine on the pneumococcal population genetic structure. The focus of this work has therefore been to provide continuing serotype-specific data on invasive strains and to compare those data to our pre-PCV7 data (12).
Sharing of the same ST among two or more serotypes is presumably indicative of past horizontal transfer events of capsular type-specific loci. Our data suggest that such capsular switching is a rare event since during the 3 years of this molecular surveillance study and the analysis of over 2,100 isolates we have detected only 11 STs associated with multiple serotypes. This is consistent with the findings of a recent study which found no evidence of capsular switching among 333 carriage isolates recovered from 100 children, including all isolates of different serotypes recovered from the same child (20). It could be argued that we did not identify more putative instances of capsular switching because only 285 of these isolates were actually submitted to MLST (data not shown). We contend that our strategy of strategic MLST of representative related clusters within serotypes and of isolates involved in all unusual PFGE associations between isolates of divergent serotypes actually allows the targeting of more unusual multilocus sequence type associations than a strategy that uniformly targets a more limited number of isolates for MLST. In only 1 of the 11 instances of a shared ST between isolates of different serotypes (STs 62923F,23A, 8123F,19F, 1386A,6B, 1569A,9V, 19919A,15BC,19F, 27119F,19A, 4606A,23F, 6446B,19F, 3766A,19A, 906B,6A, 696A,6B) was the ST (ST199) associated with both PCV7-PCV7R and PCV7NR serotypes. However, it is noteworthy that ST199 accounted for the majority of serotype 15B/C and 19A isolates during 1999, even before the introduction of PCV7 (12). The remaining 10 STs were shared only between isolates of PCV7 or PCV7R serotypes. Our data and the global data reveal relatively few instances in which identical or highly related STs are associated with both PCV7-PCV7R and PCV7NR serotypes. These findings are in marked contrast to those presented in a recent report (15) describing 11 instances of STs shared among multiple serotypes from a sample set of only 252 isolates, in which four STs were shared among PCV7-PCV7R and PCV7NR serotypes and in which three STs accounted for three to four different serotypes that included PCV7-PCV7R serotypes together with PCV7NR serotypes. In general, the data from our study indicate an extensive history of capsular switching between penicillin-nonsusceptible strains restricted to the PCV7 and the PCV7R serotypes but few deduced instances of genetic exchange involving strain pairs that represent both a PCV7-PCV7R serotype and a PCV7NR serotype. CC13, CC37, CC81, CC156, CC172, CC199, CC236, CC268, CC1269, and CC1296 are examples of clonal complexes documented here and elsewhere that are primarily associated with antibiotic-resistant strains and multiple PCV7 and PCV7R serotypes.
Of interest was the single example in which a single PCV7NR serotype (serotype 15B) PI had an ST (ST162 within CC156) that was previously reported primarily among strains with PCV7 serotypes (28). Within CC156, we found an ST162 serotype 15B isolate recovered from the blood of an infant. ST162 is primarily associated with penicillin-sensitive and intermediately penicillin-resistant strains with PCV7 serotypes 19F, 9V, and 14 (28). The recent observation of multiply resistant serotype 11A isolate derivatives of the predicted founder strain Spain9V-3 (ST156) within CC156 also supports the need for the continued monitoring of the clonal composition of serogroup 15 (29). Although such occurrences appear to be rare at present, the constant usage of PCV7 may aid the expansion of such serotype switch variants with implications for the long-term effectiveness of the current vaccine.
The clonal structures of most serotypes characterized appeared to be stable by comparison to the prevaccine clonal structure data. Only 9 of the 83 clonal sets included both PCV7-PCV7R and PCV7NR serotypes, with only 3 of these clonal sets (CC156, CC1292, and CC899) in the post-PCV7 era showing new associations not previously seen in the pre-PCV7 period. Furthermore, three clonal sets which consisted primarily of PCV7NR serotypes prior to the introduction of PCV7 showed an association with PCV7-PCV7R serotypes only after PCV7 introduction (CC62 and CC63) or single isolates prior to PCV7 introduction (CC662). The other examples of such associations with both PCV7-PCV7R and PCV7NR serotypes include CC199, CC460, CC1257, and CC199. It is likely that CC460 was common within both serotype 6A and serotype 10A isolates before and after PCV7 introduction (Fig. 2). CC1257 was seen among two serotype 20 isolates (ST1257) in 2001 and in a single serotype 9N isolate (ST1375) in 2002. Since multiple SLVs and DLVs of these STs are documented at mlst.net in association only with serotype 20 isolates, it is likely that CC1257 originated within serotype 20.
Of the nine clonal sets inclusive of both PCV7-PCV7R and PCV7NR serotypes, CC199 is currently the most important. Before and after the introduction of PCV7, CC199 accounted for the majority of both serotype19A and 15B/C isolates (Fig. 2). Recent ABCs data indicate that the rate of invasive pneumococcal disease associated with serotype 19A is on the increase (23). Also, other reports have indicated that serogroup 15 is an important component of serotype replacement occurring in vaccinated children (13). Serogroup 15 and serotype 19A showed the most pronounced increases in incidence among invasive pediatric isolates in 2002 relative to the incidence in 1999 (Table 2); and these increases were associated with a corresponding increase in the incidence of CC199, which constituted 70 to 92% of both the serogroup 15 and the serotype 19A isolate sets in 2001 and 2002 (Fig. 2). This trend has continued among the more recent year 2004 isolates analyzed (23).
Since ST199 was the most common ST from both serotype 15B/C and serotype 19A isolates recovered from 1999 to 2002, presumably serotype 15B/C and 19A CC199 strains have a recent common ancestry. In contrast to serotype 15B/C isolates, which are penicillin susceptible, the majority of serotype 19A isolates are penicillin nonsusceptible, indicating that the serotype 19A derivative of ST199 might have originated following the transformation of a type 15B/C ST199 strain with serotype 19A-specific cps locus genes. It appears that PCV7 does not protect against the PCV7-related serotype 19A, which is consistent with the findings in a recent report in which a nine-valent conjugate vaccine protected against carriage by vaccine types and vaccine-related serotype 6A, with a concomitant increase in the rate of carriage of nonvaccine types (6). Significantly, this vaccine offered reduced protection against vaccine serotype 19F and did not reduce the rate of carriage of serotype 19A strains (6). These findings are also consistent with the results found for serotypes 19A and 19F in a clinical trial assessing the efficacy of PCV7 against acute otitis media (9). PCV7 may have created an improved niche within the pediatric population for the proliferation of strains with specific nontargeted serotypes.
The cumulative data from this genotypic surveillance indicate that clonal associations remained relatively stable overall from 1999 to 2002, with PCV7 potentially having a modest, yet important, impact on the clonal composition within individual serotypes during 2001 and 2002. For example, although serotype 19A showed a constant proportion of CC199 isolates during the 3 years of surveillance, we have noted an increase in the rate of antibiotic resistance in 2003 and 2004 among isolates within this serotype (23), due in part to the continuing presence and increased incidence of clonal complexes, such as CC236, CC1296, and CC81, that first appeared in our serotype 19A surveillance isolates during 2001 and 2002 (Fig. 2; Table 4).
Among the PCV7NR serotypes, two additional clonal types have accounted for a larger proportion of pneumococcal isolates from invasive pediatric disease cases within the ABCs following PCV7 implementation. CC62 (which a showed a more than twofold increase in both years, with 14 to 20 serotype 33F isolates, respectively), and ST393 (which a showed a more than threefold increases incidence in 2002 relative to that in 1999, with 17 serotype 38 isolates) were observed to be the predominant clonal complexes within their respective serotypes even before PCV7 implementation (Fig. 2; Table 4). These data correspond to the overall increase in the incidence of disease caused by serotypes 33F and 38 in 2002 relative to the incidence in 1999 (Table 3). Extended surveillance will monitor the potential for these two clones to become common invasive pathogens in children.
Although the genetic structure of most serotypes remained relatively stable, the composition among some serotypes (e.g., serotypes 6A, 15B/C, and 19A) appeared to be altered to some degree through the occurrence of additional clonal sets when pre- and postvaccination data were compared (Fig. 2). Although clonal sets were assigned for only 72 to 77% of the serotype 6A and 19A isolates from 1999 (Fig. 2), we were unable to find matches between unassigned PFGE profiles from year 1999 isolates with PFGE profiles from the 2001 and 2002 data set. Therefore, it is unlikely that new clonal groups within these serotypes that were identified during 2001 and 2002 accounted for a major portion of unassigned year 1999 isolates. ST1292, one of the clonal sets not found within the 1999 serotype 6A isolates, shared only four allelic identities with its closest match at mlst.net at the time of its discovery within the 2001 isolate set. This ST is an SLV of ST1379 subsequently reported from a serotype 6A blood isolate recovered in Sweden in 2000 (28). It is interesting that within this nine-isolate CC1292 set, a full range of penicillin resistance phenotypes is evident (sensitive, intermediately resistant, and fully resistant), indicating that this strain might be rapidly acquiring resistance.
Although this study has documented the maintenance of established serotype-clonal group associations and has documented new associations in the post-PCV7 era, it has limitations. The findings here are limited to strains obtained immediately after the introduction of PCV7. Pneumococcal populations evolve gradually, warranting long-term surveillance. Also, the study does not account for all pneumococcal isolates collected in the ABCs database, although the majority of the pediatric isolates from each year are accounted for (this is true even for 2002, where PCV7 serotypes were largely omitted). Nonetheless, the data do serve as a continuing reference point for future molecular studies that can be used to assess the impact of the vaccine upon the spectrum of invasive isolates collected from children in the United States. Additionally, targeted analysis of specific isolates from older individuals allows assessment of the clonal types associated with important antibiotic resistance.
Genotyping of surveillance isolates led to a fairly extensive list of corrections of the phenotypic data, primarily due to the retesting of isolates with unusual serotype or resistance associations with STs. A modest degree of serotyping error has been found within other reference laboratories (17), emphasizing that additional levels of quality control are desirable. We are currently targeting all isolates with serotypes associated with unusual antibiotic resistances or genotypes for the detection of potential phenotyping errors. The pneumococcal MLST database has proved very valuable for screening of serotype associations in this regard.
Recent studies have indicated that it is possible that pneumococcal serotypes, rather than specific clonal features, are the primary predictors of pneumococcal virulence (3, 5). These studies have relied upon the relative number of isolates of specific clones and serotypes isolated from invasive infections relative to their occurrence during carriage. A contrasting report of a study that used a similar strategy indicated that specific clonal features other than the serotype could be important predictors of virulence (30). It is possible that this general approach is misleading, since in many geographic locations a specific serotype might be limited to very little clonal variation. It is also possible that insufficient numbers of isolates within specific clonal types were recovered from either invasive infections or carriers, making comparisons difficult. Other potentially confounding aspects include the potential to not detect cocolonizing carriage strains and not detect carriage strains present in the upper respiratory tract in small numbers. One recent report (22) of a study that used a mouse model clearly showed profound differences in the invasiveness of two strains of various backgrounds that shared the same serotype and showed the similar invasiveness of two different serotypes that shared an otherwise identical genotype. It is hoped that further investigations will shed light on this issue. Continued meaningful integration of genetic identifiers, phenotypic data, disease manifestations, and demographic information will help to provide a better understanding of the epidemiology of this important pathogen.
Acknowledgments
We are indebted to all of the hospitals and laboratories participating in CDC's Emerging Infections Program Network (EIP) and the EIP core program ABCs. We thank Tamara Pilishvili, Carolyn Wright, and other CDC ABCs-EIP members for providing isolate information. We thank Kathleen Shutt from the University of Pittsburgh Graduate School of Public Health and School of Medicine for statistical support. We gratefully acknowledge the laboratory support of Delois Jackson, Alma Ruth Franklin, Varja Sakota, Zhongya Li, Veena Kolli, Lynn (Lingling) Chen, Reyna Gilbert, Saundra Mathis, Tim Bailiff, and Shantia Warren. Special thanks also to Lynn Shewmaker for helping to improve the efficiency of ABCs isolate testing and information recording. We thank the pneumococcal MLST database curator, Angela Brueggemann, for designation of new alleles and allelic profiles.
This work was supported through the National Vaccine Program Office and the CDC Opportunistic Infections Working Group. Marie-Joe Medina was an Emerging Infectious Diseases Laboratory training fellow, sponsored by the Association of Public Health Laboratories and funded by CDC. Rekha Pai was an International Emerging Infectious Diseases postdoctoral fellow sponsored by Eli Lilly & Company in partnership with the Association of Public Health Laboratories, CDC, and the CDC Foundation. We acknowledge the pneumococcal MLST database, which is located at Imperial College London and which is funded by the Wellcome Trust.
The ABCs team is represented here by the following members: Monica M. Farley, Emory University School of Medicine and the Veterans Affairs Medical Center, Atlanta, Ga.; James Hadler, Connecticut Department of Public Health, Hartford; Lee H. Harrison, Johns Hopkins University Bloomberg School of Public Health, Baltimore, Md.; Nancy M. Bennett, Monroe County Department of Health and University of Rochester, Rochester, N.Y.; Ruth Lynfield and John Besser, Minnesota Department of Health, Minneapolis; Arthur Reingold, School of Public Health, University of California, Berkeley; Paul Cieslak, Oregon Department of Human Services, Portland; Allen Craig, Tennessee Department of Health; and James H. Jorgensen, University of Texas Health Science Center, San Antonio.
REFERENCES
- 1.Active Bacterial Core Surveillance. 26. December 2004, posting date. http://www.cdc.gov/ncidod/dbmd/abcs/.
- 2.Beall, B., M. C. McEllistrem, R. E. Gertz, D. J. Boxrud, J. M. Besser, L. H. Harrison, J. H. Jorgensen, C. G. Whitney, and the Active Bacterial Core Surveillance/Emerging Infections Program Network. 2002. Emergence of a novel penicillin-nonsusceptible, invasive serotype 35B clone of Streptococcus pneumoniae within the United States. J. Infect. Dis. 186:118-122. [DOI] [PubMed] [Google Scholar]
- 3.Brueggemann, A. B., D. T. Griffiths, E. Meats, T. Peto, D. W. Crook, and B. G. Spratt. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae and serotype- and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424-1432. [DOI] [PubMed] [Google Scholar]
- 4.Brueggemann, A. B., and B. G. Spratt. 2003. Geographic distribution and clonal diversity of Streptococcus pneumoniae serotype 1 isolates. J. Clin. Microbiol. 41:4966-4970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brueggemann, A. B., T. E. Peto, D. W. Crook, J. C. Butler, K. G. Kristinsson, and B. G. Spratt. 2004. Temporal and geographic stability of the serogroup-specific invasive disease potential of Streptococcus pneumoniae in children. J. Infect. Dis. 190:1203-1211. [DOI] [PubMed] [Google Scholar]
- 6.Dagan, R., N. Givon-Lavi, O. Zamir, M. Sikuler-Cohen, L. Guy, J. Janco, P. Yagupsky, and D. Fraser. 2002. Reduction of nasopharyngeal carriage of Streptococcus pneumoniae after administration of a 9-valent pneumococcal conjugate vaccine to toddlers attending day care centers. J. Infect. Dis. 185:927-936. [DOI] [PubMed] [Google Scholar]
- 7.Enright, M., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 8.Enright, M. C., and B. G. Spratt. 1999. Extensive variation in the ddl gene of penicillin-resistant Streptococcus pneumoniae results from a hitchhiking effect driven by the penicillin-binding protein 2b gene. Mol. Biol. Evol. 16:1687-1695. [DOI] [PubMed] [Google Scholar]
- 9.Eskola, J., T. Kilpi, A. Palmu, J. Jokinen, J. Haapakoski, E. Herva, A. Takala, H. Kayhty, P. Karma, R. Kohberger, G. Siber, and P. H. Makelä. 2001. Efficacy of a pneumococcal conjugate vaccine against acute otitis media. N. Engl. J. Med. 344:403-409. [DOI] [PubMed] [Google Scholar]
- 10.Farrell, D. J., S. G. Jenkins, S. D. Brown, M. Patel, B. S. Lavin, and K. P. Klugman. 2005. Emergence and spread of Streptococcus pneumoniae with erm(B) and mef(A) resistance. Emerg. Infect. Dis. 11:851-858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feil, E. J., B. C. Li, D. M. Aanensen, W. P. Hanage, and B. G. Spratt. 2004. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J. Bacteriol. 186:1518-1530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gertz, R. E., M. C. McEllistrem, D. J. Boxrud, Z. Li, V. Sakota, T. A. Thompson, R. R. Facklam, J. M. Besser, L. H. Harrison, C. G. Whitney, and B. Beall. 2003. Clonal distribution of invasive pneumococcal isolates from children and selected adults in the United States prior to 7-valent conjugate vaccine introduction. J. Clin. Microbiol. 41:4194-4216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ghaffar, F., T. Barton, J. Lozano, L. S. Muniz, P. Hicks, V. Gan, N. Ahmad, and G. H. McCracken, Jr. 2004. Effect of the 7-valent pneumococcal conjugate vaccine on nasopharyngeal colonization by Streptococcus pneumoniae in the first 2 years of life. Clin. Infect. Dis. 39:930-938. [DOI] [PubMed] [Google Scholar]
- 14.Gherardi, G., C. G. Whitney, R. R. Facklam, and B. Beall. 2000. Major sets of genetically related β-lactam resistant pneumococci in the United States based upon PFGE and pbp1a-pbp2b-pbp2x-dhf restriction profiles. J. Infect. Dis. 181:216-229. [DOI] [PubMed] [Google Scholar]
- 15.Johanna, M., C. Jefferies, A. Smith, S. C. Clarke, C. Dowson, and T. J. Mitchell. 2004. Genetic analysis of diverse disease-causing pneumococci indicates high levels of diversity within serotypes and capsule switching. J. Clin. Microbiol. 42:5681-5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaplan, S. L., E. O. Mason, Jr., E. R. Wald, G. E. Schutze, J. S. Bradley, T. Q. Tan, J. A. Hoffman, L. B. Givner, R. Yogev, and W. J. Barson. 2004. Decrease of invasive pneumococcal infections in children among 8 children's hospitals in the United States after the introduction of the 7-valent pneumococcal conjugate vaccine. Pediatrics 113:443-449. [DOI] [PubMed] [Google Scholar]
- 17.Konradsen, H. B. 2005. Validation of serotyping of Streptococcus pneumoniae in Europe. Vaccine 23:1368-1373. [DOI] [PubMed] [Google Scholar]
- 18.Leimkugel, J., A. Adams Forgor, S. Gagneux, V. Pfluger, C. Flierl, E. Awine, M. Naegeli, J. P. Dangy, T. Smith, A. Hodgson, and G. Pluschke. 2005. An outbreak of serotype 1 Streptococcus pneumoniae meningitis in northern Ghana with features that are characteristic of Neisseria meningitidis meningitis epidemics. J. Infect. Dis. 192:192-199. [DOI] [PubMed] [Google Scholar]
- 19.McEllistrem, M. C., J. Adams, E. O. Mason, and E. R. Wald. 2003. Epidemiology of acute otitis media caused by Streptococcus pneumoniae before and after licensure of the 7-valent pneumococcal protein conjugate vaccine. J. Infect. Dis. 188:1679-1684. [DOI] [PubMed] [Google Scholar]
- 20.Meats, E., A. B. Brueggemann, M. C. Enright, K. Sleeman, D. T. Griffiths, D. W. Crook, and B. G. Spratt. 2003. Stability of serotypes during nasopharyngeal carriage of Streptococcus pneumoniae. J. Clin. Microbiol. 41:386-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Medina, M. J., C. M. Greene, R. E. Gertz, R. R. Facklam, G. Jagero, M. Hamel, Y. P. Shi, L. Slutsker, D. R. Feikin, and B. Beall. 2005. Novel antibiotic-resistant pneumococcal strains recovered from the upper respiratory tracts of HIV-infected adults and their children in Kisumu, Kenya. Microb. Drug Resist. 11:9-17. [DOI] [PubMed] [Google Scholar]
- 22.Mizrachi Nebenzahl, Y., N. Porat, S. Lifzitch, S. Novick, A. Levi, E. Ling, O. Liron, S. Mordechai, R. K. Sahu, and R. Dagan. 2004. Virulence of Streptococcus pneumoniae may be determined independently of the capsular polysaccharide. FEMS Microbiol. Lett. 233:147-152. [DOI] [PubMed] [Google Scholar]
- 23.Pai, R., M. R. Moore, T. Pilishvili, R. E. Gertz, C. G. Whitney, and B. Beall. 2005. Postvaccine genetic structure of Streptococcus pneumoniae serotype 19A from children in the United States. J. Infect. Dis. 192:1988-1995. [DOI] [PubMed] [Google Scholar]
- 24.Pai, R., R. E. Gertz, C. G. Whitney, and B. Beall. 2005. Clonal association between Streptococcus pneumoniae serotype 23A, circulating within the United States, and an internationally dispersed clone of serotype 23F. J. Clin. Microbiol. 43:5440-5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pantosti, A., G. Gherardi, M. Conte, F. Faella, G. Dicuonzo, and B. Beall. 2002. A novel, multiple drug-resistant, serotype 24F strain of Streptococcus pneumoniae that caused meningitis in patients in Naples, Italy. Clin. Infect. Dis. 35:205-208. [DOI] [PubMed] [Google Scholar]
- 26.Pletz, M. W., L. McGee, J. Jorgensen, B. Beall, R. R. Facklam, C. G. Whitney, and K. P. Klugman. 2004. Levofloxacin-resistant invasive Streptococcus pneumoniae in the United States: evidence for clonal spread and the impact of conjugate pneumococcal vaccine. Antimicrob. Agents Chemother. 48:3491-3497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pneumococcal Molecular Epidemiology Network. 20. December 2004, posting date. http://www.sph.emory.edu/PMEN/index.html.
- 28.Pneumococcal Multilocus Sequence Typing. 20. December 2004, posting date. http://spneumoniae.mlst.net/#.
- 29.Porat, N., A. Arguedas, B. G. Spratt, R. Trefler, E. Brilla, C. Loaiza, D. Godoy, N. Bilek, and R. Dagan. 2004. Emergence of penicillin-nonsusceptible Streptococcus pneumoniae clones expressing serotypes not present in the antipneumococcal conjugate vaccine. J. Infect. Dis. 190:2154-2161. [DOI] [PubMed] [Google Scholar]
- 30.Sandgren, A., K. Sjostrom, B. Olsson-Liljequist, B. Christensson, A. Samuelsson, G. Kronvall, and B. Henriques Normark. 2004. Effect of clonal and serotype-specific properties on the invasive capacity of Streptococcus pneumoniae. J. Infect. Dis. 189:785-796. [DOI] [PubMed] [Google Scholar]
- 31.Schutze, G. E., N. C. Tucker, and E. O. Mason, Jr. 2004. Impact of the conjugate pneumococcal vaccine in Arkansas. Pediatr. Infect. Dis. J. 23:1125-1129. [PubMed] [Google Scholar]
- 32.van Selm, S., L. M. van Cann, M. A. Kolkman, B. A. van der Zeijst, and J. P. van Putten. 2003. Genetic basis for the structural difference between Streptococcus pneumoniae serotype 15B and 15C capsular polysaccharides. Infect. Immun. 71:6192-6198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Whitney, C. G., M. M. Farley, J. Hadler, L. H. Harrison, N. M. Bennett, R. Lynfield, A. Reingold, P. R. Cieslak, T. Pilishvili, D. Jackson, R. R. Facklam, J. H. Jorgensen, and A. Schuchat. 2003. Decline in invasive pneumococcal disease after the introduction of protein-polysaccharide conjugate vaccine. N. Engl. J. Med. 348:1737-1746. [DOI] [PubMed] [Google Scholar]