Abstract
The aim of this study was to determine the distribution of the antimicrobial resistance phenotypes (R types), the phage types and XbaI-pulsed-field gel electrophoresis (PFGE) types, the genes coding for resistance to β-lactams and to quinolones, and the class 1 integrons among a representative sample of Salmonella enterica serotype Typhimurium isolates collected from humans in 2002 through the French National Reference Center for Salmonella (NRC-Salm) network. The trends in the evolution of antimicrobial resistance of serotype Typhimurium were reviewed by using NRC-Salm data from 1993, 1997, 2000, and 2003. In 2002, 3,998 isolates of serotype Typhimurium were registered at the NRC-Salm among 11,775 serotyped S. enterica isolates (34%). The most common multiple antibiotic resistance pattern was resistance to amoxicillin, chloramphenicol, streptomycin and spectinomycin, sulfonamides, and tetracycline (ACSSpSuTe R type), with 156 isolates (48.8%). One isolate resistant to extended-spectrum cephalosporins due to the production of TEM-52 extended-spectrum β-lactamase was detected (0.3%), and one multidrug-resistant isolate was highly resistant to ciprofloxacin (MIC > 32 mg/liter). We found that 57.2% of the isolates tested belonged to the DT104 clone. The main resistance pattern of DT104 isolates was R type ACSSpSuTe (83.2%). However, evolutionary changes have occurred within DT104, involving both loss (variants of Salmonella genomic island 1) and acquisition of genes for drug resistance to trimethoprim or to quinolones. PFGE profile X1 was the most prevalent (74.5%) among DT104 isolates, indicating the need to use a more discriminatory subtyping method for such isolates. Global data from the NRC-Salm suggested that DT104 was the main cause of multidrug resistance in serotype Typhimurium from humans from at least 1997 to 2003, with a roughly stable prevalence during this period.
Food-borne diseases caused by zoonotic Salmonella enterica species represent an important public health problem worldwide. In the United States, there are an estimated 1.4 million Salmonella infections per year, and approximately 600 are fatal (27). Among the more than 2,500 serotypes of the genus Salmonella described to date (30), two, Enteritidis and Typhimurium, are predominant in many developed countries. S. enterica serotype Typhimurium was the second most prevalent serotype (Enteritidis ranked first) in Europe during the period 1998 to 2003 (13). It has a large animal reservoir, including farm animals, pets, and wild animals. Although most Salmonella infections cause mild diseases (gastroenteritis), life-threatening infections (e.g., bacteremia) may occur, particularly in cases involving patients at the extremes of age or those who are immunocompromised. An appropriate antimicrobial-drug therapy is necessary in these severe infections. Two antimicrobial classes are used for that purpose, ciprofloxacin and, when ciprofloxacin is contraindicated, extended-spectrum cephalosporins (ESC). An increase in antibiotic resistance was observed during the 1990s due to the emergence of an epidemic multidrug-resistant (MDR) strain of serotype Typhimurium of definitive phage type 104 (DT104), especially in Europe and North America (36). In these DT104 isolates, multiple antibiotic resistance was due to chromosomal integration of a 43-kb structure called Salmonella genomic island 1 (SGI1), comprised of the genes coding for resistance to ampicillin (blaPSE-1), streptomycin and spectinomycin (aadA2), chloramphenicol and florfenicol (floR), sulfonamides (sul1), and tetracycline [tet(G)] (6). Additional resistance to nalidixic acid resistance (with reduced susceptibility to ciprofloxacin) has increasingly been reported in DT104 isolates recovered from humans and animals in England (36). A few reports have also described multidrug-resistant serotype Typhimurium isolates highly resistant to ciprofloxacin (MIC > 8 mg/liter) in humans and animals from Europe and Asia (4, 7, 15, 18, 19, 29).
Emergence of serotype Typhimurium strains that are resistant to ESC due to plasmid-mediated class A extended-spectrum β-lactamase (ESBL) belonging to the TEM (TEM-3, TEM-52, and TEM-131), SHV (SHV-2, SHV-5, and SHV-9), CTX-M (CTX-M-2 to CTX-M-7 and CTX-M-15), or PER (PER-1 and PER-2) family have been reported throughout the world since 1988 (5, 21, 28, 42). More recently, serotype Typhimurium isolates producing the plasmidic class C cephamycinase CMY (CMY-2 and, to a lesser extent, CMY-7) have been described in various countries (28).
The objectives of this study were to determine the antimicrobial resistance profiles seen among a representative sample of 320 serotype Typhimurium strains isolated from humans in France in 2002 through the French National Reference Center for Salmonella (NRC-Salm) network and to describe the distribution of phage types and pulsed-field gel electrophoresis (PFGE) types. The molecular characterization of genes coding for resistance to ampicillin and to quinolones and the detection of class 1 integrons by PCR were also performed. The trends in the evolution of antimicrobial resistance of serotype Typhimurium strains isolated from humans in France were reviewed by using NRC-Salm data from 1993, 1997, 2000, and 2003.
MATERIALS AND METHODS
Bacterial strains.
In 2002, a total of 11,775 serotyped S. enterica isolates from humans were registered at the NRC-Salm (6,636 isolates received at NRC-Salm for serotyping and 5,139 laboratory-confirmed cases reported to the NRC-Salm by its network). The NRC-Salm network comprises approximately 1,500 voluntary hospital or private clinical laboratories representing approximately 30% of all French clinical laboratories.
The present study was conducted on a sample of 320 serotype Typhimurium isolates collected from humans and received at the NRC-Salm in 2002. The isolates (one per patient) were randomly selected from various geographic regions of France from January to December.
Escherichia coli ATCC 25922 was used as a control in the disk diffusion method and in MIC determinations. S. enterica serotype Braenderup H9812 was used as a molecular size marker in the PFGE experiments.
Serotyping.
Isolates were serotyped on the basis of somatic O and phase 1 and phase 2 H flagellar antigens by agglutination tests with antisera (Bio-Rad, Marnes la Coquette, France, and WHO Collaborative Centre for Reference and Research on Salmonella, Institut Pasteur, Paris, France) as specified by the White-Kauffmann-Le Minor scheme (30).
Phage typing.
Phage typing of serotype Typhimurium isolates was done following a standardized methodology using 31 phage suspensions (1). Five additional typing phages (2, 3, 18, 10, and 10 variant) were also used. Phage suspensions and the interpretive guide were kindly provided by the Health Protection Agency, Colindale, United Kingdom.
Pulsed-field gel electrophoresis.
PFGE using XbaI (Amersham Biosciences, Freiburg, Germany) was carried out with a CHEF-DR III system (Bio-Rad) as described previously (44). The running conditions were 6 V/cm at 14°C for 20 h with pulse times ramped from 2.2 to 63.8 s. XbaI-digested plugs of S. enterica serotype Braenderup H9812 were used as molecular size markers. Image normalization and construction of similarity matrices were carried out using BioNumerics 4.1 (Applied Maths, Sint-Martens-Latem, Belgium). Bands were assigned manually, and clustering was performed using the unweighted pair group method with arithmetic averages based on the Dice similarity index, utilizing an optimization parameter of 1% and a 1% band position tolerance. Each profile that differed by at least one clear band was considered a distinct profile.
Antimicrobial susceptibility testing.
Antibiotic susceptibility was determined by disk diffusion on Mueller-Hinton agar according to the guidelines of the Antibiogram Committee of the French Society for Microbiology (35). The following antimicrobials (Bio-Rad) were tested: amoxicillin (A), amoxicillin-clavulanic acid, ticarcillin, ticarcillin-clavulanic acid, piperacillin, piperacillin-tazobactam, cephalothin, cefamandole, cefoperazone, cefoxitin, ceftriaxone, ceftazidime, cefepime, aztreonam, moxalactam, imipenem, streptomycin (S), spectinomycin (Sp), kanamycin (K), tobramycin (To), netilmicin, gentamicin (G), amikacin, isepamicin, nalidixic acid (Nal), pefloxacin, ciprofloxacin (Cip), sulfonamides (Su), trimethoprim (Tmp), chloramphenicol (C), and tetracycline (T).
MICs of the β-lactams and ciprofloxacin were determined by Etest (AB Biodisk, Solna, Sweden). The ESBL phenotype was detected by using the ESBL detection Etest strips and the double disk diffusion test (20).
Multidrug resistance was defined as isolates being resistant to ≥2 separate classes of antibiotics.
PCR amplification and DNA sequencing.
Total DNA was extracted using the InstaGene matrix kit (Bio-Rad) in accordance with the manufacturer's recommendations. Table 1 lists the oligonucleotide primers synthesized by MWG-Biotech (Ebersberg, Germany). Amplification of antibiotic resistance genes, the blaTEM, blaPSE-1, blaOXA-1 group and blaSHV, was performed on all isolates resistant to amoxicillin. Amplifications of the quinolone resistance-determining regions (QRDR) of gyrA and the plasmid-mediated qnrA gene were performed on all isolates resistant to Nal. Amplifications of the QRDR of gyrB and parC were performed on the isolate highly resistant to Cip. Amplifications of left and right junctions of SGI1with the chromosome were performed on all isolates positive for the blaOXA-1 group gene. Amplification of the gene cassette of the class 1 integron was performed by using 5′-CS and 3′-CS primers on all isolates resistant to Su. Mapping of class1 integrons by using the A (5′-CS and OXA-1-R) and B (aadA1-F and 3′-CS) sets of primers was performed on all isolates positive for the blaOXA-30 gene. All amplifications were performed on 50-μl samples containing DNA (2.5 μl), primers (50 pmol each), deoxynucleoside triphosphate (200 μM), Taq DNA polymerase (1.25 U Ampli Taq Gold; Roche) and its buffer, MgCl2 (2 mM), and dimethyl sulfoxide (10%). The cycling conditions included 10 min of denaturation at 94°C (1 cycle) and 30 s of denaturation at 94°C, 30 s of annealing at 50°C (47°C for blaOXA, 54°C for qnrA, and 55°C for class 1 integrons and left and right junctions of SGI1), and 1 min (2 min for class 1 integrons) of polymerization at 72°C (35 cycles; 30 cycles for class 1 integrons), followed by 10 min of extension at 72°C.
TABLE 1.
Oligonucleotide primers used in this study
| Target | Primer designation | Oligonucleotide sequence (5′→3′)a | Reference | PCR product size (bp) |
|---|---|---|---|---|
| Antibiotic resistance genes | ||||
| blaOXA-1 group | OXA-1-F | ATGAAAAACACAATACATATC | 7 | 830 |
| OXA-1-R | AATTTAGTGTGTTTAGAATGG | |||
| blaTEM | TEM-F | ATAAAATTCTTGAAGACGAAA | 26 | 1,080 |
| TEM-R | GACAGTTACCAATGCTTAATC | |||
| blaPSE-1 | PSE-1-F | CGCTTCCCGTTAACAAGTAC | 34 | 420 |
| PSE-1-R | CTGGTTCATTTCAGATAGCG | |||
| blaSHV | SHV-F | TTATCTCCCTGTTAGCCACC | 3 | 795 |
| SHV-R | GATTTGCTGATTTCGCTCGG | |||
| Class 1 integron | 5′-CS | GGCATCCAAGCAGCAAGC | 23 | Variable |
| 3′-CS | AAGCAGACTTGACCTGAT | |||
| AadA1-F | TATCAGAGGTAGTTGGCGTCA | 32 | ||
| gyrA | GyrA-F | CTGAAGCCGGTACACCGTCG | 8 | 290 |
| GyrA-R | TCGGCCATCAGTTCGTGGGC | |||
| gyrB | GyrB-F | TTATCGATGCTGCGCGTGCC | 8 | 1,280 |
| GyrB-R | TCGCCGCTTTCAGGGCGTTC | |||
| parC | ParC-F | CGCCTACTTAAACTACTCCA | 8 | 540 |
| ParC-R | ATCAGCGTAATCGCCGCTTT | |||
| qnrA | Qnr-F | TCAGCAAGAGGATTTCTCA | 41 | 625 |
| Qnr-R | GGCAGCACTATTACTCCCA | |||
| SGI1 | ||||
| Left junction | ||||
| thdF | U7-L12 | ACACCTTGAGCAGGGCAAG | 6 | 500 |
| int | LJ-R1 | AGTTCTAAAGGTTCGTAGTCG | ||
| Right junction | ||||
| S044 | 104-RJ | TGACGAGCTGAAGCGAATTG | 6 | 515 |
| int2 | C9-L2 | AGCAAGTGTGCGTAATTTGG |
Y = C, T; R = A, G.
Sequencing of purified amplicons was performed on both strands by Genome Express (Meylan, France) using an ABI 100 DNA sequencer (Applied Biosystems, Foster City, CA).
The nucleotide sequence was analyzed with Lasergene software (Dnastar, Madison, WI). The BLASTN program of NCBI (http://www.ncbi.nlm.nih.gov) was used for database searches.
RESULTS
S. enterica serotype Typhimurium isolates from humans, NRC-Salm, France, 1993 to 2003.
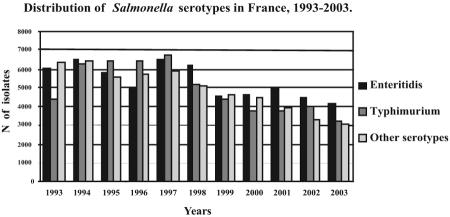
A total of 168,034 Salmonella isolates from humans were registered at the NRC-Salm from 1993 to 2003. Serotypes Enteritidis and Typhimurium represented 35% (58,766 isolates) and 32.5% (54,551 isolates) of all the Salmonella isolates, respectively. During the periods 1993 to 1994 and 1998 to 2003, serotype Typhimurium was the second most prevalent serotype (after Enteritidis), whereas between 1995 and 1997, it ranked first (Fig. 1).
FIG. 1.
Distribution of Salmonella serotypes reported by the NRC-Salm in humans in France from 1993 to 2003.
In 2002, of the 11,775 Salmonella isolates (belonging to 232 serotypes) registered at the NRC-Salm, 4,469 (38%) were serotype Enteritidis and 3,998 (34%) were serotype Typhimurium. Among the 3,998 registered isolates of serotype Typhimurium, 1,756 isolates were serotyped at the NRC-Salm and 2,242 were serotyped locally and reported to the NRC-Salm.
Antimicrobial susceptibility testing.
The antibiotics against which the 320 serotype Typhimurium isolates tested demonstrated the highest levels of resistance in 2002 were tetracycline (71%), sulfonamides (68%), amoxicillin (64.7%), streptomycin (64.5%), spectinomycin (59%), and chloramphenicol (57%) (Table 2). A single isolate resistant to ESC (MIC of ceftazidime, >256 mg/liter; MIC of ceftriaxone, 128 mg/liter) which exhibited an ESBL phenotype was detected. Another isolate (HRC) was highly resistant to Cip (MIC > 32 mg/liter). No isolates were resistant to amikacin or imipenem. The most common multiple antibiotic resistance pattern (R type) was resistance to amoxicillin, chloramphenicol, streptomycin and spectinomycin, sulfonamides, and tetracycline (R type ACSSpSuT), with 156 isolates (48.8%). The number of isolates that were pansusceptible was 69 (21.5%). Single resistance to tetracycline was found in 27 isolates (8.5%). R types ASSuT, ACSSpSuTNal (with reduced susceptibility to ciprofloxacin; MIC range, 0.25 to 0.5 mg/liter), and ACSSpSuTTmp were found in 12 (3.8%), 12, and 10 (3%) isolates, respectively. Other R types found in less than 10 isolates are indicated in Table 3. Tables 2 and 4 show the percentages of resistance to individual antimicrobials and the distribution of R types commonly associated with the DT104 clone among samples of serotype Typhimurium isolates collected from humans by NRC-Salm in 1993, 1997, 2000, and 2003.
TABLE 2.
Percentages of resistance to specific antibiotics in S. enterica serotype Typhimurium in France in 1993, 1997, 2000, 2002, and 2003
| Antimicrobial | % of isolates (95% confidence interval) resistanta
|
||||
|---|---|---|---|---|---|
| 1993 (n = 280; N = 1,593) | 1997 (n = 205; N = 2,801) | 2000 (n = 320; N = 1,613) | 2002 (n = 320; N = 1,756) | 2003 (n = 100; N = 1,489) | |
| Amoxicillin | 54.3 (48.5-60.1) | 66.3 (59.8-72.8) | 64.3 (59.1-69.5) | 64.7 (59.5-69.9) | 62.0 (52.5-71.5) |
| Ceftriaxone | 0 | 0 | 0 | 0.3 (0-0.9) | 0 |
| Streptomycin | 53.9 (48.1-59.7) | 65.9 (59.4-72.4) | 71.8 (66.9-76.7) | 64.5 (59.3-63.7) | 57.0 (47.3-66.7) |
| Kanamycin | 3.2 (1.1-5.3) | 1.0 (0-2.4) | 1.3 (0.1-2.5) | 0.9 (0-1.9) | 0 |
| Gentamicin | 0.4 (0-1.1) | 1.0 (0-2.4) | 0.9 (0-1.9) | 0.3 (0-0.9) | 0 |
| Nalidixic acid | 3.2 (1.1-5.3) | 2.9 (0.6-5.2) | 10.3 (7.0-13.6) | 4.0 (1.9-6.1) | 1.0 (0-3.0) |
| Ciprofloxacin | 0 | 0 | 0 | 0.3 (0-0.9) | 0 |
| Sulfonamides | 58.2 (52.4-64.0) | 68.8 (62.5-75.1) | 69.6 (64.6-74.6) | 68.0 (0-0.9) | 64.0 (54.6-73.4) |
| Trimethoprim | 11.8 (8.0-15.6) | 5.9 (2.7-9.1) | 8.7 (5.6-11.8) | 5.3 (2.8-7.8) | 8.0 (2.7-13.3) |
| Chloramphenicol | 43.6 (37.8-49.4) | 60.0 (53.3-66.7) | 59.0 (53.6-64.4) | 57.0 (51.6-62.4) | 46.0 (36.2-55.8) |
| Tetracycline | 69.6 (64.2-75.0) | 82.0 (76.7-87.3) | 81.2 (76.9-85.5) | 71.0 (66.0-76.0) | 67.0 (57.8-76.2) |
n, number of Salmonella isolates studied; N, number of Salmonella isolates received at NRC-Salm.
TABLE 3.
Antimicrobial resistance patterns, bla genes, class 1 integrons, phage types, and XbaI PFGE types of S. enterica serotype Typhimurium isolates under studya
| Antimicrobial resistance pattern | n (%) | β-Lactam resistance gene (n) | Integron PCR size (kb)b | Phage type (n)c | Combined-type phage type-PFGE type (n)c |
|---|---|---|---|---|---|
| ASSpSuCTe | 156 (48.8) | blaPSE-1 (147) | 1, 1.2 | DT120 (2), DT104 (135), U302 (7), NT (3) | DT120-X3 (2), DT104-X1 (23), DT104-X8 (9), DT104-X12 (1), DT104-X4 (1), U302-X5 (1), U302-X1 (3), U302-X8 (1), NT-X1 (2) |
| blaOXA-30 (9) | 2 | DT41 (1), DT120 (5), DT204c (2), RDNC (1) | DT41-X11 (1), DT120-X6 (4), DT204c-X11 (2), RDNC-X29 (1) | ||
| Pansusceptible | 69 (21.5) | DT1 (4), DT3 (1), DT8 (15), DT12 (5), DT14 (10), DT27 (1), DT40 (1), DT41 (4), DT49 (7), DT56 (1), DT68 (2), DT104 (2), DT104at (3), DT108 (1), DT135 (1), DT161 (1), DT180 (1), DT184 (1), DT193 (2), U302at (1), RDNC (5) | DT1-X42 (1), DT3-X43 (1), DT8-X23 (7), DT12-X14 (1), DT12-X45 (1), DT14-X18 (1), DT14-X36 (4), DT27-X21 (1), DT40-X33 (1), DT41-X24 (1), DT49-X19 (1), DT49-X24 (1), DT56-X34 (1), DT68-X26 (1), DT68-X31 (1), DT104-X1 (2), DT104at-X28 (1), DT104at-X44 (1), DT135-X35 (1), DT193-X55 (1), RDNC-X31 (1) | ||
| Te | 27 (8.5) | DT12 (4), DT22 (1), DT104at (4), DT120 (2), DT190 (1), DT193 (3), DT208 (10), NT (1), RDNC (1) | DT104at-X28 (1), DT120-X47 (1), DT193-X37 (1), NT-X30 (1) | ||
| ASSuTe | 12 (3.8) | blaTEM (12) | NDa | DT120 (5), U302at (6), NT (1) | DT120-X22 (2), DT120-X39 (1), DT120-X49 (1), U302at-X20 (1) |
| ASSpSuCTeNal | 12 (3.8) | blaPSE-1 (11) | 1, 1.2 | DT104 (9), NT (2) | DT104-X1 (2), NT-X1 (1) |
| blaOXA-30 (1) | 2 | DT120 (1) | DT120-X6 (1) | ||
| ASSpSuCTeTmp | 10 (3.0) | blaPSE-1 (9) | 1, 1.2 | DT104 (9) | DT104-X1 (3), DT104-X7 (1) |
| blaTEM, blaOXA-30 (1) | 2 | DT120 (1) | DT120-X6 (1) | ||
| SSpSu | 7 (2.2) | DT104 (7) | DT104-X1 (3) | ||
| ASu | 6 (1.8) | blaPSE-1 (5) | 1.2 | DT104 (5) | DT104-X1 (4) |
| blaTEM (1) | ND | DT14 (1) | DT14-X40 (1) | ||
| SuTmpTe | 3 (0.9) | DT104at (1), DT120 (1), DT186 (1) | DT104at-X48 (1), DT120-X46 (1), DT186-X14 (1) | ||
| ATe | 2 (0.6) | blaTEM (2) | ND | DT104at (1), U302at (1) | DT104at-X41 (1) |
| SSu | 2 (0.6) | DT104at (1), DT120 (1) | |||
| S | 2 (0.6) | DT12 (1), DT104at (1) | |||
| ASSpSu | 1 (0.3) | blaOXA-30 (1) | 2 | DT204c (1) | DT204c-X16 (1) |
| ASSpKSuCTe | 1 (0.3) | blaPSE-1 (1) | 1, 1.2 | DT104 (1) | DT104-X1 (1) |
| ASSpKToGSuCTe | 1 (0.3) | blaPSE-1 (1) | 1, 1.2 | DT104 (1) | DT104-X2 (1) |
| ACro (BLSE) | 1 (0.3) | blaTEM-52 (1) | ND | DT146 (1) | DT146-X38 (1) |
| ASSpSuCTeNalCip | 1 (0.3) | blaOXA-30 (1) | 2 | DT12var (1) | DT12var-X13 (1) |
| Su | 1 (0.3) | U281 (1) | |||
| STe | 1 (0.3) | RDNC (1) | RDNC-X50 (1) | ||
| SuTmp | 1 (0.3) | DT12 (1) | DT12-X14 (1) | ||
| ASSuTmpC | 1 (0.3) | blaTEM (1) | 1.5 | NT (1) | NT-X27 (1) |
| ASKSulTe | 1 (0.3) | blaTEM (1) | ND | DT99 (1) | DT99-X32 (1) |
| ASulTmpTe | 1 (0.3) | blaTEM (1) | 1.5 | DT8 (1) | DT8-X23 (1) |
| A | 1 (0.3) | blaTEM (1) | ND | U302at (1) | U302at-X14 (1) |
n, number of isolates.
ND, none detected.
NT, not typeable; at, atypical; var, variant.
TABLE 4.
Distribution of antimicrobial resistance phenotypes commonly associated with DT104 clone among S. enterica serotype Typhimurium isolates in France, 1993, 1997, 2000, 2002, and 2003
| Antimicrobial resistance profile | % (95% confidence interval) of isolates resistanta
|
||||
|---|---|---|---|---|---|
| 1993 (n = 280; N = 1,593) | 1997 (n = 205; N = 2,801) | 2000 (n = 320; N = 1,613) | 2002 (n = 320; N = 1,756) | 2003 (n = 100; N = 1,489) | |
| ACS[Sp]SuTb | 34.3 (28.7-39.9) | 54.6 (47.8-61.4) | 50.9 (45.4-56.4) | 48.8 (43.3-53.3) | 43.0 (33.3-52.7) |
| ACS[Sp]SuTNalb | 0 | 1.5 (0-3.2) | 3.8 (1.7-5.9) | 3.8 (1.7-5.9) | 1 (0-3.0) |
| ACS[Sp]SuTmpb | 2.5 (0.7-4.3) | 1.5 (0-3.2) | 2.8 (1.0-4.6) | 3.0 (1.1-4.9) | 2 (0-4.7) |
| S[Sp]Sub | 1.1 (0-0.3) | 0.5 (0-1.5) | 0.9 (0-1.9) | 2.2 (0.6-3.8) | 2 (0-4.7) |
| ASu | 0.7 (0-1.7) | 0 | 0.3 (0-0.9) | 1.8 (0.6-3.3) | 3 (0-6.3) |
| Total | 38.6 (32.9-44.3) | 58.0 (51.2-64.8) | 58.8 (53.4-64.2) | 59.6 (54.2-65.0) | 51.0 (41.2-60.8) |
n, number of Salmonella isolates studied; N, number of Salmonella isolates received at NRC-Salm.
Spectinomycin was not tested before 2002.
Phage typing.
Of the 320 serotype Typhimurium isolates phage typed, 169 (52.8%) were DT104, 16 (5%) were DT120 or DT8, 13 (4.1%) were atypical DT104 (DT104at), 11 (3.4%) were DT12 or DT14, and 8 (2.5%) were RDNC (reacts but does not conform to the scheme) or untypeable; 28 other phage types (including two atypical phage types, U302at and 12var) were identified at low frequency (in less than 10 isolates) among the remaining 71 isolates (Table 3).
Among the 69 pansusceptible isolates, there were 20 different phage types (including atypical phage types, DT104at and U302at): 15 (21.7%) were DT8, 10 (14.5%) were DT14, 7 (10.1%) were DT49, 5 (7.2%) were RDNC, and 2 (2.9%) were DT104 (Table 2).
Among the 156 isolates that were R type ACSSpSuT, 135 (86.5%) were DT104, 8 (5.1%) were DT120, 7 (4.5%) were U302, 3 (1.9%) were untypeable, 2 (1.3%) were DT204c, 1 (0.6%) was DT41, and 1 other was RDNC.
Among the 23 isolates that were R type ACSSpSuT with additional resistance to Nal (n = 12), to Tmp (n = 9), to K (n = 1), or to KToG (n = 1), 20 (87%) were DT104, 2 (8.7%) were untypeable, and 1 (4.3%) was DT120.
Among the 169 DT104 isolates, 135 (79.9%) exhibited R type ACSSpSuT, 9 (5.3%) exhibited R type ACSSpSuTNal or ACSSpSuTTmp, 7 (4.1%) exhibited R type SSpSu, 5 (3%) exhibited R type ASu, 2 (1.2%) were pansusceptible, 1 (0.6%) exhibited R type ACSSpKSuT, and 1 other exhibited R type ACSSpKToGSuT.
PFGE.
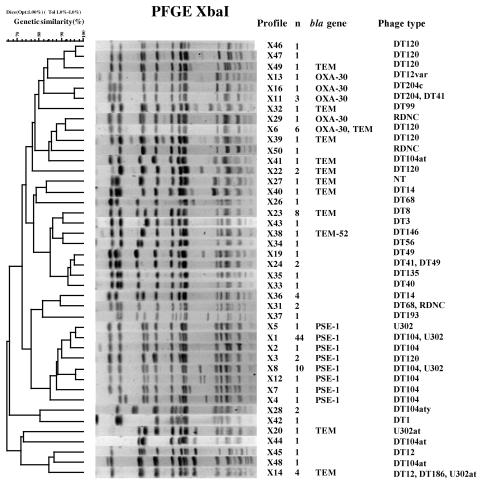
PFGE of XbaI restriction digests of DNAs from a subset of 122 isolates generated 45 PFGE profiles (Table 3 and Fig. 2). PFGE profile X1 was the most prevalent (n = 44; 36.1%). Among the 44 isolates tested with profile X1, 28 (63.6%) exhibited R type ACSSpSuT; 4 (9.1%) exhibited R type ASu; 3 (6.8%) isolates (each) of R type ACSSpSuTNal, SSpSu, and ACSSpSuTTmp were seen; 2 (4.6%) were pansusceptible; and 1 (2.3%) exhibited R type ACSSpKSuT. Of the 46 isolates of R type ACSSpSuT tested, profile X1 was found in 28 (61%); X8 in 10 (21.7%); X3 and X6 each in 2 (4.3%); and X4, X5, and X11 in 1 (2.2%) isolate. Of the nine isolates tested that were R type ACSSpSuT with additional resistance to Nal (n = 4), to Tmp (n = 3), to K (n = 1), or to KToG (n = 1), profile X1 was found in six (66.6%) and profiles X2, X6, and X7 were found in a single (11.1%) isolate. Among 30 susceptible isolates tested belonging to 15 different phage types, 20 PFGE profiles were seen. Profiles X23 and X36 were always associated with PT8 (n = 7) and PT14 (n = 4) susceptible isolates, respectively.
FIG. 2.
Dendrograms generated by BioNumerics showing the results of cluster analysis on the basis of XbaI-PFGE of S. enterica serotype Typhimurium isolates. Similarity analysis was performed using the Dice coefficient, and clustering was by the unweighted pair group method with arithmetic averages. The different PFGE profiles and corresponding numbers of isolates, the types of bla genes (if present), and the phage types are indicated.
Profile X1 was found in 38 DT104 isolates, in 3 U302 isolates, and in 3 untypeable isolates.
Among the 51 DT104 isolates tested, profile X1 was found in 38 (74.5%); X8 in 9 (17.5%); and X2, X4, X7, and X12 in 1 (2%) each. Among the 13 DT120 isolates tested, profile X6 was found in 5 (38.4%); X3 and X22 in 2 (15.4%); and X39, X46, X47, and X49 in 1 (7.7%) each. Among the five U302 isolates tested, profile X1 was found in three (60%) and X5 and X8 in one (20%) each.
A dendrogram revealed that PFGE profiles X1 to X5, X7, X8, and X12 were assigned to one cluster with a similarity percentage of 80% (Fig. 2).
Characterization of genes encoding resistance to β-lactams and quinolones and gene cassettes of class 1 integrons.
The distribution of amoxicillin resistance genes (blaPSE-1 [also called blaCARB-2], blaTEM, the blaOXA-1 group, and blaSHV) and class 1 integron gene cassettes between different antimicrobial resistance patterns is indicated in Table 3.
Among the 207 isolates resistant to amoxicillin, the blaPSE-1-like gene was found in 174 isolates (84.1%), the blaTEM gene in 20 isolates (9.7%), and the blaOXA-30 gene in 13 isolates (6.3%). One isolate was positive for both the blaTEM and blaOXA-30 genes, and the ESBL-producing isolate was positive for the blaTEM-52 gene (43). No blaSHV gene was detected.
The 174 serotype Typhimurium strains that contained the blaPSE-1-like gene were of R types ACSSpSuT (n = 147), ACSSpSuTNal (n = 11), ACSSpSuTTmp (n = 9), ASu (n = 5), ACSSpKSuT (n = 1), and ACSSpKToGSuT (n = 1). These strains belonged to phage types DT104 (n = 160), U302 (n = 7), and DT120 (n = 2) or were phage untypeable (n = 5). Among the 45 isolates positive for the blaPSE-1-like gene tested by PFGE, profile X1 was observed in 29 isolates (64.4%); X8 in 10 (22.2%); X3 in 2 (4.4%); and X2, X4, X5, and X12 in a single isolate (2.2%) each.
The 13 serotype Typhimurium isolates that contained the blaOXA-30 gene were of R types ACSSpSuT (n = 9), ACSSpSuTNal (n = 1), ACSSpSuTTmp (n = 1), ACSSpSu (n = 1), and ACSSpSuTCip (n = 1). They belonged to phage types DT120 (n = 7), DT204c (n = 3), DT41 (n = 1), and DT12var (n = 1; the HRC isolate) or were RDNC (n = 1). Among the 11 isolates tested by PFGE, profile X6 was observed in 6 isolates (54.6%), X8 in 3 (27.3%), and X13 and X16 in a single isolate (9.1%) each.
The 19 serotype Typhimurium isolates that contained only the blaTEM gene were of various R types not found in isolates that contained other bla genes. R type ASSuT was the most prevalent (n = 12, 63.2%). These isolates belonged to phage types U302at (n = 8), DT120 (n = 5), DT8 (n = 1), DT14 (n = 1), DT99 (n = 1), and DT104at (n = 1) or were untypeable (n = 2). Among the 10 isolates tested by PFGE, nine profiles (not found in isolates that contained other bla genes) were observed (X14, X20, X22, X23, X27, X32, and X39 to X41).
Class 1 integrons with classical gene cassettes of 1.0 (known to contain the aadA2 gene encoding resistance to S and Sp) and 1.2 kb (known to contain the blaPSE-1 gene) were found in all isolates that contained the blaPSE-1-like gene (Table 3). Amplicons of 1.2 kb were amplified from isolates of R type ASu. Amplicons of 1.0 kb were amplified from isolates of R type SSpSu. Both amplicons were amplified from isolates of R type ACSSpSuT and in those with additional resistance to Nal, to Tmp, to K, or to KToG. Class 1 integrons with gene cassettes of approximately 2.0 kb were found in all isolates that contained the blaOXA-30 gene. DNA sequence analyses identified blaOXA-30-aadA1 gene cassettes (encoding resistance to A and to S-Sp, respectively) in five representative isolates. PCR mapping gave amplicons of 990 bp with set A primers and 860 bp with set B primers, thus confirming the presence of such cassettes in all the remaining isolates that contained the blaOXA-30 gene. The HRC isolate also contained additional dhfrXII-orfF-aadA2 gene cassettes (encoding resistance to Tmp and to S-Sp).
Of the 19 isolates that contained the blaTEM gene, only two isolates harbored class 1 integrons of 1.5 kb. Both isolates carried the same dhfr1-aadA2 gene cassette (encoding resistance to Tmp and to S-Sp).
To identify mutations responsible for resistance to quinolones and fluoroquinolones, the QRDR of gyrA of all the isolates resistant to Nal (with decreased susceptibility to Cip) and the HRC isolate were amplified by PCR and sequenced. Nine isolates contained a change at codon aspartate 87 (Asp87) to either asparagine (Asn) (n = 5) or tyrosine (n = 4). Three isolates contained a change at codon serine 83 (Ser83) to phenylalanine (Phe). Both mutations Phe83 and Asn87 were seen in the HRC isolate. No substitutions at alanine 119 were seen in the isolates tested. The qnrA gene was absent from all strains resistant to Nal. The nucleotide sequences of the PCR-amplified QRDR of gyrB and parC were determined for the HRC isolate showing mutations in GyrB (Ser464 to Phe) and in ParC (Ser80 to arginine).
DISCUSSION
Overall, the total number of nontyphoid salmonellae from humans registered at the NRC-Salm decreased from 1997 (19,174 isolates) to 2003 (10,472 isolates). The total number of serotype Typhimurium isolates also decreased during the same period from 6,755 isolates in 1997 to 3,222 in 2003. As no change in surveillance practice had occurred in France and as similar results have been observed in Europe and North America (12, 16), this trend is most likely correct. The present comprehensive study of serotype Typhimurium isolates collected from humans in France in 2002 through a representative network comprising both private and public clinical laboratories concluded that multidrug resistance remains common within the serotype (68.8% of the isolates were resistant to ≥2 separate classes of antimicrobials) and is due mainly to the DT104 clone (82.3% of the MDR isolates belonged to the DT104 complex, as defined below).
Multiresistant (R type ACSSuTe) DT104 strains of serotype Typhimurium were first identified in the United Kingdom in the early 1980s in gulls and exotic birds. Isolations in humans started in 1989, when the clone became epidemic in cattle throughout the United Kingdom. Afterward, the DT104 clone also became common in poultry, pigs, and sheep (36). The SGI1 structure was first described in serotype Typhimurium DT104 and was well characterized (6). This structure is located between the chromosomal genes thdf and int2. The int2 gene, located upstream of the yidY gene, is part of a cryptic retron-phage sequence reported to date only in serotype Typhimurium (6, 24). In other S. enterica serotypes (Agona, Albany, Cerro, Derby, Dusseldorf, Emek, Infantis, Kiambu, Meleagridis, Newport, and Paratyphi B biotype Java), SGI1 is located between the Salmonella genes thdf and yidY (6, 10, 24). The multidrug resistance region is located at the 3′ end of SGI1 in a 13-kb region corresponding to a large class 1 integron with a complex structure named In104 (6, 24). The resistance genes floR and tet(G) are bracketed by two class 1 integrons, one carrying an aadA2 cassette (1.0 kb) and the other a blaPSE-1 cassette (1.2 kb). Transduction by phages or self-transmission of the SGI1 structure has been proposed to explain the insertion of SGI1 in S. enterica strains (10, 11). Strains containing SGI1 variants (classified as SGI1-A to SGI1-J), possibly generated by recombination between homologous regions of the MDR region and conferring different antibiotic resistance profiles, have recently been described in various serotypes of S. enterica (6, 10, 24).
In serotype Typhimurium, the SGI1 resistance gene cluster has been detected mainly in multiresistant DT104 isolates and to a lesser extent in closely related phage type U302 or DT120 and -12 (22, 40). It has been suggested that for non-DT104, the phage typing results were due to changes in phage susceptibility in a small proportion of SGI1-containing DT104 strains, possibly after acquisition of new phages or plasmids, rather than to horizontal transfer of resistance genes (22).
After analyzing the results of phage typing, XbaI-PFGE, and antimicrobial susceptibility testing and characterization of bla genes and class 1 integrons in the present study, we found that 183 isolates among the 320 tested (57.2%) belonged to the DT104 complex (DT104 and closely related phage types harboring SGI1 or variants of SGI1) in France in 2002. The use of phage typing as a single subtyping method gave us 52.8% (169/320) DT104 isolates. This proportion of DT104 isolates was relatively high compared to the data from an international study (16). In that study, for the period 2000 and 2001, the percentage of DT104 among serotype Typhimurium isolates ranged from less than 1% in Oceania (Australia and New Zealand) to approximately 52% in Eastern Europe (35.5% for North America). In Europe, there were large differences in national trends: Spain, 18.3%; Nordic countries, 22.1%; Belgium, 27%; The Netherlands, 37.2%; England and Wales, 42.3%; Germany, 44%; Scotland, 56.3%; and Hungary, 57.7%.
The main resistance pattern observed in the isolates of the DT104 complex in the present study was the pentaresistant ACSSpSuT R type (83.2%). This result is in accordance with other studies (16, 31, 37), confirming that the classical SGI1 is the most prevalent resistance mechanism among serotype Typhimurium DT104 isolates.
Analysis of the distribution of PFGE profiles among DT104 isolates of R type ACSSpSuTe confimed the low discriminatory power of the PFGE method for clonal DT104 (six profiles were seen, with one highly prevalent at 65%). Due to the high prevalence of such DT104 isolates of R type ACSSpSuTe in France since 1997, it became necessary to use a method complementary or alternative to PFGE for investigation of outbreaks due to serotype Typhimurium. Consequently, we are evaluating the multilocus variable number of tandem repeats analysis developed by Lindstedt et al. (25).
In our study, it was noteworthy that 5.8% (9/156) of isolates with R type ACSSpSuT, 8.3% (1/12) with R type ACSSpSuTNal, and 10% (1/10) with R type ACSSpSuTTmp did not belong to the DT104 complex as defined above. They did not contain SGI1, as detected by PCR of right and left junctions, and they possessed the blaOXA-30 gene coding for ampicillin resistance (instead of blaPSE-1 for SGI1-positive isolates) located with aadA1 (instead of aadA2 for SGI1-positive isolates) within a class 1 integron of 2 kb (instead of two class 1 integrons of 1.0 and 1.2 kb for SGI1-positive isolates). Without performing the detection of resistance genes, such isolates, which belong mostly to DT120 (7/11, 63.6%), closely related to DT104, and which may contain SGI1 (22), would have been classified in the DT104 complex. Serotype Typhimurium isolates of R type ACSSuTe containing the plasmid-mediated blaOXA-30 gene (also located with aadA1 within a class 1 integron of 2.0 kb) were identified in 2002 and 2003 in Portugal, and the authors suggested that pigs were the source of the isolates (2).
Isolates exhibiting the ACSSpSuT R type with additional resistance to relevant drugs, such as Nal or Tmp, were found in 6% and 4.9% of the DT104 complex isolates. Our results are in accordance with the data from the international study, indicating that Nal resistance and Tmp resistance were found in 6% and 6.6% of the MDR serotype Typhimurium DT104 isolates tested in 2001, respectively (16). There was an exception for the United Kingdom, where serotype Typhimurium isolates of R types ACSSpSuTNal and ACSSpSuTTmp were more prevalent in humans. In the United Kingdom, Tmp resistance in MDR DT104 isolates began in 1992 (0.9%), peaked in 1995 (27.2%), and then decreased until 2000 (13%), while Nal resistance began in 1992 (0.1%), peaked in 1996 (13%), and then decreased until 2000 (9%) (37, 39).
Although quinolone resistance can be caused by different mechanisms of chromosomal origin, mutations within gyrA resulting in amino acid substitutions in the QRDR of the A subunit of DNA gyrase play a major role in Nal resistance in Salmonella (17). The mutations occur most frequently at codons Ser83 and Asp87 (17). In the present study, we found three different mutations among 12 isolates resistant to Nal (and with reduced susceptibility to Cip): Ser83 to Phe (25%), Asp87 to Asn (41.7%), and Asp87 to Tyr (33.3%). In serotype Typhimurium, Asp87 to Asn was the most commonly isolated mutation in human DT104 isolates in the United Kingdom between 1994 and 1997 (33, 38), and all three mutations were observed with a different distribution (Ser83 to Phe, 60%; Asp87 to Asn, 34.3%; and Asp87 to Tyr, 5.7%) among a panel of 40 veterinary isolates collected between 1997 and 2000 (12), also in the United Kingdom. The presence of these different mutations within the chromosomal gyrA gene may be used to confirm epidemiological relationships between Nal-resistant serotype Typhimurium isolates, particularly if they belong to the DT104 clone. High-level resistance to Cip in salmonellae is of great concern but has been found rarely in serotype Typhimurium isolates (4, 7, 15, 18, 19, 29). Such isolates were collected from humans and animals (cattle, pigs, and pets) in various countries (Germany, Belgium, France, United Kingdom, Japan, and Taiwan). The prevalence of Cip resistance (MIC > 2 mg/liter) in serotype Typhimurium was low (0.3%; 1/320) in France in 2002. Other NRC-Salm surveys conducted in 1993, 1997, 2000, and 2003 did not detect Cip-resistant serotype Typhimurium isolates. The single isolate detected in 2002 had mutations in GyrA, GyrB, and ParC. Other resistance mechanisms, such as overexpression of multidrug efflux pumps or decreased expression of outer membrane proteins, were not investigated. This MDR isolate not related to the DT104 clone (different antibiotic resistance genes and a different PFGE profile) shared common features (QRDR mutations and blaOXA-30-aadA1 and dhfr12-orfF-aadA2 gene cassettes) with some other isolates reported in the literature. In France, two human cases due to DT12var (DT12 reclassified as DT12var) were reported by Casin et al. (8) in 2003. The source of the contamination was not identified for any of the French isolates.
Additional resistance to Tmp for DT104 isolates may be explained by different mechanisms: (i) the presence of a plasmid harboring a Tmp resistance gene (9, 38), (ii) the presence of an independent chromosomally located class 1 integron containing the dfrA1 gene (9), (iii) or the presence of a variant of SGI, SGI1-A, which comprises a dfrA10 gene between the two class I integrons of SGI1 (6). However, SGI1-A was identified in serotypes Agona, Kiambu, and Infantis but not yet in serotype Typhimurium (24). Other R types associated with variants of SGI1 were found in DT104 isolates in the present study; 3.8% (7/183) were of R type SSpSu (SGI1-C), and 3.3% (6/183) were of R type ASu (SGI1-A). These results also confirmed the observation by Threlfall et al. (38) that evolutionary changes have occurred within DT104 involving both loss (variants of SGI1) and acquisition of drug resistance genes (plasmid-located or chromosomally located due to a mutation[s] in the QRDR).
Interestingly, only 1.1% (2/183) of the DT104 isolates were pansusceptible. We found 13 isolates with R types or PFGE profiles different from those of the DT104 clone which could have been classified as DT104 on the basis of the 31 selected routine phage suspensions and the interpretive guide. However, there were unexpected susceptibilities to additional phages 2 and 3, and thus, they have been reclassified as DT104-atypical to avoid confusion with the MDR DT104 clone.
Analysis of previous NRC-Salm antimicrobial resistance surveys revealed that the DT104 clone emerged in France before 1993 (no susceptibility data were available before 1993). In 1993, 38.6% of the isolates displayed R types commonly associated with the DT104 clone, and a study conducted on 86 ampicillin-resistant strains isolated in 1994 through a hospital-based network found that 76.7% of the strains belonged to the DT104 clone (7). It became prevalent between 1993 and 1997 and was still the main cause of multidrug resistance in 2003 without significant decrease.
The prevalence of resistance to ESC was low in our study. A single isolate was detected in 2002 (prevalence, 0.3%), and no isolates were detected in 1993, 1997, 2002, and 2003. This isolate produced the plasmid-mediated TEM-52 ESBL. Three other TEM-52-producing S. enterica isolates of different serotypes were isolated from human cases (without prior treatment with ESC and without recent hospitalization) in France in 2002 and 2003. A common plasmid carrying blaTEM-52 was found in three of the four isolates (43). As TEM-52 has been identified increasingly in various serotypes of Salmonella in poultry in The Netherlands (14) and in Belgium (A. Cloeckaert, unpublished results) since 2001, we can hypothesize that poultry is the source for human contamination.
In conclusion, multidrug resistance is the rule for French serotype Typhimurium isolates collected from humans from 1993 until 2003. This is due mainly to the DT104 clone, which seems well established in France and which has undergone changes in antimicrobial resistance phenotypes. The emergence of serotype Typhimurium isolates resistant to ESC and Cip have been reported since 2002. These trends should be followed carefully by the use of adequate subtyping methods and by the detection of resistance genes and class 1 integrons.
Acknowledgments
We thank all the corresponding laboratories of the French National Reference Center Salmonella network.
This work was partially supported by grant AQS 2002/S24 from the “Direction générale de l'alimentation.”
REFERENCES
- 1.Anderson, E. S., L. R. Ward, M. J. Saxe, and J. D. de Sa. 1977. Bacteriophage-typing designations of Salmonella typhimurium. J. Hyg. (London) 78:297-300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antunes, P., J. Machado, J. C. Sousa, and L. Peixe. 2004. Dissemination amongst humans and food products of animal origin of a clone expressing an integron-borne OXA-30 beta-lactamase. J. Antimicrob. Chemother. 54:429-434. [DOI] [PubMed] [Google Scholar]
- 3.Arlet, G., M. Rouveau, and A. Philippon. 1997. Substitution of alanine for aspartate at position 179 in the SHV-6 extended-spectrum β-lactamase. FEMS Microbiol. Lett. 152:163-167. [DOI] [PubMed] [Google Scholar]
- 4.Baucheron, S., H. Imberechts, E. Chaslus-Dancla, and A. Cloeckaert. 2002. The AcrB multidrug transporter plays a major role in high-level fluoroquinolone resistance in Salmonella enterica serovar Typhimurium phage type DT204. Microb. Drug Resist. 8:281-289. [DOI] [PubMed] [Google Scholar]
- 5.Bonnet, R. 2004. Growing group of extended-spectrum β-lactamases: the CTX-M enzymes. Antimicrob. Agents Chemother. 48:1-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyd, D., G. A. Peters, A. Cloeckaert, K. S. Boumedine, E. Chaslus-Dancla, H. Imberechts, and M. R. Mulvey. 2001. Complete nucleotide sequence of a 43-kilobase genomic island associated with the multidrug resistance region of Salmonella enterica serovar Typhimurium DT104 and its identification in phage type DT120 and serovar Agona. J. Bacteriol. 183:5725-5732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Casin, I., J. Breuil, A. Brisabois, F. Moury, F. Grimont, and E. Collatz. 1999. Multidrug-resistant human and animal Salmonella typhimurium isolates in France belong predominantly to a DT104 clone with the chromosome- and integron-encoded β-lactamase PSE-1. J. Infect. Dis. 179:1173-1182. [DOI] [PubMed] [Google Scholar]
- 8.Casin, I., J. Breuil, J. P. Darchis, C. Guelpa, and E. Collatz. 2003. Fluoroquinolone resistance linked to GyrA, GyrB, and ParC mutations in Salmonella enterica Typhimurium isolates in humans. Emerg. Infect. Dis. 9:1455-1457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Daly, M., J. Buckley, E. Power, and S. Fanning. 2004. Evidence for a chromosomally located third integron in Salmonella enterica serovar Typhimurium DT104b. Antimicrob. Agents Chemother. 48:1350-1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Doublet, B., F. X. Weill, L. Fabre, E. Chaslus-Dancla, and A. Cloeckaert. 2004. Variant Salmonella genomic island 1 antibiotic resistance gene cluster containing a novel 3′-N-aminoglycoside acetyltransferase gene cassette, aac(3)-Id, in Salmonella enterica serovar Newport. Antimicrob. Agents Chemother. 48:3806-3812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Doublet, B., D. Boyd, M. R. Mulvey, and A. Cloeckaert. 2005. The Salmonella genomic island 1 is an integrative mobilizable element. Mol. Microbiol. 55:1911-1924. [DOI] [PubMed] [Google Scholar]
- 12.Eaves, D. J., L. Randall, D. T. Gray, A. Buckley, M. J. Woodward, A. P. White, and L. J. Piddock. 2004. Prevalence of mutations within the quinolone resistance-determining region of gyrA, gyrB, parC, and parE and association with antibiotic resistance in quinolone-resistant Salmonella enterica. Antimicrob. Agents Chemother. 48:4012-4015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fisher, I. S., and Enter-net Participants. 2004. International trends in Salmonella serotypes 1998-2003—a surveillance report from the Enter-net international surveillance network. Euro Surveill. 9:45-47. [PubMed] [Google Scholar]
- 14.Hasman, H., D. Mevius, K. Veldman, I. Olesen, and F. M. Aarestrup. 2005. Beta-lactamases among extended-spectrum beta-lactamase (ESBL)-resistant Salmonella from poultry, poultry products and human patients in The Netherlands. J. Antimicrob. Chemother. 56:115-121. [DOI] [PubMed] [Google Scholar]
- 15.Heisig, P., B. Kratz, E. Halle, Y. Graser, M. Altwegg, W. Rabsch, and J. P. Faber. 1995. Identification of DNA gyrase A mutations in ciprofloxacin-resistant isolates of Salmonella typhimurium from men and cattle in Germany. Microb. Drug Resist. 1:211-218. [DOI] [PubMed] [Google Scholar]
- 16.Helms, M., S. Ethelberg, K. Mølbak, and the DT104 Study Group. 2005. International Salmonella Typhimurium DT104 infections, 1992-2001. Emerg. Infect. Dis. 11:859-867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hopkins, K. L., R. H. Davies, and E. J. Threlfall. 2005. Mechanisms of quinolone resistance in Escherichia coli and Salmonella: recent developments. Int. J. Antimicrob. Agents 25:358-373. [DOI] [PubMed] [Google Scholar]
- 18.Hsueh, P. R., L. J. Teng, S. P. Tseng, C. F. Chang, J. H. Wan, J. J. Yan, C. M. Lee, Y. C. Chuang, W. K. Huang, D. Yang, J. M. Shyr, K. W. Yu, L. S. Wang, J. J. Lu, W. C. Ko, J. J. Wu, F. Y. Chang, Y. C. Yang, Y. J. Lau, Y. C. Liu, C. Y. Liu, S. W. Ho, and K. T. Luh. 2004. Ciprofloxacin-resistant Salmonella enterica Typhimurium and Choleraesuis from pigs to humans, Taiwan. Emerg. Infect. Dis. 10:60-68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Izumiya, H., K. Mori, T. Kurazono, M. Yamaguchi, M. Higashide, N. Konishi, A. Kai, K. Morita, J. Terajima, and H. Watanabe. 2005. Characterization of isolates of Salmonella enterica serovar Typhimurium displaying high-level fluoroquinolone resistance in Japan. J. Clin. Microbiol. 43:5074-5079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jarlier, V., M. H. Nicolas, G. Fournier, and A. Philippon. 1988. Extended broad-spectrum beta-lactamases conferring transferable resistance to newer beta-lactam agents in Enterobacteriaceae: hospital prevalence and susceptibility patterns. Rev. Infect. Dis. 10:867-878. [DOI] [PubMed] [Google Scholar]
- 21.Kruger, T., D. Szabo, K. H. Keddy, K. Deeley, J. W. Marsh, A. M. Hujer, R. A. Bonomo, and D. L. Paterson. 2004. Infections with nontyphoidal Salmonella species producing TEM-63 or a novel TEM enzyme, TEM-131, in South Africa. Antimicrob. Agents Chemother. 48:4263-4270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lawson, A. J., M. U. Dassama, L. R. Ward, and E. J. Threlfall. 2002. Multiply resistant (MR) Salmonella enterica serotype Typhimurium DT 12 and DT 120: a case of MR DT 104 in disguise? Emerg. Infect. Dis. 8:434-436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lévesque, C., L. Piché, C. Larose, and P. H. Roy. 1995. PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39:185-191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Levings, R. S., D. Lightfoot, S. R. Partridge, R. M. Hall, and S. P. Djordjevic. 2005. The genomic island SGI1, containing the multiple antibiotic resistance region of Salmonella enterica serovar Typhimurium DT104 or variants of it, is widely distributed in other S. enterica serovars. J. Bacteriol. 187:4401-4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lindstedt, B. A., T. Vardund, L. Aas, and G. Kapperud. 2004. Multiple-locus variable-number tandem-repeats analysis of Salmonella enterica subsp. enterica serovar Typhimurium using PCR multiplexing and multicolor capillary electrophoresis. J. Microbiol. Methods 59:163-172. [DOI] [PubMed] [Google Scholar]
- 26.Mabilat, C., and S. Goussard. 1993. PCR detection and identification of genes for extended-spectrum β-lactamases, p. 553-559. In D. H. Persing, T. F. Smith, F. C. Tenover, and T. J. White (ed.), Diagnostic molecular microbiology: principles and applications. American Society for Microbiology, Washington, D.C.
- 27.Mead, P. S., L. Slutsker, V. Dietz, L. F. McCaig, J. S. Bresee, C. Shapiro, P. M. Griffin, and R. V. Tauxe. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607-625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miriagou, V., P. T. Tassios, N. J. Legakis, and L. S. Tzouvelekis. 2004. Expanded-spectrum cephalosporin resistance in non-typhoid Salmonella. Int. J. Antimicrob. Agents 23:547-555. [DOI] [PubMed] [Google Scholar]
- 29.Murray, A., J. E. Coia, H. Mather, and D. J. Brown. 2005. Ciprofloxacin resistance in non-typhoidal Salmonella serotypes in Scotland, 1993-2003. J. Antimicrob. Chemother. 56:110-114. [DOI] [PubMed] [Google Scholar]
- 30.Popoff, M. Y. 2001. Antigenic formulas of the Salmonella serovars, 8th ed. WHO Collaborating Center for Reference and Research on Salmonella, Institut Pasteur, Paris, France.
- 31.Rabatsky-Ehr, T., J. Whichard, S. Rossiter, B. Holland, K. Stamey, M. L. Headrick, T. J. Barrett, F. J. Angulo, and the NARMS Working Group. 2004. Multidrug-resistant strains of Salmonella enterica Typhimurium, United States, 1997-1998. Emerg. Infect. Dis. 10:795-801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Randall, L. P., S. W. Cooles, M. K. Osborn, L. J. Piddock, and M. J. Woodward. 2004. Antibiotic resistance genes, integrons and multiple antibiotic resistance in thirty-five serotypes of Salmonella enterica isolated from humans and animals in the UK. J. Antimicrob. Chemother. 53:208-216. [DOI] [PubMed] [Google Scholar]
- 33.Ridley, A., and E. J. Threlfall. 1998. Molecular epidemiology of antibiotic resistance genes in multiresistant epidemic Salmonella typhimurium DT 104. Microb. Drug Resist. 4:113-118. [DOI] [PubMed] [Google Scholar]
- 34.Sandvang, D., F. M. Aarestrup, and L. B. Jensen. 1997. Characterisation of integrons and antibiotic resistance genes in Danish multiresistant Salmonella enterica Typhimurium DT104. FEMS Microbiol. Lett. 157:177-181. [DOI] [PubMed] [Google Scholar]
- 35.Soussy, C. J., G. Carret, J. D. Cavallo, H. Chardon, C. Chidiac, P. Choutet, P. Courvalin, H. Dabernat, H. Drugeon, L. Dubreuil, F. Goldstein, V. Jarlier, R. Leclercq, M. H. Nicolas-Chanoine, A. Philippon, C. Quentin, B. Rouveix, and J. Sirot. 2000. Comité de l'Antibiogramme de la Société Française de Microbiologie. Communiqué 2000-2001. Pathol. Biol. 48:832-871. [PubMed] [Google Scholar]
- 36.Threlfall, E. J. 2000. Epidemic Salmonella typhimurium DT 104—a truly international multiresistant clone. J. Antimicrob. Chemother. 46:7-10. [DOI] [PubMed] [Google Scholar]
- 37.Threlfall, E. J., C. J. Teale, R. H. Davies, L. R. Ward, J. A. Skinner, A. Graham, C. Cassar, and K. Speed. 2003. A comparison of antimicrobial susceptibilities in nontyphoidal salmonellas from humans and food animals in England and Wales in 2000. Microb. Drug Resist. 9:183-189. [DOI] [PubMed] [Google Scholar]
- 38.Threlfall, E. J., K. L. Hopkins, and L. R. Ward. 2005. Diversification in Salmonella Typhimurium DT 104. Emerg. Infect. Dis. 11:980-981. [Google Scholar]
- 39.Threlfall, E. J., L. R. Ward, and B. Rowe. 1997. Increasing incidence of resistance to trimethoprim and ciprofloxacin in epidemic Salmonella typhimurium DT104 in England and Wales. Euro Surveill. 2:81-84. [DOI] [PubMed] [Google Scholar]
- 40.Walker, R. A., E. Lindsay, M. J. Woodward, L. R. Ward, and E. J. Threlfall. 2001. Variation in clonality and antibiotic-resistance genes among multiresistant Salmonella enterica serotype typhimurium phage-type U302 (MR U302) from humans, animals, and foods. Microb. Drug Resist. 7:13-21. [DOI] [PubMed] [Google Scholar]
- 41.Wang, M., J. H. Tran, G. A. Jacoby, Y. Zhang, F. Wang, and D. C. Hooper. 2003. Plasmid-mediated quinolone resistance in clinical isolates of Escherichia coli from Shanghai, China. Antimicrob. Agents Chemother. 47:2242-2248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Weill, F. X., J. D. Perrier-Gros-Claude, M. Demartin, S. Coignard, and P. A. D. Grimont. 2004. Characterization of extended-spectrum-β-lactamase (CTX-M-15)-producing strains of Salmonella enterica isolated in France and Senegal. FEMS Microbiol. Lett. 238:353-358. [DOI] [PubMed] [Google Scholar]
- 43.Weill, F. X., M. Demartin, L. Fabre, and P. A. D. Grimont. 2004. Extended-spectrum-β-lactamase (TEM-52)-producing strains of Salmonella enterica of various serotypes isolated in France. J. Clin. Microbiol. 42:3359-3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weill, F. X., M. Demartin, D. Tandé, E. Espié, I. Rakotoarivony, and P. A. D. Grimont. 2004. Extended-spectrum-β-lactamase (SHV-12 like)-producing strains of Salmonella enterica serotypes Babelsberg and Enteritidis isolated in France among infants adopted from Mali. J. Clin. Microbiol. 42:2432-2437. [DOI] [PMC free article] [PubMed] [Google Scholar]